Abstract
The diversity and dynamics of Legionella species along a French river watershed subject to different thermal and wastewater discharges during an annual cycle were assessed by 16S rRNA gene sequencing and by a fingerprint technique, single-strand conformation polymorphism. A high diversity of Legionella spp. was observed at all the sampling sites, and the dominant Legionella clusters identified were most closely related to uncultured bacteria. The monthly monitoring revealed that Legionella sp. diversity changes were linked only to season at the wastewater site whereas there was some evidence for anthropogenic effects on Legionella sp. diversity downstream of the thermal bath. Quantification of Legionella pneumophila and Legionella spp. by culture and quantitative PCR (qPCR) was performed. Whereas only L. pneumophila was quantified on culture media, the qPCR assay revealed that Legionella spp. were ubiquitous and abundant from the pristine source of the river to the downstream sampling sites. These results suggest that Legionella spp. may be present at significant concentrations in many more freshwater environments than previously thought, highlighting the need for further ecological studies and culturing efforts.
Since the discovery of the human pathogen Legionella pneumophila in 1976, about 50 species of Legionella have been described. About one-half of them have been associated with human Legionnaires' disease (20), which occurs after inhalation of aerosolized water contaminated with virulent Legionella strains. Numerous cases of legionellosis after exposure to contaminated water from the water distribution systems of hotels, hospitals, and cooling towers have been reported (8, 9, 36). The major reservoirs of Legionella spp. are freshwater environments such as lakes, rivers, groundwater, and hot springs, but they can also survive in seawater and water from wastewater treatment plants (WWTPs) (12, 14, 22, 34, 38). Their presence in these different reservoirs demonstrates their ability to grow, or at least persist, under a wide range of different environmental conditions (e.g., temperature, pH). They can use a number of different strategies to survive in these different environments, including their use of free-living amoebae as hosts for intracellular replication, the protection of the cells in amoebal cysts (31), their persistence in biofilms (16), and their ability to enter a viable but nonculturable state (18, 47). These strategies have complicated the detection of Legionella, and there is limited knowledge of Legionella species diversity in the natural environment essentially due to the reliance on culture-based methods for Legionella sp. detection, which select for L. pneumophila (7, 28, 39). Further insights into Legionella ecology have been gained by the development of cultivation-independent techniques using cellular approaches (immunofluorescence or fluorescence in situ hybridization) (6, 45) and, more recently, molecular approaches based on PCR (34, 43, 48). The construction of 16S rRNA gene clone libraries and DNA sequencing were used to study Legionella diversity in sand filters (10), in acidic biofilm communities in Yellowstone National Park (38), in drinking water (46), and more recently in river water in Brazil (11). However, the construction of clone libraries is labor-intensive and expensive and is not usually applied to numerous samples. A more in-depth analysis of the diversity of natural Legionella species requires the analysis of many samples by high-throughput screening methods. One such genetic fingerprinting technique, single-strand conformation polymorphism (SSCP), is well suited to bacterial diversity analysis of large sample sets due to its rapidity, reproducibility, and low cost and was also used recently to study the dynamics of Legionella spp. in water from a cooling tower plant (44).
The majority of studies have focused on the source of Legionella contamination in the man-made systems where they were proliferating. However, the increasing incidence of legionellosis highlights the need to better understand the origin of the Legionella pneumophila and non-L. pneumophila species in the major freshwater reservoirs and how different Legionella species may be impacted by environmental or anthropogenic effects.
The main objectives of this study were to better characterize Legionella diversity in natural water samples and to determine if there were seasonal or anthropogenic effects on Legionella diversity and composition. To meet these objectives, Legionella abundance and diversity were investigated along a river before and after thermal bath and wastewater discharges during an annual cycle. L. pneumophila and Legionella spp. were quantified by the standard culture method and by quantitative PCR (qPCR). Legionella diversity at the different sampling sites was characterized by cloning and sequencing of 16S rRNA gene fragments, and the dynamics of Legionella diversity was followed throughout the year by SSCP analysis of 16S rRNA genes.
MATERIALS AND METHODS
Strains.
Bacterial strains used in this study and their sources are indicated in Table S1 in the supplemental material. Legionella strains were grown on buffered charcoal-yeast extract (αBCYE) agar (Oxoid, France) at 37°C for 48 to 72 h. Non-Legionella strains were grown on nutrient agar at 37°C for 24 h.
Study sites and sample collection.
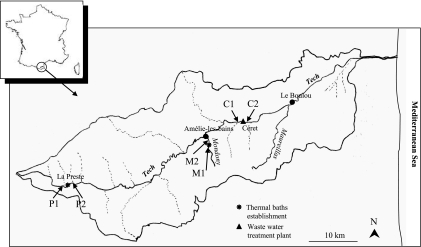
Water was sampled from the Tech River, located in the south of France, which is 84 km long from its source (Costabonne, 2,345 m high) to its estuary in the Mediterranean Sea (Fig. 1). The three sampling sites were located near to the source of the river at La Preste in a wooded mountainous area, where the water system was turbulent, from the tributary river Mondony close to Amélie-les-bains, and from Céret, 42 km downstream in an important urban area. At each sampling site, samples were taken upstream and downstream of the discharge from a thermal bath establishment and a wastewater treatment plant (Fig. 1).
FIG. 1.
Locations of sampling sites within the Tech River watershed at La Preste (P), Amélie-les-bains (M), and Céret (C), upstream (1) and downstream (2) of anthropogenic discharges.
For each sample, 1 liter surface water was collected in duplicate in sterile plastic bottles. Samples were treated the same day of collection or were stored at 4°C for analysis 24 h later.
Samples were collected each month, from October 2007 to September 2008, and were given designations identifying the sample site (P, La Preste; C, Céret; M, Amélie-les-bains), the sample location (1, upstream; 2, downstream), and the month. For example, P1OCT is the October upstream sample from La Preste. The water temperature was measured in each sample using a Checktemp thermometer (Hanna Instruments) (see Table S2 in the supplemental material).
Sample concentration and culture of Legionella species.
Legionella species were enumerated and identified according to the Association Française de Normalisation (AFNOR) standard (AFNOR T90-431) for each sample collected (see above) (4), which is equivalent to ISO 11731. Briefly 0.1- to 1-liter water samples were concentrated by filtration through 0.45-μm-pore-size polycarbonate filters (Sartorius). The filtered volume was dependent on the sample filterability. Membranes were transferred into 5 ml of sterile milli-Q water and sonicated twice for 1 min (42 kHz). Volumes (100 μl) of the concentrate were plated onto GVPC medium (Oxoid) after heat treatment (50°C for 30 min) and acid treatment (0.2 M HCl, 0.2 M KCl, pH 2, for 5 min) to eliminate non-Legionella organisms. Plates were incubated at 37 ± 2°C, and colonies were counted after 3, 5, and 10 days. Colonies were examined for fluorescence under a Wood lamp. Colonies exhibiting Legionella morphology were transferred to αBCYE medium, αBCYE medium without cysteine, and blood agar medium (Oxoid) as controls. At least five colonies per sample were identified by Legionella-specific latex reagents (Oxoid). When 1 liter of water was filtered, the detection limit (DL) of the method was one colony per plate, equivalent to 50 CFU·liter−1, whereas the statistically significant quantification limit (QL) was based on the AFNOR recommendation of counting 5 colonies per plate, equivalent to 250 CFU·liter−1 (4).
DNA isolation and purification from water samples.
For each sample, 0.1 to 1 liter of surface water (according to sample filterability) was filtered in duplicate through 0.45-μm-pore-size polycarbonate filters (Sartorius). DNA was extracted directly from filters with the Aquadien extraction kit, according to the manufacturer's instructions (Bio-Rad). For each sample, qPCR and SSCP analyses were carried out with the same DNA extract.
Real-time quantitative PCR.
Quantitative PCR was performed using two different iQ-Check Quanti Legionella kits (Bio-Rad) containing specific primers targeting the mip and the 5S rRNA genes for the detection of all 15 serogroups of L. pneumophila and Legionella spp., respectively. The protocol was carried out according to the French standard AFNOR XP T90-471 (5) and used the LightCycler instrument (1.0; Roche Diagnostics). Standard DNA curves were generated from genomic DNA of L. pneumophila ATCC 33152 provided in the kit. PCR mixtures contained 13 μl of amplification mix, 2 μl of the fluorescent probes per sample, and 5 μl of sample or negative control or genomic DNA standard. The PCR protocol consisted of 2 min at 50°C, 15 min at 95°C for enzyme activation, and then 50 cycles of 15 s at 95°C, 30 s at 57°C, and 30 s at 72°C and a final elongation at 72°C for 10 min. All samples lacking a fluorescent signal were reanalyzed after being spiked with a known quantity of a genomic DNA standard provided in the kit (300 genome units [GU] of L. pneumophila per capillary) and compared with a positive control. Equal signals indicated an absence of inhibition of the amplification reaction.
Analyses were performed in duplicate, and results were expressed as GU per liter. When filtering one liter of sample, the DLs of qPCR methods were 128 GU·liter−1 and 64 GU·liter−1 for L. pneumophila and Legionella species, respectively. The QL was 800 GU·liter−1 for both methods.
PCR and capillary electrophoresis SSCP (CE-SSCP) analyses of Legionella spp.
Two sets of primers, which targeted two different variable regions of the 16S rRNA gene of Legionella, were tested. The first set, JRP (phosphoramidite [TET]-5′-AGGGTTGATAGGTTAAGAGC-3′) and JFP (5′-CCAACAGCTAGTTGACATCG-3′) (25), amplified a 386-bp fragment of the V3-V4-V5 region, and the second set of primers, TempF (5′-CCTGGCTCAGATTGAACG-3′) and TempR (TET-5′-AGGCTAATCTTAAAGCGCC-3′) (41), amplified a 218-bp fragment of the V1-V2 region. The specificity of the primers was tested in silico (http://rdp.cme.msu.edu/index.jsp) and on Legionella and non-Legionella cultured strains (see Table S1 in the supplemental material). Single colonies were suspended in 100 μl sterile physiological saline water, and cells were lysed at 95°C for 15 min.
PCR mixtures (25 μl) contained approximately 3 to 10 ng DNA, 1 μM primers, 0.8 mM deoxynucleoside triphosphates (dNTPs), and 1 to 1.5 U of Pfu DNA polymerase (Promega). The PCR protocol included an initial denaturation of 5 min at 95°C, followed by 30 cycles of 1 min at 95°C, 1 min at 55°C (TempF/TempR) or 57°C (JFP/JRP), and 1 min at 72°C, with a final extension of 10 min at 72°C. Amplification products were verified by agarose gel electrophoresis, and their concentrations were estimated by comparison with molecular size markers. Dilutions of the PCR products were made in molecular grade water, and 1 μl of each dilution was mixed with 18.8 μl of Hi-Di formamide (Applied Biosystems) and 0.2 μl of the internal size standard GeneScan-400HD (ROX) (Applied Biosystems). Samples were denatured at 94°C for 5 min and cooled rapidly in a water-ice bath for 10 min. Fragments were separated by CE-SSCP as described previously (17) using an ABI310 genetic analyzer (Applied Biosystems). To check the reproducibility of the SSCP peak profiles, duplicate PCRs from duplicate extractions were carried out for 6 samples (P1FEB, P2FEB, C1FEB, C2FEB, M1FEB, and M2FEB) and then analyzed by CE-SSCP. Identical profiles were generated for each duplicate sample (data not shown), and therefore single reactions were carried out for the remaining samples.
The raw data generated by the Genescan analysis software of the ABI310 genetic analyzer were exported using Chromagna (19) into csv files. These files were then imported into the SAFUM program (49), which normalizes the total area of the SSCP profiles and the mobilities between different runs using the internal standard. The similarities of the SSCP profiles were compared by first calculating a Bray-Curtis resemblance matrix and then by constructing dendrograms with the unweighted-pair group method using average linkages (UPGMA) or by plotting nonmetric multidimensional scaling (nMDS) graphs using Primer Enterprises (Primer-E) software (13).
Identification of Legionella species by cloning and sequencing.
The primers TempF and TempR were used to amplify genomic DNA from samples M2JAN, P2DEC, and C2JUN based on the method described previously (1). For each DNA sample, 10 replicate PCRs were made to reduce PCR bias. Each 25 μl of PCR mixture contained approximately 2 ng DNA together with 0.3 μM primers, 0.5 mM dNTPs, 1 U of Super-Taq polymerase, and 1× Super-Taq buffer (HT Biotechnology, Cambridge, United Kingdom). The reactions were cycled using the following parameters: an initial denaturation of 5 min at 95°C followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, with a final extension of 10 min at 72°C. The PCR products were pooled and gel purified (gel extraction kit; Qiagen) before cloning the same day using the TOPO TA cloning kit (pCR2.1) according to the manufacturer's instructions.
Plasmid DNA (96 clones for each of the 3 libraries) was sequenced with the BigDye Terminator kit and the 3730xl automatic sequencer (Macrogen, Seoul, South Korea) using the M13 primer. Sequences were manually corrected using the sequence editing software Bioedit (23) and aligned using the MEGA software (40) and the ARB program (30). Clone sequences were submitted to Blastn (2) to identify the closest relatives. Clusters or operational taxonomic units (OTUs) were defined from the aligned sequences at 99% similarity by using the software Clusterer with the UPGMA setting (26).
Statistical tests.
The ANOSIM (analysis of similarities) routine of Primer-E was used to test the null hypothesis that there were no differences in Legionella diversity between the sampling sites or that there were no differences in diversity between the different seasons.
ANOSIM is a nonparametric test of significant difference between two or more groups that uses the underlying Bray-Curtis resemblance matrix to rank similarities among a priori-defined groups. ANOSIM calculates a global R statistic from the fraction of the difference between average rank similarity within groups and average rank similarity between groups as a function of the number of samples. The resulting R value usually ranges between 0 and 1, with high values indicating a large degree of discrimination among groups whereas values near 0 indicate that there is little difference among the groups (13). The significance level or P value is computed by permutation of group membership, with 1,000 replicates. The observed value of R is then referred to its permutation distribution. If the observed value is unlikely to come from the null distribution, the null hypothesis can be rejected.
To test for significant differences in cell concentrations, the data were analyzed by the nonparametric Wilcoxon test. All statistical tests were done using XL stat software (Addinsoft, France).
Nucleotide sequence accession numbers.
The sequence data from this study have been submitted to the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under the accession numbers GU180682 to 180770 (Céret), GU180771 to 180840 (Amélie-les-bains), and GU180841 to 180919 (La Preste).
RESULTS
Detection and quantification of Legionella spp. in river water samples.
The quantification of Legionella spp. present in the water samples collected along the Tech River (Fig. 1) was conducted by the standard culture method and by quantitative PCR. After being cultured on the GVPC medium, only 15 samples out of 72 (20.8%) were positive for Legionella spp. Among them, 9 (12.5%) were collected at P2 and 6 (8.3%) at M2, whereas there were no Legionella spp. detected at Céret. All selected colonies were identified as L. pneumophila. Concentrations at P2 ranged from 143 to 500 CFU·liter−1 all year except in December, April, and May. At M2, the concentrations ranged from 100 to 583 CFU·liter−1 in the winter months, June, and July. For all the positive samples, plate count results were between the detection limit (DL; 50 to 250 CFU·liter−1) and the quantification limit (QL; 250 to 1,250 CFU·liter−1) of the method, except for the P2SEP sample, where the plate counts were higher than the QL (250 CFU·liter−1). Concentrations at P2 ranged from 143 to 500 CFU·liter−1 all year except in December, April, and May. At M2, the concentrations ranged from 100 to 583 CFU·liter−1 in the winter months, June, and July.
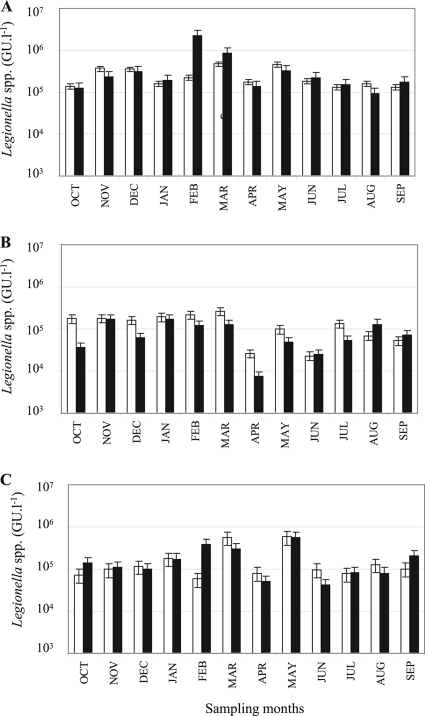
For the qPCR analyses, PCR inhibitors were detected in all the water samples from nondiluted DNA extracts. This problem was resolved by repeating the analyses with 10-fold dilutions of the DNA extracts. However, this decreased the sensitivity of the qPCR assays and increased the quantification limits of the qPCR method from 8 × 103 to 8 × 104 GU·liter−1 (Fig. 2). No L. pneumophila organisms were detected by qPCR in any of the samples, but Legionella spp. were detected above the QL of the method for all the different sampling sites analyzed throughout the year (Fig. 2). The highest concentrations of Legionella spp. were detected at the Céret sampling site and ranged from 1.24 × 105 GU·liter−1 to 9.36 × 105 GU·liter−1. Slightly lower concentrations were found at the Amélie-les-bains site (5.01 × 104 GU·liter−1 to 5.82 × 105 GU·liter−1) and at the La Preste site (7.39 × 103 GU·liter−1 to 2.62 × 105 GU·liter−1). Pairwise Wilcoxon tests revealed that for the Céret and Amélie-les-bains sampling sites the Legionella sp. concentrations of the upstream and downstream sample sites were not significantly different (P > 0.05), whereas there was a significant difference for La Preste (P < 0.05). Over the year, mean temperature variations between water taken upstream and downstream of anthropogenic discharges varied according to the sampling site location. The average temperature variations were 2.7°C, 6°C, and 0°C for the La Preste, Amélie-les-bains, and Céret sampling sites, respectively (see Table S2 in the supplemental material).
FIG. 2.
Temporal change of Legionella sp. concentrations at the three sampling sites, Céret (A), La Preste (B), and Amélie-les-bains (C) upstream (white) and downstream (black) of discharge, as measured by qPCR.
Specificity testing of Legionella-specific PCR primers.
Two sets of primers for the amplification of fragments located in two different hypervariable regions of the 16S RNA of Legionella were tested. The first set (25) allowed the amplification of fragments located in the hypervariable regions V3, V4, and V5, whereas the second set (41) amplified fragments located in the hypervariable regions V1 and V2. The primer sets were first tested with a range of Legionella strains and non-Legionella strains (see Table S1 in the supplemental material) to verify their specificity. All the Legionella strains tested were amplified by both sets of primers (TempR/TempT and JRP/JFP), whereas the non-Legionella species were not (data not shown). The amplification products from the two sets of primers producing two different fragment sizes were then analyzed by CE-SSCP to determine which primer set allowed better discrimination of the 42 different Legionella strains tested (27 Legionella spp. and 15 Legionella pneumophila strains comprising serogroups 1 to 15). The primer set TempR/TempF, which gave the shortest fragment, allowed better discrimination of the Legionella species than the other primer set, JRP/JFP, for which the SSCP profiles were not clearly distinguishable due to significant overlapping (see Fig. S2 in the supplemental material). These differences can be explained by the low resolution of large fragments with the applied electrophoretic conditions (3). Consequently, all further analyses of Legionella diversity in the different environmental samples were done using the TempR/TempF primer set.
Diversity of Legionella 16S rRNA gene fragments.
To characterize the Legionella diversity and to verify the specificity of the PCR primers, SSCP PCR products obtained from 3 different sampling sites after water discharge (M2JAN, P2DEC, and C2JUN) were cloned and sequenced.
A total of 238 sequences were obtained: 89 sequences from Céret, 79 from La Preste, and 70 from Amélie-les-bains. All sequences had the greatest similarity to Legionella spp., confirming the specificity of the primers used in this study. Similarities between the 16S rRNA gene sequences and the Legionella sequences deposited in the GenBank database ranged from 87% to 100% for the three sampling sites (Table 1). However, even the sequences with lower similarities had Legionella species as their nearest relatives. Sequences were then clustered into operational taxonomic units (OTUs) at 99% similarity, giving 29 clusters of 163 sequences (68% of the total sequences) and 75 singletons. The 99% threshold was chosen since this permitted different cultured Legionella spp. to be placed into different OTUs. The closest relatives of the representative sequences from each cluster together with the relative abundances of sequences in a given cluster at each of the sampling sites are shown in Table 1.
TABLE 1.
Relative abundances of clone sequences in each cluster (defined at 99% similarity) for the individual libraries and out of all sequences, presented with the closest relatives in GenBank
| Cluster no. | % clones/cluster/library for sitea: |
% clones out of all sequences | Closest relative in GenBank | % identity | Accession no. | ||
|---|---|---|---|---|---|---|---|
| C | M | P | |||||
| 3 | 23.6 | 15.7 | 12.7 | 17.6 | Uncultured gammaproteobacterium clone CRO-2 | 98 | AF141558 |
| 8 | 7.9 | 1 | 11.4 | 9.7 | “Candidatus L. jeonii” | 96 | AY598719 |
| 2 | 4.5 | 5.7 | 2.5 | 4.2 | L. pneumophila | 100 | M59157 |
| 16 | 2.2 | 1.4 | 7.6 | 3.8 | Uncultured bacterium clone EV818EB5CPSAJJ36 | 96 | DQ337039 |
| 13 | 3.4 | 5.7 | 1.3 | 3.4 | Uncultured bacterium clone EV818SWSAP64 | 96 | DQ337087 |
| 9 | 6.7 | 1.4 | 2.9 | Uncultured bacterium clone IC-85 | 97 | AB255083 | |
| 19 | 4.5 | 1.4 | 2.1 | LLAP14 | 97 | U66104 | |
| 23 | 1.1 | 2.9 | 2.5 | 2.1 | Uncultured gammaproteobacterium clone WCB127 | 96 | AY217479 |
| 6 | 3.4 | 1.4 | 1.7 | Uncultured soil bacterium clone 1_H3 | 96 | EU589279 | |
| 7 | 1.4 | 3.8 | 1.7 | Uncultured bacterium clone EV818SWSAP64 | 97 | DQ337087 | |
| 11 | 1.1 | 2.5 | 1.3 | LLAP8 | 94 | X97361 | |
| 14 | 2.9 | 1.3 | 1.3 | L. cincinnatiensis (ATCC 43753) | 100 | X73407 | |
| 15 | 1.1 | 2.5 | 1.3 | Uncultured bacterium clone 300G-C11 | 99 | AY662002 | |
| 17 | 1.1 | 1.4 | 1.3 | 1.3 | LLAP11 | 96 | X97362 |
| 18 | 3.8 | 1.3 | Uncultured bacterium clone EV818SWSAP64 | 97 | DQ337087 | ||
| 21 | 1.4 | 2.5 | 1.3 | L. feeleii (ATCC 35072) | 99 | X73395 | |
| 28 | 3.4 | 1.3 | Uncultured gammaproteobacterium clone ADK-HDe02-55 | 92 | EF520570 | ||
| 29 | 1.1 | 2.9 | 1.3 | Uncultured Legionella sp. clone CL1.B267 | 95 | FM175311 | |
| 1 | 2.9 | 0.8 | Legionella oakridgensis (ATCC 33761) | 96 | X73397 | ||
| 4 | 2.2 | 0.8 | Uncultured bacterium clone IC-85 | 97 | AB255083 | ||
| 5 | 2.5 | 0.8 | Uncultured gammaproteobacterium clone ADK-BTh02-70 | 92 | EF520563 | ||
| 10 | 1.4 | 1.3 | 0.8 | Legionella beliardensis | 96 | AF122884 | |
| 12 | 1.1 | 1.4 | 0.8 | Uncultured bacterium clone P7X3b1C06 | 87 | EU491023 | |
| 20 | 1.1 | 1.3 | 0.8 | Uncultured bacterium clone TSAF31 | 97 | AB186839 | |
| 22 | 1.1 | 1.3 | 0.8 | Legionella wadsworthii (ATCC 33877) | 97 | X73401 | |
| 24 | 1.4 | 1.3 | 0.8 | Legionella beliardensis | 94 | AF122884 | |
| 25 | 1.1 | 1.4 | 0.8 | Legionella jordanis (ATCC 33623) | 95 | X73396 | |
| 26 | 2.2 | 0.8 | Uncultured gammaproteobacterium clone ADK-BTh02-70 | 94 | EF520563 | ||
| 27 | 2.2 | 0.8 | Uncultured gammaproteobacterium clone CRO-2 | 98 | AF141558 | ||
C, Céret; M, Amélie-les-bains; P, La Preste.
For all sampling sites, the major clusters were cluster 3 and cluster 8, accounting for 17.6% and 9.7%, respectively, of the total sequences, and for Céret, cluster 3 accounted for nearly one quarter of the sequences. The closest relative of the most dominant cluster, cluster 3, was an uncultured Gammaproteobacteria sequence isolated from the Columbia River (15), and the nearest Legionella species was only 94% similar. For cluster 8, the closest identified sequence was “Candidatus Legionella jeonii,” at 96% similarity. This species was previously described as an obligate endosymbiotic bacterium in Amoeba proteus (35) and was placed apart from Legionella-like amoebal pathogens (LLAP) in phylogenetic analyses. The representative sequence from cluster 2 was 100% identical to L. pneumophila, and this cluster was the third most abundant when all sequences were considered. Interestingly, L. pneumophila sequences were recovered from all sites, even at the most pristine site at La Preste. Clusters 13, 16, and 9 accounted for 3.4%, 3.8%, and 2.9%, respectively, of the total sequences, and the closest relatives identified by Blastn were uncultured clones isolated from surface water in South Africa and from a corroded sewer system (32). The other clusters represented between 0.8% and 2.1% of the total sequences. Among them, clusters 14 and 21 had high similarities (99% to 100%) with the already-described species Legionella cincinnatiensis and Legionella feeleii. Cluster 15 also included sequences with high similarity (99%) to an uncultured clone isolated from groundwater (M. W. Fields et al., unpublished data). It is interesting to note that, out of 29 clusters, 17 were most closely related to uncultured bacteria, and these included the major OTUs, highlighting the large diversity of clones representing not yet described Legionella species. The high diversity of Legionella OTUs was also observed in rarefaction curves (see Fig. S3 in the supplemental material), which had not reached a plateau.
Seasonal and anthropogenic effects on Legionella diversity.
Considering all the water samples, a high number of SSCP peaks were observed, varying from 6 to 15, for all sampling sites throughout the year. The SSCP peak profiles obtained for June and December for each sampling site before and after anthropogenic discharge are presented in Fig. 3 and show clear differences in the number of peaks and in their relative abundances for the different sampling sites and for the different seasons for a given sampling site. The similarities of the SSCP profiles, taking into account the areas under the profiles, were compared to determine if the distribution of Legionella species was site specific. Considering all profiles from the whole year, the Legionella populations were not separated according to sample site, as indicated by the overlap in the similarities of many of the SSCP profiles (see Fig. S1 in the supplemental material). An ANOSIM test also gave an R value close to 0 (R = 0.103, P < 0.001), indicating that the Legionella diversities at the sample sites were not significantly different. However, the wide separation of many of the Amélie-les-bains samples and some Céret samples in the nMDS plot (see Fig. S1 in the supplemental material) suggests that, for some time points, the Legionella diversities at these two sampling sites were different. Interestingly, the similarities of the monthly profiles for La Preste were more closely clustered than those for Céret and Amélie-les-bains (see Fig. S1 in the supplemental material), which could suggest that there was less variability in Legionella diversity at this pristine site.
FIG. 3.
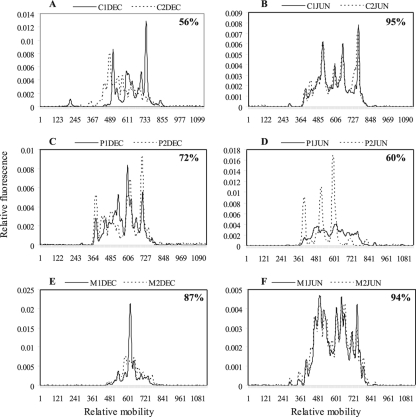
Examples of temporal and spatial variation of Legionella CE-SSCP profiles for the three sampling sites, Céret (A and B), La Preste (C and D), and Amélie-les-bains (E and F), upstream (solid lines) and downstream (dashed lines) of the water discharges. Percent similarities between the upstream and downstream profiles are indicated.
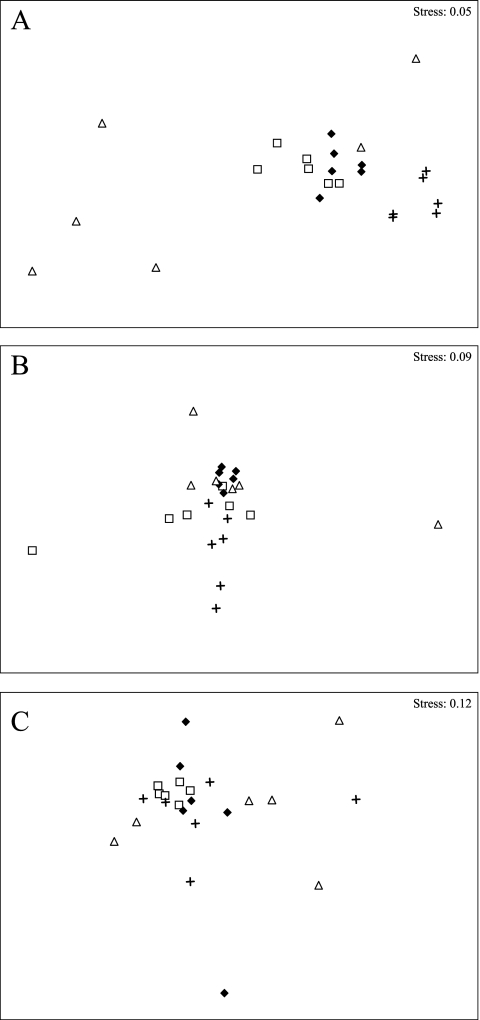
nMDS plots for the monthly samples for the individual sample sites (Fig. 4A to C) and the dendrograms (Fig. 5) showed that for Céret the samples appeared to be clustered roughly according to season whereas such a clustering was less clear for La Preste and Amélie-les-bains. ANOSIM tests confirmed the good separation of the seasonal groups for Céret (R = 0.512, P < 0.001) but less separation for La Preste (R = 0.277, P < 0.001) and Amélie-les-bains (R = 0.152, P < 0.001). In addition, at Céret, pairwise ANOSIM tests indicated that, whereas the spring and the summer profiles were not well separated from each other (R = 0.1, P < 0.17), they were very different from the winter and fall profiles (Rspring/winter = 0.957, P < 0.002; Rsummer/winter = 0.954, P < 0.002; Rfall/winter = 0.607, P < 0.002).
FIG. 4.
nMDS plots showing the definition of seasonal groups from the CE-SSCP profiles for Céret (A) but not for La Preste (B) or Amélie-les-bains (C). ▵, fall; +, winter; □, spring; ⧫, summer.
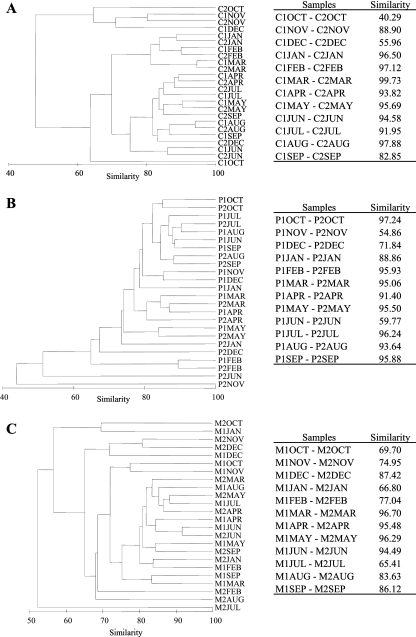
FIG. 5.
Dendrograms and similarity comparisons of CE-SSCP profiles before (1) and after (2) wastewater (Céret [C]) or thermal bath (La Preste [P] and Amélie-les-bains [M]) discharge for all monthly samples from the 3 sampling sites. (A) Céret; (B) La Preste; (C) Amélie-les-bains.
To determine the impacts of anthropogenic discharges on Legionella population structure, the CE-SSCP profiles obtained from samples collected upstream and downstream of water discharges were compared in dendrograms and by calculating the similarity of the profiles for each time point (Fig. 5). At the Céret sampling site, 10 of 12 CE-SSCP profiles obtained during the year were very similar (more than 82% similarity). Only two, obtained in October and December, showed quite different peak patterns (40 and 56% similarity), suggesting that different Legionella species were present at different relative abundances. At La Preste in November and June, the CE-SSCP profiles after the discharge of the thermal baths were very different from the ones obtained upstream. Indeed three major peaks dominated the SSCP fingerprint, whereas the upstream pattern possessed the usual 12 to 14 peaks (June) (Fig. 3D). Percentages of similarity obtained at these 2 dates were the lowest of the year (55 and 60% similarity), followed by December (72% similarity), which showed similarity differences more in the relative abundance of peaks than in the absence or presence of peaks. When considering all profiles of the year, we could observe that the discharge of the thermal baths of La Preste appeared to have an occasional impact on Legionella population, especially in the fall (November and December) and June. At the Amélie-les-bains sampling site, only 4 profiles (March to June) were very similar (>94%) before and after the thermal bath discharge, whereas the other profiles obtained upstream and downstream of the thermal baths for the rest of the year were less similar (<80 to 87%). Indeed, in fall and winter, the upstream and downstream profiles were very different (<80% similarity). Therefore, whereas there appeared to be an occasional effect of the water discharges from Céret and La Preste on Legionella diversity, this effect was more marked for the thermal baths on the Mondony River (Amélie-les-bains).
DISCUSSION
Although a few limited studies have revealed the presence of Legionella spp. in different aquatic environments (11, 38, 46), little is known about how these natural populations are affected by environmental and anthropogenic factors. Therefore, research into Legionella sp. ecology is essential to better understand their sources in the natural environment, the mechanism of their entry into man-made water systems, and the factors enabling their survival and growth in aquatic habitats. To address this need, this work investigated the dynamics of natural Legionella sp. populations along a river before and after water discharges from thermal bath establishments and a wastewater treatment plant during an annual cycle. To our knowledge this is the first time that such a comprehensive study of natural Legionella diversity and abundance has been performed during a whole year using both culture and culture-independent methods.
Whereas no Legionella spp. other than Legionella pneumophila were quantified on culture media, a real-time qPCR assay enabled us to detect and to quantify Legionella spp. in all samples throughout the year. Indeed, it is widely recognized that some Legionella species, such as L. lytica, and other Legionella-like amoebal pathogens (LLAPs) remain unculturable on the medium normally used to grow Legionella and on other media tested (29, 39). Although L. pneumophila could be quantified by the culture method, this species could not be quantified reliably by qPCR because the concentrations fell below the QL. The QL for the qPCR method was higher than the culture one, and qPCR was therefore less sensitive because the DNA samples had to be diluted 10-fold to overcome PCR inhibition. However, for Legionella spp., qPCR abundances were as high as 1 × 105 GU·liter−1 for the majority of the sampling dates for all sampling sites along the Tech River, even close to the source of the river, where there is no human activity. This could suggest that the Legionella spp. originate from the source of the river or that they could come from groundwater and/or atmospheric inputs, as has been proposed previously (14, 37). Very few studies have quantified Legionella abundance in their natural reservoirs, and the majority of these used culturing methods. Studies using qPCR quantification of Legionella have been mostly restricted to man-made systems such as drinking water distribution systems (hot sanitary) and cooling towers implicated in legionellosis outbreaks. Surprisingly, the Legionella abundances found close to the source of the Tech River, which could be considered a pristine environment, were comparable to those found in cooling towers (44).
Another important result from this study was the high diversity of Legionella for all sampling sites, shown both by sequencing of the 16S rRNA gene V1-V2 fragment and by SSCP analysis, which showed a high number of different OTUs and peaks, respectively.
This is in contrast to a phylogenetic study of Legionella species performed from a hypervariable short fragment of 16S rRNA gene (V3-V4-V5 region) in a river in the Brazilian rain forest that found a low diversity of Legionella species (11). Only L. pneumophila was detected in the pristine sampling site, and a dozen species of Legionella were found downstream, where the anthropogenic effect was greater (11). In addition to finding high Legionella diversity, this study found that the majority of the OTUs were most closely related to clones of uncultured bacteria than to already-described Legionella strains.
A high number of sequences affiliated with not yet described Legionella species were also found in cooling towers (44), suggesting that cultured representatives are lacking for Legionella not only in natural reservoirs but also in man-made systems. Interestingly, although the Legionella community was initially diverse in these cooling towers, the Legionella sp. diversity was dramatically reduced during the proliferation of L. pneumophila. The quantification data and the clone sequences in our study all show that L. pneumophila was not the most abundant member of the Legionella community in the Tech River. This may suggest that this species is able to proliferate and dominate the Legionella community only under specific conditions (e.g., higher temperatures characteristic of man-made systems), as already reported by Wéry et al. (44).
To better understand the dynamics of the Legionella populations, SSCP was used to analyze monthly samples at the different sampling sites before and after the anthropogenic discharges. No changes in Legionella populations were observed in relation to their position upstream or downstream in the river. Indeed, the high diversity was preserved throughout the year along the Tech River, and no SSCP profile was specific to a given station. Consequently, we could conclude that a large number of Legionella species were able to survive along the river in spite of the environmental pressure. Nevertheless, SSCP analyses of the dynamics of Legionella populations during a year revealed that some changes in the Legionella diversity could be related to the season. This was observed especially in Céret, where fingerprints of Legionella detected in fall and winter were different from those obtained in spring and summer. We did not find the same seasonal effect at La Preste and Amélie-les-bains, where different groups could be identified but were unrelated to the seasons. We suggest that seasonal variations were less marked at La Preste than at Céret and may be explained by different environmental factors. Biotic and abiotic factors (e.g., grazing, UV radiation, temperature, sedimentation, etc.) (21) may have had more impact at Céret, where the river bed was wider and shallower than at La Preste, where the river was narrow and the flow turbulent.
Comparison of the SSCP profiles upstream and downstream of the water discharges throughout the year suggested that the WWTP discharges had little impact on Legionella diversity. However, further studies should be carried out to determine the impact of other WWTPs on Legionella diversity and abundance to confirm that the effect of the wastewater discharge is limited. In contrast, at the thermal bath establishment sampling sites, the differences in Legionella sp. diversity between the upstream and downstream sites were more marked for a greater number of sampling dates (3 sampling dates for La Preste and 5 to 8 for Amélie-les-bains). The change in Legionella sp. diversity could be due to a direct addition of different Legionella spp. into the river population from the natural hot spring water or the water distribution system in the thermal bath establishment (24, 33). The thermal discharge water could also modify the Legionella sp. populations in the river by altering the physicochemical properties of the river water. The average temperature variations were 2.7°C and 6°C for La Preste and Amélie-les-bains, respectively, whereas no temperature difference was observed at Céret (see Table S2 in the supplemental material). The more marked changes in Legionella sp. diversity observed downstream of the thermal bath stations could be explained by the higher temperatures since temperature changes can influence the distribution of Legionella species (38). In addition to modifying Legionella sp. diversity via temperature effects, the discharge water could modify diversity via other abiotic and biotic factors such as pH or the presence of amoebae and biofilms, which could improve the survival of Legionella in the river (16, 27, 42).
However, we cannot rule out the possibility that the discharge waters of the two sites could be differently contaminated (in terms of Legionella diversity and abundance), explaining the different impacts on the Legionella population. In terms of Legionella sp. abundance, the qPCR data indicated little difference between the upstream and downstream sites except for La Preste, where 7 out of 12 monthly samples gave lower concentrations of Legionella spp. downstream of the site. Possible explanations of this observation could be a greater dilution effect from the discharge water (and assuming a low concentration of Legionella spp. in this water) at this site or perhaps an inhibitory effect of the discharge water on the natural Legionella sp. populations.
In order to confirm anthropogenic effects on Legionella populations, further studies involving higher numbers of replicate samples upstream and downstream of the site of interest and the sampling of the discharge water itself will be required.
In conclusion, our results showed a high abundance and diversity of Legionella species at all the sampling sites, even near the source of the river. This is an important result because it suggests that Legionella reservoirs may be much more widespread than previously thought. The dominance of sequences most closely related to uncultured Legionella demonstrates the urgent need to isolate these different Legionella species to better understand their persistence in aquatic environments and to determine their pathogenicity. A necessary step will be to study the interaction of Legionella with the protozoa, which are often essential for their proliferation and survival in the environment.
Supplementary Material
Acknowledgments
This work was supported by grants from Electricité De France (Research and Development Department, Laboratoire National d'Hydraulique et d'Environnement).
We thank Bio-Rad for providing iQ-Check Quanti kits and Marion Mottier for her technical support.
Footnotes
Published ahead of print on 22 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., R. Sarma-Rupavtarm, V. Klepac-Ceraj, and M. F. Polz. 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71:8966-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakawa, H., S. Nakashiro, M. Maeda, and A. Tsuji. 1996. Analysis of single-strand DNA conformation polymorphism by capillary electrophoresis. J. Chromatogr. A 722:359-368. [DOI] [PubMed] [Google Scholar]
- 4.Association Française de Normalisation. 2003. Water quality-detection and enumeration of Legionella spp. and L. pneumophila. Method by direct inoculation and after concentration by membrane filtration or centrifugation. AFNOR NF T90-431. Association Française de Normalisation, Paris, France.
- 5.Association Française de Normalisation. 2005. Détection et quantification des Legionella spp. et/ou Legionella pneumophila par concentration et amplification génique par réaction de polymérisation en chaîne (PCR). XP T 90-471. Association Française de Normalisation, Paris, France.
- 6.Aurell, H., P. Catala, P. Farge, F. Wallet, M. Le Brun, J. H. Helbig, S. Jarraud, and P. Lebaron. 2004. Rapid detection and enumeration of Legionella pneumophila in hot water systems by solid-phase cytometry. Appl. Environ. Microbiol. 70:1651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartie, C., S. N. Venter, and L. H. Nel. 2003. Identification methods for Legionella from environmental samples. Water Res. 37:1362-1370. [DOI] [PubMed] [Google Scholar]
- 8.Bernander, S., K. Jacobson, J. H. Helbig, P. C. Luck, and M. Lundholm. 2003. A hospital-associated outbreak of Legionnaires' disease caused by Legionella pneumophila serogroup 1 is characterized by stable genetic fingerprinting but variable monoclonal antibody patterns. J. Clin. Microbiol. 41:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borella, P., M. T. Montagna, S. Stampi, G. Stancanelli, V. Romano-Spica, M. Triassi, I. Marchesi, A. Bargellini, D. Tato, C. Napoli, F. Zanetti, E. Leoni, M. Moro, S. Scaltriti, G. Ribera D'Alcala, R. Santarpia, and S. Boccia. 2005. Legionella contamination in hot water of Italian hotels. Appl. Environ. Microbiol. 71:5805-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo-Bado, L. A., J. A. W. Morgan, M. Sergeant, T. R. Pettitt, and J. M. Whipps. 2003. Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops. Appl. Environ. Microbiol. 69:533-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho, F., R. Vazoller, A. Foronda, and V. Pellizari. 2007. Phylogenetic study of Legionella species in pristine and polluted aquatic samples from a tropical Atlantic forest ecosystem. Curr. Microbiol. 55:288-293. [DOI] [PubMed] [Google Scholar]
- 12.Catalan, V., F. Garcia, C. Moreno, M. J. Vila, and D. Apraiz. 1997. Detection of Legionella pneumophila in wastewater by nested polymerase chain reaction. Res. Microbiol. 148:71-78. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, K., and R. Gorley. 2006. PRIMER v6: user manual/tutorial. PRIMER Enterprises, Plymouth, United Kingdom.
- 14.Costa, J., I. Tiago, M. S. da Costa, and A. Verissimo. 2005. Presence and persistence of Legionella spp. in groundwater. Appl. Environ. Microbiol. 71:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Declerck, P., J. Behets, V. van Hoef, and F. Ollevier. 2007. Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res. 41:3159-3167. [DOI] [PubMed] [Google Scholar]
- 17.Delbès, C., R. Moletta, and J. J. Godon. 2000. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 18.Dusserre, E., C. Ginevra, S. Hallier-Soulier, F. Vandenesch, G. Festoc, J. Etienne, S. Jarraud, and M. Molmeret. 2008. A PCR-based method for monitoring Legionella pneumophila in water samples detects viable but noncultivable legionellae that can recover their cultivability. Appl. Environ. Microbiol. 74:4817-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekete, R., M. Miller, and D. Chattoraj. 2003. Fluorescently labeled oligonucleotide extension: a rapid and quantitative protocol for primer extension. Biotechniques 35:90-98. [DOI] [PubMed] [Google Scholar]
- 20.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fliermans, C. 1996. Ecology of Legionella: from data to knowledge with a little wisdom. Microb. Ecol. 32:203-228. [DOI] [PubMed] [Google Scholar]
- 22.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 24.Hsu, B.-M., C.-H. Chen, M.-T. Wan, and H.-W. Cheng. 2006. Legionella prevalence in hot spring recreation areas of Taiwan. Water Res. 40:3267-3273. [DOI] [PubMed] [Google Scholar]
- 25.Jonas, D., A. Rosenbaum, S. Weyrich, and S. Bhakdi. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J. Clin. Microbiol. 33:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klepac-Ceraj, V., I. Ceraj, and M. Polz. 2006. Clusterer: extendable Java application for sequence grouping and cluster analyses. Online J. Bioinform. 7:15-21. [Google Scholar]
- 27.Lau, H. Y., and N. J. Ashbolt. 2009. The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J. Appl. Microbiol. 107:368-378. [DOI] [PubMed] [Google Scholar]
- 28.Lee, T. C., R. M. Vickers, V. L. Yu, and M. M. Wagener. 1993. Growth of 28 Legionella species on selective culture media: a comparative study. J. Clin. Microbiol. 31:2764-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leoni, E., and P. P. Legnani. 2001. Comparison of selective procedures for isolation and enumeration of Legionella species from hot water systems. J. Appl. Microbiol. 90:27-33. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molmeret, M., M. Horn, M. Wagner, M. Santic, and Y. Abu Kwaik. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okabe, S., M. Odagiri, T. Ito, and H. Satoh. 2007. Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems. Appl. Environ. Microbiol. 73:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada, M., K. Kawano, F. Kura, J. Amemura-Maekawa, H. Watanabe, K. Yagita, T. Endo, and S. Suzuki. 2005. The largest outbreak of legionellosis in Japan associated with spa baths: epidemic curve and environmental investigation. Kansenshogaku Zasshi 79:365-374. [DOI] [PubMed] [Google Scholar]
- 34.Palmer, C. J., Y. L. Tsai, C. Paszko-Kolva, C. Mayer, and L. R. Sangermano. 1993. Detection of Legionella species in sewage and ocean water by polymerase chain reaction, direct fluorescent-antibody, and plate culture methods. Appl. Environ. Microbiol. 59:3618-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, M., S. Yun, M. Kim, J. Chun, and T. Ahn. 2004. Phylogenetic characterization of Legionella-like endosymbiotic X-bacteria in Amoeba proteus: a proposal for “Candidatus Legionella jeonii” sp. nov. Environ. Microbiol. 6:1252-1263. [DOI] [PubMed] [Google Scholar]
- 36.Sabria, M., J. Alvarez, A. Dominguez, A. Pedrol, G. Sauca, L. Salleras, A. Lopez, M. A. Garcia-Nunez, I. Parron, and M. P. Barrufet. 2006. A community outbreak of Legionnaires' disease: evidence of a cooling tower as the source. Clin. Microbiol. Infect. 12:642-647. [DOI] [PubMed] [Google Scholar]
- 37.Sakamoto, R., A. Ohno, T. Nakahara, K. Satomura, S. Iwanaga, Y. Kouyama, F. Kura, N. Kato, K. Matsubayashi, K. Okumiya, and K. Yamaguchi. 2009. Legionella pneumophila in rainwater on roads. Emerg. Infect. Dis. 15:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ta, A. C., J. E. Stout, V. L. Yu, and M. M. Wagener. 1995. Comparison of culture methods for monitoring Legionella species in hospital potable water systems and recommendations for standardization of such methods. J. Clin. Microbiol. 33:2118-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 41.Templeton, K. E., S. A. Scheltinga, P. Sillekens, J. W. Crielaard, A. P. van Dam, H. Goossens, and E. C. J. Claas. 2003. Development and clinical evaluation of an internally controlled, single-tube multiplex real-time PCR assay for detection of Legionella pneumophila and other Legionella species. J. Clin. Microbiol. 41:4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadowsky, R. M., R. Wolford, A. M. McNamara, and R. B. Yee. 1985. Effect of temperature, pH, and oxygen level on the multiplication of naturally occurring Legionella pneumophila in potable water. Appl. Environ. Microbiol. 49:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wery, N., V. Bru-Adan, C. Minervini, J.-P. Delgenes, L. Garrelly, and J.-J. Godon. 2008. Dynamics of Legionella spp. and bacterial populations during the proliferation of L. pneumophila in a cooling tower facility. Appl. Environ. Microbiol. 74:3030-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wullings, B., R. Voogt, H. Veenendaal, and D. van der Kooij. 2002. The fluorescent in situ hybridization test in comparison with culture for detection of Legionella pneumophila in water samples, p. 263-266. In M. Reinhard (ed.), Legionella. ASM Press, Washington, DC.
- 46.Wullings, B. A., and D. van der Kooij. 2006. Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15°C. Appl. Environ. Microbiol. 72:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, H., Y. Hashimoto, and T. Ezaki. 1996. Study of nonculturable Legionella pneumophila cells during multiple-nutrient starvation. FEMS Microbiol. Ecol. 20:149-154. [Google Scholar]
- 48.Yaradou, D. F., S. Hallier-Soulier, S. Moreau, F. Poty, Y. Hillion, M. Reyrolle, J. Andre, G. Festoc, K. Delabre, F. Vandenesch, J. Etienne, and S. Jarraud. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl. Environ. Microbiol. 73:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zemb, O., B. Haegeman, J. P. Delgenes, P. Lebaron, and J. J. Godon. 2007. SAFUM: statistical analysis of SSCP fingerprints using PCA projections, dendrograms and diversity estimators. Mol. Ecol. Notes 7:767-770. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.