Abstract
Quaternary ammonium compounds such as benzalkonium chloride (BC) are widely used as disinfectants in both food processing and medical environments. BC-resistant strains of Listeria monocytogenes have been implicated in multistate outbreaks of listeriosis and have been frequently isolated from food processing plants. However, the genetic basis for BC resistance in L. monocytogenes remains poorly understood. In this study, we have characterized a plasmid (pLM80)-associated BC resistance cassette in L. monocytogenes H7550, a strain implicated in the 1998-1999 multistate outbreak involving contaminated hot dogs. The BC resistance cassette (bcrABC) restored resistance to BC (MIC, 40 μg/ml) in a plasmid-cured derivative of H7550. All three genes of the cassette were essential for imparting BC resistance. The transcription of H7550 BC resistance genes was increased under sublethal (10 μg/ml) BC exposure and was higher at reduced temperatures (4, 8, or 25°C) than at 37°C. The level of transcription was higher at 10 μg/ml than at 20 or 40 μg/ml. In silico analysis suggested that the BC resistance cassette was harbored by an IS1216 composite transposon along with other genes whose functions are yet to be determined. The findings from this study will further our understanding of the adaptations of this organism to disinfectants such as BC and may contribute to the elucidation of possible BC resistance dissemination in L. monocytogenes.
Listeria monocytogenes is a food-borne pathogen associated with severe illness (listeriosis) in at-risk individuals, including those in extremes of age, pregnant women and their fetuses, and those with compromised immunity. Environmental contamination with this pathogen plays a key role in the eventual contamination of ready-to-eat foods and subsequent food-borne illness (16, 19, 30). Biofilm formation and persistence, resistance to disinfectants, resistance to Listeria-specific viruses, and the ability to replicate at low temperatures are among the attributes contributing to the organism's prevalence and persistence in food processing environments (7, 18, 19).
Resistance to quaternary ammonium disinfectants such as benzalkonium chloride (BC) is especially relevant to Listeria's adaptations in food-related environments, as these compounds are used extensively in food processing, in retail, and for household or personal use (24, 26). BC resistance of L. monocytogenes isolated from foods and from the processing plant environment has been found to range from 10% (1) to as much as 42 to 46% (25, 27, 37). A study of strains from turkey processing plants revealed that resistance to BC was especially high among those of serotype 1/2a (or 3a) and 1/2b (or 3b) (60% and 51%, respectively) and that all BC-resistant strains were also resistant to the heavy metal cadmium (27).
Mechanisms underlying BC resistance in L. monocytogenes remain poorly understood. Several studies have provided evidence for chromosomal determinants (6, 34, 35, 37, 38), and evidence for plasmid-mediated resistance to BC also exists (21, 34, 35), even though plasmid-associated genes mediating BC resistance were not identified. Genome sequencing of several L. monocytogenes strains has identified numerous efflux systems on the chromosome as well as on plasmids (8, 9, 28; http://www.broadinstitute.org/annotation/genome/listeria_group/MultiHome.html), and some of these are potential candidates for BC resistance.
In this study, we have identified a plasmid-associated BC resistance system on an IS1216 composite transposon in L. monocytogenes H7550, implicated in the 1998-1999 multistate outbreak of listeriosis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1 . L. monocytogenes strain H7550 was isolated from the 1998-1999 hot dog outbreak (4). Its pulsed-field gel electrophoresis profile with AscI and ApaI was indistinguishable from that of the sequenced strain H7858 (28; R. M. Siletzky and S. Kathariou, unpublished data). Both H7550 and H7858 are resistant to BC and to cadmium (27; data not shown). H7550-Cds is a cadmium-susceptible, plasmid-free derivative of L. monocytogenes H7550. The plasmid-cured derivative was obtained following repeated passages of the bacteria at 42°C. H7550-CdsS is a spontaneous mutant of H7550-Cds with resistance to streptomycin and was isolated on plates containing streptomycin (150 μg/ml). L. monocytogenes was grown either in Trypticase soy broth with 0.6% yeast extract (TSBYE) (Becton, Dickinson and Company, Sparks, MD) or in brain heart infusion (BHI) broth (Becton, Dickinson and Company). Agar media were BHI agar (1.2% Bacto agar; Becton, Dickinson and Company) or TSBYE with 1.2% agar (1.2% Bacto agar). Escherichia coli strains were grown at 30°C in Luria Bertani (LB) broth (Becton, Dickinson and Company) or on LB broth supplemented with 1.2% Bacto agar.
TABLE 1.
Bacterial strains used in this studya
| Strain | BCr | Source or reference |
|---|---|---|
| Listeria monocytogenes (serotype 4b) | ||
| H7550 | + | 1998-1999 hot dog outbreak (4) |
| H7550-Cds | − | Plasmid-cured derivative of H7550 |
| H7550-CdsS | − | Streptomycin-resistant mutant of H7550-Cds |
| H7550-CdsS(pDS195) | + | This study |
| H7550-CdsS(pDS202) | + | This study |
| H7550-CdsS(pDS201) | − | This study |
| H7550-CdsS(pBEC59) | − | This study |
| H7550-CdsS(pBEC57) | − | This study |
| H7550-CdsS(pCON-1) | − | This study |
| Escherichia coli | ||
| SM10 thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km | 36 |
BCr, resistance to BC determined by growth on MHA plates with an added BC concentration of 20 μg/ml as described in Materials and Methods. +, growth; −, no growth.
BC susceptibility and determinations of MIC.
BC susceptibility of L. monocytogenes was assessed as described previously (27). Strains H7550 and H7550-Cds were used as resistant and susceptible controls, respectively. For MIC determinations, a single colony from a blood agar plate (Remel, Lenexa, KS) was suspended in 100 μl of Mueller Hinton broth (Becton, Dickinson and Company), 5 μl of the suspension was spotted in duplicate on Mueller Hinton agar (1.2% agar) plates with 1.2% defibrinated sheep blood (Becton, Dickinson and Company) and variable concentrations of BC (0, 5, 10, 15, 20, 25, 30, 35, and 40 μg/ml), and the plates were incubated at 30°C for 48 h. MIC was defined as the lowest assessed concentration of BC that prevented growth. All MICs were determined in at least two independent trials.
Recombinant plasmid constructs.
Primers used to construct the recombinant plasmids are listed in Table 2, and their locations are indicated in Fig. 1. PCR analysis was performed with the Takara Ex Taq kit (Takara, Madison, WI) and a T1 thermal cycler (Biometra, Göttingen, Germany). Primers BcF and BcR were used to produce a PCR fragment containing the entire bcrABC cassette along with the ca. 800-nucleotide (nt) upstream intergenic region. Primers BcF2 and BcR were employed to amplify the bcrABC cassette with 105 nt upstream of the putative start codon of bcrA, including the putative promoter region. The bcrABC cassette without the putative promoter was amplified with primers BcF5 and BcR. A PCR fragment containing the ca. 800-nt upstream intergenic region, bcrA and bcrB, but lacking bcrC was amplified with BcF and BcR2 (Table 2 and Fig. 1). The PCR fragments obtained with BcF and BcR, BcF2 and BcR, BcF5 and BcR, and BcF and BcR2 were digested with BamHI and EcoRI (New England Biolabs, Beverly, MA), ligated into the temperature-sensitive shuttle vector pCON-1 (3), and similarly digested with BamHI and EcoRI, resulting in pDS195, pDS202, pBEC59, and pDS201, respectively. The recombinant plasmids were electroporated into E. coli SM10 (36), and transformants were selected on LB agar supplemented with ampicillin (100 μg/ml). For the plasmid construct pBEC57 harboring an in-frame deletion of bcrB, pDS202 was used as template for inverse PCR analysis with primers BcF7 and BcR3 (both harboring a KpnI restriction site) (Fig. 1). The PCR amplicon was digested with KpnI, self-ligated with T4 DNA ligase (Promega, Madison, WI), and electroporated into E. coli SM10.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| BcF | GAATGGATCCTTCAATTAGATCGAGGCACG |
| BcR | GTATGAATTCGTATAATCCGGATGCTGCCC |
| BcR2 | GTATGAATTCCATCAACCTTCCTTTAATGAGG |
| BcF2 | GACTGGATCCGATTCTGGAACATCCCTATC |
| BcF5 | GAATGGATCCGGAGGGTAATCATGTCAG |
| BcF7 | GACTGGTACCCTCATTAAAGGAAGGTTGATGG |
| BcR3 | GACTGGTACCCATATGGATTCATGTG |
| p1 | CATTAGAAGCAGTCGCAAAGCA |
| p2 | GTTTTCGTGTCAGCAGATCTTTGA |
| p3 | GTTATCAGGATCTACGACTGTC |
| p4 | GTAATTCCTGCATTACGCATAACTG |
| p5 | GTCATAGATCGCTGCCGTAATTTCAG |
| p6 | CAATATGGCTATTGTATTCCCATGCTG |
| p7 | CAGAGTGAACGTGATCAGCATC |
| s1 | ATTACAGATGTGAGATTACGACG |
| s2 | ACGTTTACTTGGCATAGCTAC |
| c1 | ACAAGTTAGATCAAAAGAGTCTTTTATTAAACG |
| c2 | ATCTTCTTCATTTAGTGTTCCTGCAAATACTTC |
Underlined sequences correspond to restriction enzyme sites as follows: GGATCC, BamHI; GAATTC, EcoRI; GGTACC, KpnI.
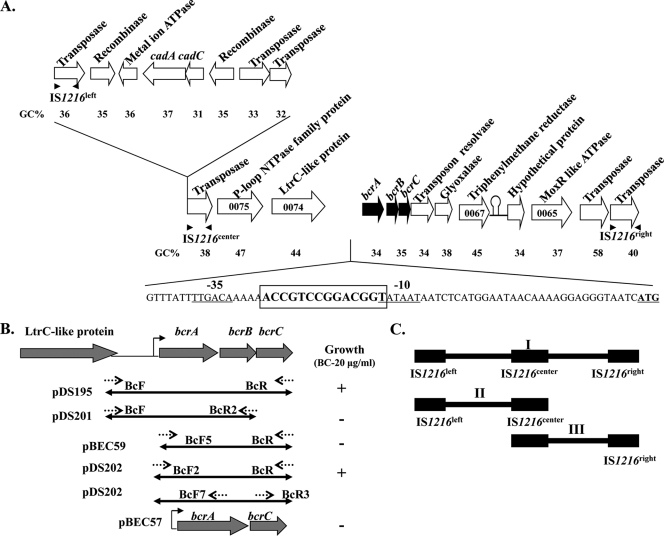
FIG. 1.
BC resistance cassette bcrABC in L. monocytogenes H7858. (A) Genetic organization and putative annotation of the bcrABC resistance cassette region in plasmid pLM80. Arrows indicate the direction of transcription. Genes implicated in BC resistance (bcrABC) are in black. The 67-nt sequence upstream of the start codon of the bcrA gene harboring a canonical promoter is shown along with the −10 and −35 regions (underlined). A palindromic sequence between the −10 and −35 regions, and partially overlapping with the −10 region, is shown in bold and boxed. Three copies of IS1216 are indicated as IS1216left, IS1216center, and IS1216right with solid arrows indicating the orientation of IS1216 inverted repeats. (B) Recombinant plasmids harboring different bcrABC segments. Dotted arrows indicate the position and orientation of the primers used for recombinant plasmid construction. The plasmids were introduced in the pLM80-cured derivative H7550-CdSS, and the resulting strains harboring these plasmids were tested for growth on BC (20 μg/ml), as described in Materials and Methods. Growth or lack of growth are indicated by + and −, respectively. (C) Schematic diagram of the possible transposable units harboring both cadmium and BC resistance genes (I), cadmium resistance genes (II), and BC resistance genes (III).
Recombinant plasmids were mobilized into H7550-CdsS via conjugation with E. coli SM10 harboring the recombinant plasmids, as described previously (20). Transconjugants were selected on BHI agar plates supplemented with chloramphenicol (6 μg/ml) and streptomycin (1,200 μg/ml) at 30°C for 2 to 3 days and confirmed using PCR. BC susceptibility of the transconjugants was assessed as described above.
RNA isolation and RT-PCR.
L. monocytogenes H7550 was grown in TSBYE at 4, 8, 25, and 37°C until the mid- to late logarithmic phase (optical density at 600 nm [OD600], ∼0.7 to 0.9), with the growth phase being monitored with a spectrophotometer (SmartSpec 3000; Bio-Rad, Hercules, CA). Cultures were then divided into two portions, one of which was treated at the indicated temperature for 30 min with a sublethal concentration (10 μg/ml) of BC or other concentrations as indicated. The other portion (control) remained untreated at the indicated temperature for 30 min. Total RNA was isolated using the SV total RNA isolation system (Promega). RNA was then subjected to DNase treatment using Turbo DNA-free (Ambion, Austin, TX). The concentration and quality of the RNA were determined by measuring the absorbance at 260 nm (NanoDrop, Wilmington, DE). RNA was stored at −80°C when necessary. Reverse transcription-PCR (RT-PCR) analysis for cadA involved a similar approach with cells treated for 30 min with sublethal concentrations (10 μg/ml) of cadmium chloride. Reverse transcription was carried out with primer c2, and PCR was done with primers c1 and c2 (Table 2).
RT-PCR experiments included spoVG as a housekeeping gene control, as previous studies indicated that expression of this gene was constitutive in L. monocytogenes, including at low temperatures (22). Total RNA was reverse transcribed to produce cDNA using 200 ng of RNA and the ImProm-II reverse transcription system (Promega) according to the manufacturer's protocol, using extension temperatures of 46°C and 43°C for bcrABC and spoVG transcripts, respectively. Self-priming controls (reverse transcription without a gene-specific primer) and negative controls (reverse transcription with no RNA) were included with all RT-PCR analyses. Sense and antisense primers for bcrABC RT-PCR were p1 and p2, respectively (Table 2), whereas those for spoVG RT-PCR were s1 and s2, respectively (Table 2). Each RT-PCR analysis was done in duplicate and in at least two independent experiments. For band quantifications we used the image processing software ImageJ (http://rsbweb.nih.gov/ij/). In order to calculate the fold increase in the transcript levels of the genes in response to presence of BC or cadmium, the gel density values obtained from the RT-PCR gel images using ImageJ were normalized to those of the control (culture without BC or cadmium). The transcript levels of the reference gene spoVG in the presence and absence of BC or cadmium were similarly normalized. The ratio of the normalized bcrABC levels to that of the normalized spoVG levels corresponded to the fold increase. To determine the impact of temperature, bcrABC levels were first normalized to those of spoVG from the same culture, and the fold increase upon addition of BC was determined as the ratio of the normalized levels in the absence or presence of BC.
RESULTS
Identification of BC resistance determinants on L. monocytogenes plasmid pLM80.
L. monocytogenes strain H7550 from the 1998-1999 hot dog outbreak was resistant to both cadmium and BC and harbored a large, ca. 80-kb plasmid, pLM80 (28). Plasmid curing of H7550 rendered the bacteria susceptible to the heavy metal cadmium. This was expected since pLM80 harbored genes for cadmium resistance (28). Testing of H7550-Cds for BC resistance revealed that the derivative was also susceptible to BC (Table 1). These findings suggested that, in addition to cadmium resistance determinants, pLM80 also harbored determinants for resistance to BC. However, unlike BC-resistant derivatives of originally BC-susceptible strains obtained following prolonged exposure to BC, which were resistant both to BC and to ethidium bromide (34, 35), the ethidium bromide MICs for H7550 and H7550-Cds were the same (both strains grew at 32 but not at 64 μg/ml).
Analysis of the annotation of pLM80 (RefSeq accession no. NZ_AADR00000000), identified three open reading frames (ORFs) as possible determinants for BC resistance (Fig. 1). These included a putative transcriptional regulator of the TetR family harboring a helix-turn-helix (HTH) DNA binding motif (bcrA; Pfam, PF00440). This ORF was followed by two putative small multidrug resistance (SMR) genes, bcrB and bcrC (Pfam: PF00893). Sequence analysis of the region suggested a long (850 bp; GC, 36%) intergenic region between bcrA and its upstream ORF (LtrC-like protein). Upstream (31 nt) of the putative start codon of bcrA we identified a canonical promoter (−10, TATAAT; −35, TTGACA) (Fig. 1). A perfect palindrome (ACCGTCCGGACGGT) was identified between the −10 and −35 promoter sequences (Fig. 1), suggesting a possible binding site for a transcriptional regulator. Highly similar (99 to 100% identity) bcrABC sequences (including the upstream region harboring the putative promoter) were detected in two L. monocytogenes strains among those with sequenced genomes: J0161, of serotype 1/2a (NZ_AARW02000017.1) and FSL F2-515, also of serotype 1/2a (NZ_AARI02001718). The genes were located on a large plasmid in J0161 (19a); even though strain FSL F2-515 harbored a plasmid, the genes were absent from that plasmid (19a), suggesting a chromosomal location in that strain.
With the exception of J0161 and FSL F2-515, no other sequences with significant homology to bcrABC were detected in the nucleotide database. The deduced BcrB and BcrC polypeptides, however, exhibited similarity (44 to 73%) to multiple proteins of the SMR family in diverse Gram-positive and Gram-negative bacteria. Furthermore, a chromosomal cassette (SugE1-E2) with 33 to 34% identity at the amino acid sequence level was identified in the genomes of all screened L. monocytogenes strains, including H7858 (data not shown).
Sequence analysis of the bcrABC region provided evidence for a composite transposon flanked by two IS1216 elements (IS1216 center and IS1216 right, each containing a transposase [75.3% nt identity; 83.2% amino acid identity to each other] flanked by IS1216 inverted repeats [5′ GGTTCTGTTGCAAAGTTT 3′]) (Fig. 1) (12). The putative composite transposon was ca. 12.4 kb long. In addition to the two IS1216 elements and the bcrABC cassette, the transposon harbored genes with putative conjugative functions: plm80_0074 and plm80_0075 were annotated as an LtrC-like protein and a member of the P-loop NTPase domain superfamily, respectively. Downstream of the bcrABC cassette we identified one ORF with 87 to 99% identity to a putative resolvase gene in plasmids of Bacillus cereus and in Tn1546 of Enterococcus faecium. The putative resolvase gene was followed by four ORFs encoding a putative glyoxalase superfamily protein, a putative triphenylmethane reductase, a hypothetical protein, and a MoxR-like protein, respectively. A stem-loop structure (ΔG = −18.7 kcal/mol) was identified 110 nt downstream of the plm80_0067 stop codon, possibly corresponding to a Rho-independent transcriptional terminator (Fig. 1).
The overall GC content of the ca. 12.4-kb region is 40%. However, pronounced diversity in GC content was observed within this region: the GC content of the bcrABC cassette was 34%, significantly lower than the average for the genome of L. monocytogenes (38%); other ORFs had widely variable GC contents, ranging from 34 to 58% (Fig. 1). It was of interest that plm80_0067, downstream of the putative glyoxalase and encoding a putative triphenylmethane reductase (TMR), had an unusually high GC content (45%) and 99 to 100% identity to putative TMR genes from Pseudomonas sp., Aeromonas sp., and Citrobacter sp.
The IS1216 composite transposon harboring bcrABC was immediately downstream of another putative transposon harboring the cadmium resistance cassette cadAC and flanked by IS1216 left and IS1216 center (Fig. 1A). The genomic region in pLM80 suggests the presence of three possible transposable units, with one carrying both cadmium and BC resistance genes (Fig. 1C, I) and others carrying either cadmium (Fig. 1C, II) or BC (Fig. 1C, III) resistance genes.
bcrABC cassette confers BC resistance to a plasmid-cured derivative of L. monocytogenes H7550.
Subcloning of the entire bcrABC cassette along with 900 nt upstream of the bcrA start codon in pCON-1 (including the 850-bp intergenic region between bcrA and pLM80_0074) resulted in pDS195, whereas pDS202 harbored bcrABC and 105 nucleotides upstream of bcrA, including the canonical promoter (Fig. 1B). Transfer of pDS195 and pDS202 into H7550-CdsS rendered the bacteria able to grow at 35 μg/ml, whereas MICs of BC were markedly lower for H7550-CdsS harboring the empty vector (Table 3). Constructs pDS201 (lacking bcrC), pBEC57 (lacking bcrB), and pBEC59 (bcrABC with only 11 nt upstream of bcrA and thus lacking the canonical promoter) (Fig. 1B) were not able to restore BC resistance (Table 3). In spite of repeated efforts, a construct harboring bcrB and bcrC, but lacking bcrA, could not be obtained. These data suggested that BC resistance could be conferred by the bcrABC cassette and that the 105-nt upstream region that included the canonical −10 and −35 promoter sequences was both required and sufficient for expression.
TABLE 3.
Benzalkonium chloride (BC) MICs for L. monocytogenes strain H7550 and its derivatives used in this study
| Strain | MICa (μg/ml) |
|---|---|
| H7550 | 40 |
| H7550-Cds | 10 |
| H7550-CdsS | 10 |
| H7550-CdsS(pCON-1) | 10 |
| H7550-CdsS(pDS195) | 40 |
| H7550-CdsS(pDS201) | 10 |
| H7550-CdsS(pDS202) | 40 |
| H7550-CdsS(pBEC59) | 10 |
| H7550-CdsS(pBEC57) | 10 |
MIC was defined as the lowest assessed concentration of BC that prevented growth. MICs were determined as described in Materials and Methods at 30°C. Strains with MICs of 10 μg/ml had impaired growth at 2.5 and 5 μg/ml and did not grow at all at 10 μg/ml.
Cotranscription of bcrABC with downstream genes and increase in bcrABC transcript levels in response to BC.
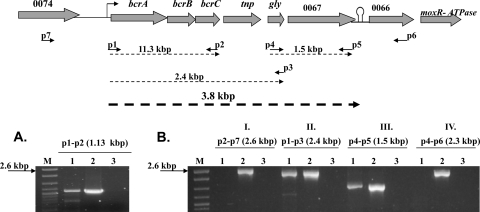
RT-PCR data suggested that bcrA, bcrB, and bcrC were cotranscribed and were also transcribed together with the downstream ORFs encoding a putative transposon resolvase, a putative glyoxalase, and a putative triphenylmethane reductase (Fig. 2). The ORF upstream of bcrA, plm80_0074, was not in this transcriptional unit and neither was plm80_0066, downstream of the putative triphenylmethane reductase (Fig. 2). These results were confirmed in five independent trials and are in congruence to the GC-rich stem-loop structure found downstream of plm80_0067 (Fig. 1). Thus, bcrABC was part of a polycistronic message of ca. 3.8 kb, with the putative triphenylmethane reductase gene being the last gene in the operon.
FIG. 2.
Cotranscription of BC resistance cassette and downstream genes assessed by reverse transcription-PCR in L. monocytogenes strain H7550 grown at 25°C. (A) Total RNA was reverse transcribed into cDNA using primer p2, and PCR analysis was performed using a cDNA template and primers p1 and p2. (B) Reverse transcription with primer p2 and PCR with cDNA template and primers p7 and p2 (I); reverse transcription with primer p3 and PCR with cDNA template and primers p1 and p3 (II); reverse transcription with primer p5 and PCR with cDNA template and primers p4 and p5 (III); and reverse transcription with primer p6 and PCR with cDNA template and primers p4 and p6 (IV). Lanes: 1, RT-PCR of H7550 exposed to BC (10 μg/ml); 2 and 3, H7550 genomic DNA and total RNA, respectively, used as positive and negative controls for RT-PCR; and M, 100- to 2,686-bp DNA molecular marker XIV (Roche, Indianapolis, IN).
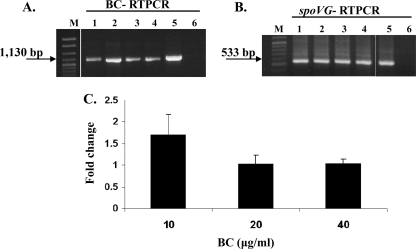
RT-PCR data also suggested that levels of the transcript that included bcrABC were higher in the presence of BC (10 μg/ml) than without the disinfectant (Fig. 3). To further assess the impact of BC on expression of bcrABC, transcript levels were assessed via RT-PCR at different concentrations of BC (0, 10, 20, and 40 μg/ml) using the housekeeping gene spoVG as a reference. RT-PCR data suggested that transcript levels of bcrABC were increased in the presence of 10 μg/ml BC (ca. 1.4- to 2-fold) but were similar to baseline levels at higher BC concentrations (20 and 40 μg/ml) (Fig. 3). RT-PCR with spoVG suggested that there were no significant changes in levels of spoVG transcripts when cells were grown in the presence of different concentrations of BC (0, 10, 20, and 40 μg/ml) (Fig. 3).
FIG. 3.
Impact of BC concentration on transcription of bcrABC. (A) RT-PCR of bcrABC using primers p2 for cDNA and p1 and p2 for PCR. Lanes: 1 to 4, H7550 exposed to 0, 10, 20, and 40 μg/ml of BC, respectively, at 25°C for 30 min; 5 and 6, H7550 genomic DNA and total RNA, respectively, used as positive and negative controls for RT-PCR; and M, 100- to 2,686-bp DNA molecular marker XIV (Roche). The arrow points to the expected bcrABC PCR product of 1,130 bp. (B) Transcript levels of housekeeping gene spoVG in the same cultures as those described for panel A. RT-PCR using primer s2 (for cDNA) and s1 and s2 for PCR. Lanes are as described for panel A. The arrow points to the expected spoVG PCR product of 533 bp. (C) Fold change in the transcript levels of bcrABC after exposure to 10, 20, and 40 μg/ml of BC. Fold change was determined as described in Materials and Methods, and the data are averages from two independent trials.
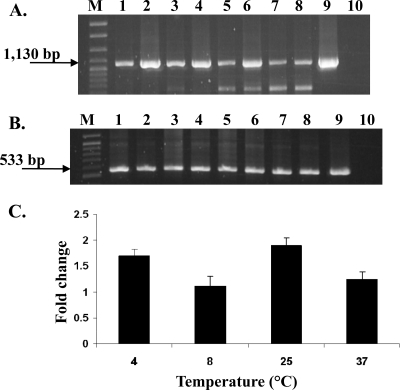
Transcription of bcrABC is higher at lower temperatures (4, 8, and 25°C) than at 37°C.
Temperature was found to impact bcrABC transcript levels, with levels at 4 to 25°C being greater than those at 37°C. Following normalization (ratio of bcrABC band density to the spoVG band from the same sample), levels in the absence of BC (baseline levels) at 37°C were ca. 57% those at 4°C (Fig. 4). The impact of temperature was even more noticeable in the presence of BC, with normalized levels at 8, 25, and 37°C being ca. 83, 92, and 42% those at 4°C, respectively (Fig. 4). Comparison of normalized values in the absence or presence of BC revealed that the presence of the disinfectant resulted in higher transcript levels at all tested temperatures. However, the impact of BC was greater at 4 and 25°C (ca. 1.8- and 2.0-fold increase, respectively) than at 8 and 37°C (ca. 1.2-fold increase each) (Fig. 4). The observed impact of temperature on increased and baseline levels of bcrABC transcription was consistently observed in independent experiments (data not shown).
FIG. 4.
Temperature regulation of bcrABC. (A) RT-PCR of bcrABC using primers p2 for cDNA and p1 and p2 for PCR. Lanes: 1, 3, 5, and 7, H7550 grown at 4, 8, 25, and 37°C, respectively; 2, 4, 6, and 8, H7550 grown at 4, 8, 25, and 37°C and exposed to BC (10 μg/ml) for 30 min as described in Materials and Methods; 9 and 10, H7550 genomic DNA and total RNA, respectively, used as positive and negative controls for RT-PCR; and M, 100- to 2,686-bp DNA molecular marker XIV (Roche). The arrow points to the expected bcrABC PCR product of 1,130 bp. Bottom bands in lanes 5 to 8 represent unspecific PCR products (confirmed with Southern blotting). (B) Transcript levels of housekeeping gene spoVG in the same cultures as those described for panel A. RT-PCR using primers s2 for cDNA and s1 and s2 for PCR. Lanes are as described for panel A. The arrow points to the expected spoVG PCR product of 533 bp. (C) Fold change in transcript levels of bcrABC at 4, 8, 25, and 37°C after exposure to BC (10 μg/ml). Fold change was determined as described in Materials and Methods, and the data are averages from two independent trials.
Lack of cross-induction of pLM80 genes mediating resistance to BC and to cadmium.
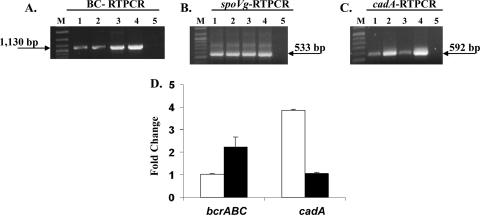
As described above, pLM80 harbors genes mediating resistance to BC (bcrABC) as well as to cadmium (cadAC), and the cadAC cassette (ORFs plm80_0082 and plm80_0083) is in the vicinity of bcrABC (Fig. 1A). To determine whether bcrABC expression may also be enhanced by cadmium, and reversely whether BC may increase expression of cadAC, RT-PCR was employed using spoVG as a reference. The data clearly indicated that bcrABC expression was increased (ca. 2-fold) by BC but not by sublethal exposure to cadmium (10 μg/ml CdCl2), and similarly cadA expression was enhanced by cadmium (ca. 4-fold) but not by BC (Fig. 5). RT-PCR data indicated that spoVG expression was stable under the conditions tested (Fig. 5).
FIG. 5.
Increased transcript levels of bcrABC and cadA by BC and cadmium in L. monocytogenes H7550 grown at 25°C. (A) RT-PCR of bcrABC using primer p2 for cDNA and primers p1 and p2 for PCR. Lanes: 1, H7550 in the absence of BC or cadmium; 2, H7550 exposed to cadmium; 3, H7550 exposed to BC; 4 and 5, H7550 genomic DNA and total RNA, respectively, used as positive and negative controls for RT-PCR; and M, 100- to 2,686-bp DNA molecular marker XIV (Roche). The arrow points to the expected bcrABC PCR product of 1,130 bp. Cadmium and BC exposures were for 30 min as described in Materials and Methods. (B) Transcript levels of housekeeping gene spoVG in the same cultures as those described for panel A. RT-PCR using primer s2 for cDNA and s1 and s2 for PCR. Lanes are as described for panel A. The arrow points to the expected spoVG PCR product of 533 bp. (C) RT-PCR of cadA using primer c2 for cDNA and primers c1 and c2 for PCR. Lanes are as described for panel A. The arrow points to the expected cadA PCR product of 592 bp. (D) Fold change in the transcript levels of bcrABC and cadA after exposure to BC (black bars) and cadmium (white bars). Fold change was determined as described in Materials and Methods, and the data are averages from two independent trials.
DISCUSSION
In this study we have described a BC resistance mechanism of L. monocytogenes H7550 (1998-1999 multistate outbreak strain) associated with a gene cassette harbored on the plasmid of this strain, pLM80. This gene cassette was also detected in two other sequenced Listeria genomes. Evidence for chromosomal efflux pumps implicated in quaternary ammonium compound resistance was provided for L. monocytogenes (6, 34, 35, 37, 38). Even though there was also evidence for plasmid-mediated resistance to BC (21, 34, 35), the plasmid-associated genes responsible for such resistance have not yet been identified.
The BC-resistance cassette described here was composed of one TetR family transcriptional regulator (bcrA) with a helix-turn-helix (HTH) DNA binding motif and two SMR genes (bcrB and bcrC). SMR proteins are proton-dependent multidrug efflux systems that typically require coexpression of two genes (2, 31) and have been characterized in several Gram-positive and Gram-negative bacteria, including E. coli EmrE and Bacillus subtilis EbrA and EbrB (14, 17, 29).
Members of the TetR family of regulators control transcription of multidrug efflux systems (33), suggesting a likely role for bcrA in transcriptional control of the bcrABC cassette. The palindrome between the putative −10 and −35 promoter regions of bcrABC may serve as a recognition sequence for the repressor. This palindrome overlaps with the putative −10 promoter sequence, thus possibly interfering with the transition of the RNA polymerase-promoter complex into a transcribing state. A palindrome overlapping the −10 region (but of different length and sequence content) was also described upstream of qacR, implicated in transcriptional control of the multidrug efflux pump qacA of Staphylococcus aureus (10, 32). Further studies are needed to confirm that bcrA functions as a repressor by binding to this region of dyad symmetry. A deletion mutant of this gene could not be obtained, suggesting that constitutive overexpression of bcrBC might be lethal to the cells.
Transcription of bcrABC was increased by sublethal levels of BC. No increase in the gene transcript level was noted in response to treatment with cadmium. Inversely, an increase in the gene transcript level of cadA was observed upon exposure to cadmium, but not BC. Thus, in spite of their genomic proximity on pLM80, the cassettes mediating resistance to BC and to cadmium are not regulated by the same substrates.
In spite of the increased bcrABC transcript levels upon exposure to BC, transcripts were also readily detected even in the absence of the disinfectant. This may suggest either that repression of transcription is weak or that other, currently unidentified molecules result in the observed baseline transcription levels in the absence of BC. Further studies are needed to determine how transcription is enhanced in the presence of sublethal levels of BC (e.g., by binding to BcrA and prevention of repression, as described in other efflux systems) (13, 40) and to assess whether additional molecules also increase transcript levels. Preliminary data suggest that, besides BC, other quaternary ammonium disinfectants (benzethonium chloride, cetyl-trimethyl ammonium bromide) also enhance the transcription of bcrABC in H7550 (V. Dutta and S. Kathariou, unpublished), but it is not known whether transcript levels can also be increased by other, structurally unrelated molecules that may be exported by this efflux system.
Transcript levels of bcrABC were increased at sublethal concentrations (10 μg/ml) of BC, but not at higher concentrations (e.g., 20 or 40 μg/ml). It is possible that BcrA conformation and the ability to bind to operator sites is different at low concentrations (10 μg/ml) compared to that at high concentrations (20 or 40 μg/ml) of BC. It is also possible that general toxicity associated with exposure to high concentrations of BC (even with a relatively short exposure time of 30 min) resulted in the observed loss of transcript level increase. However, we think this is less likely, as bcrABC transcripts were at baseline levels and spoVG levels were not impacted.
Resistance of Listeria species to BC is of special relevance to the pathogen's ecology in food processing plants, where BC and other quaternary ammonium disinfectants are used extensively. As noted before, two classes of BC-resistant strains can be recognized in L. monocytogenes, namely, resistant derivatives of previously BC-susceptible strains, resulting from adaptation to sublethal BC levels, and strains that are naturally resistant to BC, such as H7550 (34, 35, 38). Resistant derivatives of previously susceptible strains appear to involve chromosomal efflux systems, such as mdrL, that also mediate efflux of ethidium bromide and that are inhibited by the efflux inhibitor reserpine (35). The adapted derivatives may result from mutations leading to overexpression of these efflux systems, even though such mutations still need to be identified. On the other hand, in strains such as H7550, resistance to high levels of BC is mediated by bcrABC, a plasmid-associated system which shows increased transcript levels following exposure to BC and at low temperatures (4 to 25°C). In naturally resistant strains, resistance to BC is not accompanied by resistance to ethidium bromide, and neither is it inhibited by reserpine (35). We have indeed found that in strain H7550 the BC MIC was not affected by this efflux inhibitor, whereas MICs of the plasmid-cured strains were reduced in the presence of reserpine (M. Rakik-Martinez and S. Kathariou, unpublished).
The impact of temperature on transcription of bcrABC may be of special relevance to environmental adaptations of Listeria species in food processing plants where low temperatures often prevail. Temperature has a profound impact on the gene expression of L. monocytogenes, including genes involved in virulence and in environmental adaptations (5, 11, 15, 23, 39). Further studies are needed to characterize the mechanism responsible for temperature-dependent expression of bcrABC in L. monocytogenes and to also determine whether a similar impact of temperature is observed at the proteomic level.
Given that quaternary ammonium compounds such as BC are manmade reagents, it will be of interest to identify compounds in nature that can be exported by bcrABC and conditions responsible for acquisition and retention of this gene cassette by L. monocytogenes. The cassette was found to be on putative composite transposable units flanked by IS1216 inverted repeats. In addition to genes mediating resistance to BC, these putative composite transposable units harbor a cadmium resistance cassette, a putative glyoxalase, and a putative triphenylmethane reductase. The diversity of GC content within this region of pLM80 suggests acquisition of these genes through horizontal gene transfer, possibly from varied sources. The role of IS1216 transposition in gene transfer has been well described in another Gram-positive bacterium, Enterococcus faecium (12).
The presence of bcrABC on a putative IS1216 composite transposon harbored by pLM80 suggests possible mechanisms for the transfer of these genes among different Listeria genomes via transposition or plasmid mobilization. The presence of bcrABC in the vicinity of, and possibly in the same transposable unit as, cadAC may also partially explain the finding that BC-resistant strains of L. monocytogenes were also resistant to cadmium (27). Further work is needed to assess the ability of bcrABC to disseminate from H7550 to BC-susceptible strains of different serotypes of L. monocytogenes and different Listeria species.
In conclusion, we have characterized a plasmid-based BC resistance gene cassette on a putative composite transposon in a strain of L. monocytogenes associated with the 1998-1999 multistate outbreak. Such a resistance cassette draws attention to the possible role of disinfectants in adaptations and persistence of L. monocytogenes in food processing and other environments with frequent disinfectant use. Further studies are needed to elucidate the evolution and dissemination of these BC resistance genes in L. monocytogenes and to assess the possible role of these genes in environmental persistence as well as in the virulence of this pathogen.
Acknowledgments
This work was partially supported by a grant from the American Meat Institute Foundation and USDA grant 2006-35201-17377.
We thank Mira Rakic-Martinez for information on MICs for ethidium bromide and on the impact of reserpine. We are thankful to all other members of our laboratory for their support and encouragement.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Aase, B., G. Sundheim, S. Langsrud, and L. V. Rørvik. 2000. Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62:57-63. [DOI] [PubMed] [Google Scholar]
- 2.Bay, D. C., K. L. Rommens, and R. J. Turner. 2008. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778:1814-1838. [DOI] [PubMed] [Google Scholar]
- 3.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1998. Multistate outbreak of listeriosis—United States, 1998. MMWR Morb. Mortal. Wkly. Rep. 47:1085-1086. [PubMed] [Google Scholar]
- 5.Chan, Y. C., S. Raengpradub, K. J. Boor, and M. Wiedmann. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earnshaw, A. M., and L. M. Lawrence. 1998. Sensitivity to commercial disinfectants, and the occurrence of plasmids within various Listeria monocytogenes genotypes isolated from poultry products and the poultry processing environment. J. Appl. Microbiol. 84:642-648. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi, M., and M. L. Chikindas. 2007. Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113:1-15. [DOI] [PubMed] [Google Scholar]
- 8.Gilmour, M. W., M. Graham, G. Van Domselaar, S. Tyler, H. Kent, K. M. Trout-Yakel, O. Larios, V. Allen, B. Lee, and C. Nadon. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 10.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 11.Gründling, A., L. S. Burrack, H. G. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. U. S. A. 101:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heaton, M. P., L. F. Discotto, M. J. Pucci, and S. Handwerger. 1996. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene 171:9-17. [DOI] [PubMed] [Google Scholar]
- 13.Hinrichs, K., C. Kisker, M. Duvel, A. Muller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 14.Jack, D. L., M. L. Storms, J. H. Tchieu, I. T. Paulsen, and M. H. Saier, Jr. 2000. A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J. Bacteriol. 182:2311-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 16.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 17.Kikukawa, T., T. Nara, T. Araiso, S. Miyauchi, and N. Kamo. 2006. Two-component bacterial multidrug transporter, EbrAB: mutations making each component solely functional. Biochim. Biophys. Acta 1758:673-679. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J. W., and S. Kathariou. 2009. Temperature-dependent phage resistance of Listeria monocytogenes epidemic clone II. Appl. Environ. Microbiol. 75:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornacki, J. L., and J. B. Gurtler. 2007. Incidence and control of Listeria in food processing facilities, p. 681-766. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety, 3rd ed. CRC Press, Boca Raton, FL.
- 19a.Kuenne, C., S. Voget, J. Pischimarov, S. Oehm, A. Goesmann, R. Daniel, T. Hain, and T. Chakraborty. 2010. Comparative analysis of plasmids in the genus Listeria. PLoS One 5:e12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauer, P., M. Y. N. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaître, J. P., H. Echchannaoui, G. Michaut, C. Divies, and A. Rousset. 1998. Plasmid-mediated resistance to antimicrobial agents among listeriae. J. Food Prot. 61:1459-1464. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y., and A. Ream. 2008. Gene expression profiling of Listeria monocytogenes strain F2365 during growth in ultrahigh-temperature-processed skim milk. Appl. Environ. Microbiol. 74:6859-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh, E., O. Dussurget, J. Gripenland, K. Vaitkevicius, T. Tiensuu, P. Mandin, F. Repoila, C. Buchrieser, P. Cossart, and J. Johansson. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770-779. [DOI] [PubMed] [Google Scholar]
- 24.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mereghetti, L., R. Quentin, N. Marquet-Van Der Mee, and A. Audurier. 2000. Low sensitivity of Listeria monocytogenes to quaternary ammonium compounds. Appl. Environ. Microbiol. 66:5083-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merianos, J. J. 1991. Quaternary ammonium antimicrobial compounds, p. 225-255. In S. S. Block (ed.), Disinfection, sterilization and preservation, 4th ed. Lea & Feigner, Malvern, PA.
- 27.Mullapudi, S., R. M. Siletzky, and S. Kathariou. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 74:1464-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, and D. O. Bayles. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninio, S., D. Rotem, and S. Schuldiner. 2001. Functional analysis of novel multidrug transporters from human pathogens. J. Biol. Chem. 276:48250-48256. [DOI] [PubMed] [Google Scholar]
- 30.Painter, J., and L. Slutsker. 2007. Listeriosis in humans, p. 85-109. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety, 3rd ed. CRC Press, Boca Raton, FL.
- 31.Paulsen, I. T., R. A. Skurray, R. Tam, M. H. Saier, Jr., R. J. Turner, J. H. Weiner, E. B. Goldberg, and L. L. Grinius. 1996. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 19:1167-1175. [DOI] [PubMed] [Google Scholar]
- 32.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, B. A. Mitchell, and R. A. Skurray. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. U. S. A. 93:3630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos, J. L., M. Martínez-Bueno, A. J. Molina-Henares, W. Terán, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanova, N., S. Favrin, and M. W. Griffiths. 2002. Sensitivity of Listeria monocytogenes to sanitizers used in the meat processing industry. Appl. Environ. Microbiol. 68:6405-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanova, N. A., P. F. Wolffs, L. Y. Brovko, and M. W. Griffiths. 2006. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl. Environ. Microbiol. 72:3498-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 37.Soumet, C., C. Ragimbeau, and P. Maris. 2005. Screening of benzalkonium chloride resistance in Listeria monocytogenes strains isolated during cold smoked fish production. Lett. Appl. Microbiol. 41:291-296. [DOI] [PubMed] [Google Scholar]
- 38.To, M. S., S. Favrin, N. Romanova, and M. W. Griffiths. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl. Environ. Microbiol. 68:5258-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo-Arana. A., O. Dussurget, G. Nikitas, N. Sesto, H. Guet-Revillet, D. Balestrino, E. Loh, J. Gripenland, T. Tiensuu, K. Vaitkevicius, M. Barthelemy, M. Vergassola, M. A. Nahori, G. Soubigou, B. Régnault, J. Y. Coppée, M. Lecuit, J. Johansson, and P. Cossart. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950-956. [DOI] [PubMed] [Google Scholar]
- 40.Wang, L., B. Jeon, O. Sahin, and Q. Zhang. 2009. Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Appl. Environ. Microbiol. 75:5064-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]