Abstract
There is considerable interest in the use of psychrotrophic bacteria for food biopreservation and in the understanding of cold adaptation mechanisms. The psychrotrophic biopreservative Lactococcus piscium strain CNCM I-4031 was studied for its growth behavior and proteomic responses after cold shock and during cold acclimation. Growth kinetics highlighted the absence of growth latency after cold shock, suggesting a very high promptness in cold adaptation, a behavior that has never been described before for lactic acid bacteria (LAB). A comparative proteomic analysis was applied with two-dimensional gel electrophoresis (2-DE), and upregulated proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Both cold shock and cold acclimation triggered the upregulation of proteins involved in general and oxidative stress responses and fatty acid and energetic metabolism. However, 2-DE profiles and upregulated proteins were different under both conditions, suggesting a sequence of steps in cold adaptation. In addition, the major 7-kDa Csp protein was identified in the L. piscium CNCM I-4031 genome but was not cold regulated. The implication of the identified cold shock proteins and cold acclimation proteins in efficient cold adaptation, the possible regulation of a histidyl phosphocarrier protein, and the roles of a constitutive major 7-kDa Csp are discussed.
Lactic acid bacteria (LAB) usually become the dominant flora during the storage of refrigerated preserved meat or fish products. Some of them have been widely described for their beneficial role in food preservation (42), and psychrotrophic strains present a real advantage for a biopreservation strategy (29). Matamoros et al. (35) previously isolated psychrotrophic lactic acid bacteria from seafood products and examined them for their ability to inhibit pathogenic and spoiling bacteria in a model medium. One of these strains, Lactococcus piscium CNCM I-4031, previously named EU2241, has been shown to extend the sensory shelf-life of vacuum-packed shrimp and cold-smoked salmon and to limit the development of Listeria monocytogenes, Staphylococcus aureus, and Brochothrix thermosphacta (12, 13, 34). This bacterium shows an uncommon temperature-related growth profile. Unlike most of the LAB, the growth of L. piscium CNCM I-4031 occurs from 0°C to 29°C, with an optimum at 26°C, and is inhibited at temperatures higher than 29°C, suggesting a specific cold adaptation behavior.
Low temperatures usually result in a wide range of molecular and cellular disruptions: decreases in membrane fluidity and in enzymatic activity (Arrhenius), inefficient folding of some proteins, and increases in RNA and DNA secondary structures, leading to a decreased efficiency of transcription and translation (51). Cold environments are widespread on Earth, and under these conditions, the microbial communities are dominated by various psychrophilic and psychrotrophic bacteria that have developed diverse cold adaptation strategies (38). Proteomic analysis is a powerful tool to investigate strategies for adaptation to various environmental changes for numerous species, including LAB (7). According to current knowledge, after a downshift in temperature, psychrotrophic and mesophilic bacteria synthesize a set of cold shock proteins (CSPs) in amounts related to the severity of the shock (22). In Escherichia coli, Bacillus subtilis, and Lactococcus lactis, the major upregulated Csp belongs to a 7-kDa protein family (19, 47, 51). These proteins present a highly conserved nucleic acid binding domain and are implicated in numerous cellular functions, including gene regulation, freeze protection, and RNA chaperones. Seven-kilodalton CSP-like proteins have been reported for the genomes of over 50 bacterial species (51).
Despite their impact on foodstuffs, the cold shock and cold acclimation responses of psychrotrophic LAB are poorly documented. Bacteria in technological starters face a great variety of stresses throughout food processing. A better understanding of the L. piscium response to refrigerated temperatures could be helpful to optimize its application as a biopreservation agent in chilled products. The aim of this study was to describe the growth behavior of the bioprotective strain L. piscium CNCM I-4031 at a chilled temperature and after cold shock and to analyze protein responses to both cold shock and cold acclimation using two-dimensional gel electrophoresis (2-DE). The principal cold-induced proteins, detected by comparing gel electrophoresis profiles between the optimal growth of L. piscium CNCM I-4031 at 26°C and growth after cold acclimation or cold shock at 5°C, were identified and are discussed according to their molecular and cellular functions.
MATERIALS AND METHODS
Bacterial strain, growth conditions, and cold shock treatments.
Lactococcus piscium EU2241, deposited in the national collection of microorganisms of the Pasteur Institute (Paris, France) under the name CNCM I-4031, was stored at −80°C in Elliker (ELK) broth with 10% glycerol. Subcultures were performed for 20 h at an optimal temperature (26°C) in 10 ml common LAB complex nutrient-rich medium ELK broth (Biokar Diagnostics, Beauvais, France). One liter of ELK contains 20 g tryptone, 5 g yeast extract, 2.5 g gelatin, 5 g glucose, 5 g sucrose, 5 g lactose, 1.5 g Na acetate, 4 g NaCl, and 0.5 g ascorbic acid, and the pH was adjusted to 7 (11). Cultures were obtained by inoculating 1% (vol/vol) of the subculture into 200 ml ELK and incubating without shaking at 26°C.
Growth kinetics were investigated by using the automated Bioscreen C microbial growth analyzer system (Labsystem, Helsinki, Finland) with 96-well microtiter plates with 400 μl per well of the bacterial culture and without shaking. Cells were harvested from cultures at 26°C at the mid-exponential phase (optical density at 600 nm [OD600] of 0.5) and then diluted to an OD600 of 0.2 with Elliker broth at the appropriate temperature. Growth kinetic experiments were performed with Elliker broth at 5°C for acclimation experiments or at 26°C under standard conditions. For cold shock experiments, diluted cultures were submitted to 1 or 2 h of incubation at 0°C or 5°C, and growth kinetic experiments were performed at 26°C. All experiments were carried out in triplicate. Growth curves were fitted with the Gompertz model for the estimation of μmax (54).
For the proteomic study, 200 ml of mid-exponential-phase (OD600 of 0.5) cultures were obtained as described above at either 26°C under standard conditions or 5°C under acclimation conditions. For cold shock experiments, cells were cultivated at 26°C and submitted to 1 h of cold shock treatment at 5°C in fresh Elliker broth. Each culture sample was centrifuged at 10,000 × g for 20 min at 26°C or 5°C, according to the temperature of the culture. Each pellet was washed in a solution containing 10 mM Tris-HCl (pH 7.2) and 5 mM EDTA, resuspended in 10 ml of the same buffer containing 200 mM glycine, and quickly frozen in liquid nitrogen before storage at −80°C. Three independent cultures were performed under each condition.
Protein extraction.
Under each condition (standard, acclimation, and cold shock), cytosoluble proteins were extracted from frozen cell pellets. Cells were pulse sonicated by using a Vibra-Cell 72434 sonicator (Bioblock, Illkirch, France) in an ice bath during five cycles of 6 min in the presence of a protease inhibitor (Complete tablets; Roche Diagnostics, Mannheim, Germany). After ultracentrifugation (188,000 × g for 60 min), extracts were treated with RNase and DNase solutions and dialyzed against water using a 2-kDa-cutoff membrane (Sigma-Aldrich, Saint-Quentin Fallavier, France).
2-DE.
Each extract was analyzed on analytic 2-DE gels using a method adapted from that described previously by O'Farrell (39). A pH gradient of 4 to 7 was chosen for isoelectric focusing (IEF), as a large proportion of the putative hydrosoluble proteins from the draft genome sequence of Lactococcus lactis has been observed in this range of pHs (7) and confirmed for L. piscium in IEF experiments at pH 3 to 10 in our laboratory (data not shown). The second-dimension electrophoresis was performed on 13% SDS-polyacrylamide gels to optimize the separation of proteins with a molecular mass ranging from 5 to 150 kDa. Aliquots containing 50 μg of protein for analytic gels and 600 μg for preparative gels were concentrated in a 15-μl final volume and diluted in 275 μl of rehydration buffer containing 6 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% Biolyte 3/10, 0.01% bromophenol blue, and 5% dithiothreitol (DTT). After 18 h of active rehydration of immobilized pH gradient (IPG) dry strips at pH 4 to 7 (Bio-Rad, Marnes-la-Coquette, France) at 50 V, IEF was performed by using a Bio-Rad Protean IEF cell at 66,000 V·h. The strips were next treated with buffer containing 6 M urea, 2% SDS, 0.05 M Tris-HCl (pH 8.8), and 30% glycerol and supplemented with 2% DTT and then treated again with the same buffer containing 4% iodoacetamide. Finally, proteins were visualized by a silver staining method for analytic gels and Bio-Safe colloidal Coomassie blue (Bio-Rad, Marnes-la-Coquette, France) for preparative gels. Under each condition, at least two replicates from each of the three independent cultures were carried out.
Image and statistical analysis.
Images of analytic gels were recorded with a Bio-Rad GS800 densitometer. Gels were analyzed with the Progenesis SameSpots, version 3.0, software (Nonlinear Dynamics Ltd., Newcastle, United Kingdom). The quality of the gels was ensured by using the quality control (QC) of the software. This includes an analysis of image compression, image saturation (spot exposition), image bit depth, available dynamic range and intensity levels in use, alteration due to an edited image, and intensity level resolution. Only those images that passed the available quality control were included in the analysis. Available dynamic range and intensity levels were at least 95% and 96% for all gels, respectively. Automatic pixel level geometric alignments of the gels, followed by manual corrections, were performed with the vector alignment tool of the SameSpots workflow. In addition, an automatic management of spots and spot volume measurement were carried out. The background-corrected abundance of each spot was calculated, and the abundance ratio was determined by dividing the sample abundance by the reference abundance. Spot volumes were normalized to calibrate data between different sample runs, and normalized spots were then analyzed statistically by using the statistical module implemented in SameSpots. Principal component analysis (PCA) was used to separate the gels according to variations in the normalized volume of the spots. After that, data under each cold-induced condition (cold adaptation and cold shock) were compared to those for the control at 26°C using an analysis of variance (ANOVA), which was reduced to a t test by the software for comparison of one independent factor. Significantly overexpressed spots were detected at a 5% significance level (P value of <0.05). Finally, these significantly overexpressed spots were refined by using a q value of <0.05 to discard false positives, a power of >0.8 to ensure reproducibility among gels under the same conditions, and a fold value of >1.8 for the biological significance.
Protein identification.
Detectable upregulated spots were excised from preparative gels. In-gel digestion was performed with the Progest system (Genomic Solution) according to a standard trypsinolysis protocol. Gel plugs were first washed twice with 10% (vol/vol) acetic acid and 40% (vol/vol) ethanol in water and then washed with acetonitrile. They were further washed with 25 mM NH4CO3 and dehydrated in acetonitrile (two alternating cycles). Digestion was performed for 6 h at 37°C with 125 ng of modified trypsin (Promega) dissolved in 20% (vol/vol) methanol in 20 mM NH4CO3. Tryptic peptides were first extracted with 50% (vol/vol) acetonitrile and 0.5% trifluoroacetic acid in water and then extracted with pure acetonitrile. Both peptide extracts were pooled, dried in a vacuum-speed concentrator, and suspended in 25 μl of 2% (vol/vol) acetonitrile and 0.08% (vol/vol) trifluoroacetic acid in water.
Liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis was performed with an Ultimate 3000 LC system (Dionex, Voisins-le-Bretonneux, France) connected to an LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher) via a nanoelectrospray ion source. After 4 min, the precolumn was connected to the separating Pepmap C18 nanocolumn (0.075 by 15 cm, 100 Å, and 3 μm), and the linear gradient was started from 2 to 36% of buffer B (80% acetonitrile, 0.1% formic acid) in buffer A (2% acetonitrile, 0.1% formic acid) at 300 nl/min over 50 min. The double- and triple-charged precursor ions were subjected to MS/MS fragmentation with a 3-min exclusion window and with classical peptide fragmentation parameters (Qz = 0.22; activation time = 50 ms; collision energy = 35%).
The raw mass data were first converted to mzXML format with ReAdW software (http://tools.proteomecenter.org/software.php). In the absence of genomic data for L. piscium, protein identification was performed by querying MS/MS data against a Firmicutes protein database (UniProtKB, version 2009.06.08), together with an in-house contaminant database, using X!Tandem software (X!Tandem Tornado 2008.02.01.3 [http://www.thegpm.org]) with the following parameters: one trypsin missed cleavage allowed, alkylation of cysteine and conditional oxidation of methionine, and precursor and fragment ions set at 10 ppm and 0.5 Da, respectively.
A refined search was added with similar parameters except that semitryptic peptides and possible N-terminal acetylation of proteins were searched for.
All peptides that matched with an E value lower than 0.05 were parsed with an in-house program (http://PAPPSO.inra.fr/bioinformatique.html). Proteins identified with at least three unique peptides and a log E value lower than 10−8 were validated.
For the samples matching fewer than three unique peptides per protein, mass spectra with a quality score higher than 0.2 were interpreted by de novo sequencing, which was performed by using PepNovo software (version 3). Enzymatic cleavage and modifications are indicated above.
Generated sequence tags were submitted to homology searches in Lactococcus, Firmicutes, and bacterial protein databases (UniProtKB, version 2009.06.08) with Fasts software (v.34.26) (31) using the MDM20 matrix in an iterative process. Only homologies with a protein E value lower than 10−5, identified with at least two different peptides, were selected.
In all cases, the automatic de novo interpretation of MS/MS spectra was manually validated.
Identification of a 7-kDa CSP gene.
The nucleic sequence of a putative 7-kDa CSP identified by LC-MS/MS was investigated. A degenerated nucleotide sequence obtained by reverse transcription of the amino acid sequence was compared to the gene sequences of other bacterial 7-kDa CSPs retrieved from online databases. The following PCR primers were designed from the conserved regions of these sequences: CspD1-f (5′-GGCAAATGGAACAGTAAAATGG-3′) and CspD3-r (5′-TCCATCAGATTGGATTGCTGAG-3′). PCRs were performed in a total volume of 50 μl containing 1× PCR buffer, 1.5 mM MgCl2, 50 ng DNA, 0.8 μM each primer, 0.2 mM each deoxynucleoside triphosphate (dNTP), and 1 U Taq DNA polymerase. Amplification consisted of a first cycle of 5 min at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C and a final extension step for 5 min at 72°C. The nucleotide sequence of the amplified gene fragment was determined with an ABI 3730 automated sequencer using the Taq Dye-Deoxy terminator cycle sequencing method (Genome Express Company, Meylan, France) and primers cspD1-f and cspD3-r. Anticipated errors of PCR and sequencing reactions were avoided by sequencing both DNA strands. The sequence was then compared to sequences in the databases by using the nucleotide-nucleotide BlastN program (http://www.ncbi.nlm.nih.gov/BLAST/).
Contiguous parts of the gene were obtained by using an inverse-PCR strategy. Briefly, separate samples containing 10 μg of total DNA were digested using 12 restriction enzymes, EcoRI, SalI, EcoRV, BsmAI, NcoI, DpnI, BspHI, XhoI, MseI, HindIII, HaeIII, and HhaI (Ozyme, Saint-Quentin-en-Yvelines, France), as recommended by the supplier. None of these enzymes cut inside the previously identified gene fragment. Recircularization of the digested products (40 μl) was performed with 800 U of T4 DNA ligase in a final volume of 400 μl. PCR amplification was then applied as described above by using reverse primers Icq1f (5′-CGAAGCCTTTTTCAGCGTTA-3′) and Icq2r (5′-TCATCACTGGTCAGACGGAA-3′) and recircularized DNA as a template. PCR products were cloned with the Invitrogen pcrII-TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced as described above by using primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′).
RESULTS
Effect of temperature on bacterial growth.
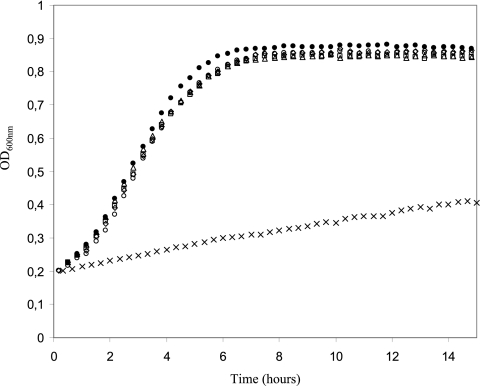
Growth kinetic studies were performed at 26°C and 5°C, and the μmax was 0.6 h−1 and 0.06 h−1, respectively (Fig. 1). These results are in accordance with those described previously by Matamoros et al. (35). It is noteworthy that at 5°C, the μmax was reached instantly without a lag time. No difference was observed in the growth rates at 26°C (μmax = 0.6 h−1) between the control cultures and the cultures submitted to cold shock during 1 or 2 h at either 0°C or 5°C. Moreover, no lag time was observed whatever the severity of the cold shock.
FIG. 1.
Effects of cold shock and cold acclimation at 5°C on the growth of Lactococcus piscium CNCM I-4031 in Elliker broth. Cells were harvested from standard cultures at 26°C at mid-exponential phase and grown at 5°C (×), at 26°C without cold shock (•), and at 26°C after 1 h at 0°C (○), 2 h at 0°C (□), 1 h at 5°C (⋄), and 2 h at 5°C (▵). Standard deviation values were lower than 0.03 and are not indicated on the graph (n = 9; three replicates of three independent cultures).
Differential proteome analysis.
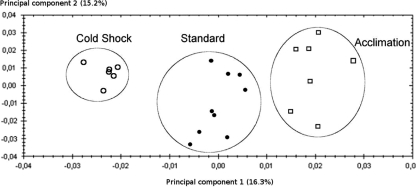
In order to characterize protein expression variation in response to L. piscium cold acclimation and cold shock, 2-DE profiles of cytosoluble protein contents of cultures at 26°C (standard culture) and at 5°C (cold acclimation) and cultures transferred from 26°C to 5°C for 1 h (cold shock) were compared. The PCAs of the complete data set (22 gels) clearly showed three distinct clusters of gels corresponding to biological reproductions of the three experimental conditions (Fig. 2), indicating specific responses of L. piscium CNCM I-4031 to cold shock and cold acclimation.
FIG. 2.
Principal component analysis performed on the complete data set of the 22 2-DE gels according to variations in the normalized volume of the spots. •, standard temperature conditions; ○, cold shock; □, cold acclimation.
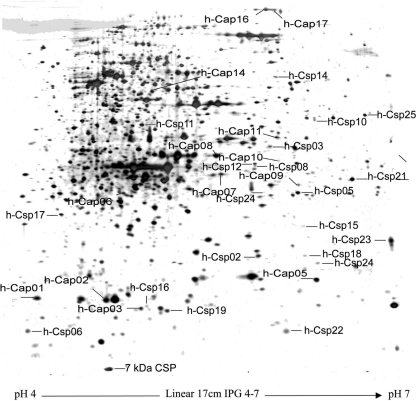
Figure 3 displays the 2-DE gels of cytosoluble silver-stained proteins extracted from L. piscium under the three temperature conditions. Under cold shock conditions, 28 spots were significantly overexpressed, and three were down-expressed compared to those under standard conditions (26°C). Under cold acclimation conditions, 31 spots were significantly overexpressed, and 32 were down-expressed. Among these spots, four were overexpressed and two were down-expressed under both cold shock and cold acclimation conditions. Overexpressed spots after cold shock and during cold acclimation were named hypothetical Csp (h-Csp) proteins and hypothetical Cap (h-Cap) proteins, respectively, and overexpressed spots under both cold conditions were named early Cap proteins according to the definition described previously by Hébraud and Potier (22).
FIG. 3.
Two-dimensional electrophoresis gel of cytosoluble silver-stained proteins from Lactococcus piscium EU2241. Proteins were extracted from exponential-phase cultures at 5°C. Identified cold shock proteins (h-Csp), cold acclimation proteins (h-Cap), and the non-cold-induced 7-kDa Csp are localized.
Identification of overexpressed proteins.
Thirty-three spots, including 20 h-Csp proteins, one early Cap protein, and 12 h-Cap proteins, were detected in preparative gels and submitted to identification by using LC-MS/MS. Overall, 23 proteins were identified from the MS/MS data (Table 1). Among them, six proteins were identified with several spot occurrences. Differences in migration for the same identified protein may be due to posttranslational modifications, isoforms, or truncated or degraded forms. Moreover, only truncated forms were identified for three proteins (glyceraldehyde-3-phosphate dehydrogenase [GAPDH], elongation factor Tu [EF-Tu], and DnaK). Identified proteins were classified into four metabolic groups.
TABLE 1.
Cold-inducible proteins of L. piscium CNCM I-4031
| Gene and functiona | Spot | UniProtKB accession no.b | Log E valuec | No. of unique peptides | Molecular mass (kDa) |
Fold change (P value)d |
||
|---|---|---|---|---|---|---|---|---|
| Theoretical | Observed | Cold shock/standard | Cold acclimation/standard | |||||
| Group 1 (energetic metabolic process) | ||||||||
| PGK | h-Csp08 | A2RHX1 | −49.3 | 12 | 42.0 | 27.0 | 2.3 (0.002) | 1.2 |
| h-Csp12 | A2RHX1 | −52.2 | 15 | 42.0 | 30.0 | 2 (<0.001) | 1.3 | |
| h-Csp21 | A2RHX1 | −47.5 | 10 | 42.0 | 27.0 | 1.8 (0.004) | 0.9 | |
| Fba | h-Csp11 | A2RN46 | −155.7 | 29 | 31.9 | 38.0 | 2.1 (0.004) | 0.8 |
| GAPDH | h-Csp04 | B8YDH3 | −69.3 | 11 | 35.8 | 18.0 | 2.7 (<0.001) | 1.3 |
| HPr | h-Cap03 | Q9ZAD9 | −9.7 | De novo synthesis | 9.1 | 14.0 | 0.9 | 3.9 (<0.001) |
| h-Csp16 | Q5LZ93 | −9.9 | De novo synthesis | 9.1 | 14.0 | 1.9 (0.006) | 1.3 | |
| AdhE | h-Cap16 | Q837E3 | −35.3 | De novo synthesis | 92.4 | 200.0 | 2 | 2.7 (0.007) |
| h-Cap17 | Q837E3 | −38.3 | De novo synthesis | 92.4 | 200.0 | 2 (0.001) | 5.1 (<0.001) | |
| Group 2 (fatty acid biosynthesis) | ||||||||
| FabF | h-Csp03 | B9DVE4 | −27.0 | De novo synthesis | 43.7 | 37.0 | 3.2 (<0.001) | 1.1 |
| FabH | h-Cap11 | Q9CHG0 | −24.0 | De novo synthesis | 34.8 | 35.0 | 1.8 (<0.001) | 2.4 (<0.001) |
| FabG | h-Cap05 | Q65J08 | −38.5 | 7 | 27.1 | 15.0 | 1.5 | 2.9 (<0.001) |
| h-Cap07 | Q65J08 | −44.1 | 7 | 27.1 | 25.0 | 1.1 | 1.8 (<0.001) | |
| h-Cap08 | Q65J08 | −129.7 | 18 | 27.1 | 27.0 | 1.1 | 1.9 (<0.001) | |
| h-Cap10 | Q65J08 | −19.8 | De novo synthesis | 27.1 | 27.0 | 1.4 | 2.4 (<0.001) | |
| Group 3 (gene expression) | ||||||||
| UPP | h-Csp05 | Q8DST6 | −46.5 | 13 | 23.0 | 26.0 | 2.8 (<0.001) | 1.1 |
| h-Csp06 | Q8DST6 | −27.0 | 3 | 23.0 | 12.0 | 2.6 (<0.001) | 0.7 | |
| h-Csp24 | P36399 | −18.2 | 6 | 22.9 | 26.0 | 1.8 (0.004) | 0.7 | |
| DAHP synthetase | h-Csp25 | B1SEM3 | −53.0 | De novo synthesis | 38.9 | 45.0 | 1.8 (0.004) | 1.6 |
| Tgt | h-Csp14 | B6XS84 | −21.0 | De novo synthesis | 50.6 | 60.0 | 2 (<0.001) | 1.4 |
| TrmU | h-Cap14 | A2RH05 | −22.6 | 6 | 41.7 | 50.0 | 0.9 | 1.8 (0.003) |
| RplE | h-Csp23 | A2RNP3 | −15.7 | 5 | 19.9 | 19.0 | 1.8 (<0.001) | 1.1 |
| RsmG | h-Csp15 | A2RKW7 | −5.6 | De novo synthesis | 19.5 | 20.0 | 1.0 | 1.4 |
| RhlB | h-Csp10 | Q03JE4 | −24.8 | 5 | 58.9 | 40.0 | 2.4 (<0.001) | 1.4 |
| EF-G | h-Cap06 | Q3D5T1 | −14.0 | De novo synthesis | 49.5 | 25.0 | 1.2 | 2.2 (<0.001) |
| EF-Tu | h-Csp02 | A2RMT1 | −19.0 | De novo synthesis | 43.2 | 17.0 | 4.2 (<0.001) | 0.8 |
| Putative sigma 54 modulation protein | h-Cap09 | A2RIX0 | −11.24 | De novo synthesis | 21 | 26.0 | 1.4 | 4.3 (<0.001) |
| Group 4 (general stress proteins) | ||||||||
| AhpF | h-Csp17 | Q9CIL9 | −26.9 | De novo synthesis | 20.6 | 21.0 | 1.9 (0.008) | 0.5 |
| Trx | h-Cap01 | Q3D1V5 | −16.5 | De novo synthesis | 11.7 | 15.0 | 0.7 | 1.8 (<0.001) |
| OsmC-like protein | h-Csp18 | Q3XZP9 | −18.1 | De novo synthesis | 14.6 | 18.0 | 1.9 (0.009) | 1.7 |
| h-Csp19 | Q3XZP9 | −5.6 | De novo synthesis | 14.6 | 13.0 | 1.9 (0.009) | 1.7 | |
| DnaK | h-Csp22 | B7CDT6 | −20.6 | 6 | 65.1 | 12.0 | 1.8 (0.003) | 0.4 |
| UspA | h-Cap02 | A2RMQ7 | 41.7 | De novo synthesis | 15.7 | 15.0 | 1.4 | 2.1 (<0.001) |
Protein functions identified by homology searches of the Lactococcus, Firmicutes, and Bacteria protein databases.
UniProtKb identification of matched proteins.
Peptide mass matching and de novo identification scorings. Protein functions identified with fewer than three unique peptides were validated by the de novo approach.
Fold changes were calculated with average values of normalized spot volumes. Fold change values are in boldface type if upregulation was significant (P values determined by a Student's t test are in parentheses).
In the energetic metabolic processes, the histidyl phosphocarrier protein (HPr) (h-Cap03 and h-Csp16) was upregulated after cold shock and during cold acclimation. Fba, GAPDH, and Pgk, implicated successively in glycolysis, were upregulated after cold shock. The enzyme of heterolactic acid fermentation and pyruvate metabolism, alcohol-acetaldehyde dehydrogenase (AdhE), was upregulated 2-fold after cold shock and upregulated 5.1-fold during cold acclimation. Three ubiquitous bacterial enzymes involved in fatty acid metabolism were identified: beta-ketoacyl acyl carrier protein (ACP) synthase III (FabH), involved in the initiation phase of type II fatty acid synthesis (27), and beta-ketoacyl ACP synthase II (FabF) and beta-ketoacyl ACP reductase (FabG), involved in fatty acid elongation (52). FabF was upregulated after cold shock, FabG was upregulated during cold acclimation, and FabH was upregulated under both cold shock and acclimation conditions.
Proteins implicated in several steps of gene expression, including nucleoside and amino acid biosynthetic pathways (uracil phosphoribosyltransferase [UPP] and Tyr-sensitive phospho-2-dehydro-deoxyheptonate aldolase [DAHP synthetase]), tRNA modifications (queuine tRNA-ribosyltransferase [Tgt] and tRNA-specific 2-thiouridylase MnmA [TrmU]), the RNA metabolic pathway (ATP-dependent RNA helicase [RhlB] and a putative sigma 54 modulation protein), the formation and methylation of ribosomes (50S ribosomal protein L5 [RplE] and putative rRNA methyltransferase [RsmG]), and peptide elongation (elongation factor G [EF-G] and EF-Tu), were identified. UPP, DAHP synthetase, Tgt, RplE, RsmG, RhlB, and EF-Tu were upregulated after cold shock, and TrmU and EF-G were upregulated during cold acclimation.
Among the five identified proteins involved in prokaryotic general stress responses, three are associated with cell redox homeostasis. OsmC-like protein and alkyl hydroperoxide reductase (AhpF) were induced after cold shock, and thioredoxin was overexpressed during cold acclimation. The ubiquitous heat shock protein chaperone DnaK was upregulated after cold shock, and universal stress protein A (UspA) was upregulated during cold acclimation.
Furthermore, exploring the major low-molecular-mass proteins, a non-cold-regulated protein migrating at 7 kDa was identified as a putative 7-kDa CSP.
Identification of a putative 7-kDa CSP (HM114314).
PCR amplification with primers CspD1-f and CspD3-r using DNA from strain L. piscium CNCM I-4031 as a template resulted in the amplification of a fragment of approximately 100 bp, as expected. Sequencing of this fragment and comparison of the nucleotide sequence with data in an online database confirmed the presence of a putative csp gene. PCR amplification with reverse PCR primers Icq1f and Icq2r, designed upon this sequence using the DNA from strain CNCM I-4031 digested by the enzyme HhaI and ligated as a template, resulted in the amplification of a 350-bp fragment. Analysis of this fragment with the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) revealed the presence of a 162-bp open reading frame (ORF) comprising the original PCR fragment. This ORF encodes a partial protein of 54 amino acids (Fig. 4) that shares 85, 84, and 81% identities with cold shock proteins from Enterococcus faecium, Enterococcus faecalis, and L. lactis subsp. lactis, respectively. The protein also contains the two RNA binding domains RNP1 and RNP2. Because of its similarity to the L. lactis 7-kDa CSPE protein (closest protein of a bacterium from the same genus), the protein from L. piscium CNCM I-4031 was named CspE.
FIG. 4.
Alignment of the Lactococcus piscium EU2241 CSPE protein with close 7-kDa CSPs from various bacteria. Consensus symbols: %, For Y; #, D, N, Q, or E. RNP1 and RNP2 motifs are boxed.
DISCUSSION
Psychrotrophic bacteria, like Pseudomonas fragi, L. monocytogenes, and Arthrobacter globiformis, respond to temperature downshifts by a lag period before growth according to the amplitude of the cold shock (5, 6, 37). For psychrophilic Vibrio sp. and the mesophilic bacteria L. lactis, Vibrio vulnificus (36), and B. subtilis, a lag period was not observed; however, the growth rate measured during the first hours was lower than the maximal growth rate (2, 30, 40). L. piscium CNCM I-4031 is able to endure cold shock within the permissive growth range without a lag period or intermediate growth rate. These original results suggest that the determinant factors of cold shock protection are constitutively expressed. Because comparative analysis of 2-DE gel profiles highlighted specific responses to cold shock and cold acclimation versus those under standard conditions, a molecular regulation should be required when L. piscium CNCM I-4031 is submitted to a cold shock, although no effect on growth was observed. Mesophilic bacteria react to a sudden downshift in temperature by a halt in general protein expression, whereas psychrotrophic bacteria do not. In L. piscium CNCM I-4031, most of the spots on 2-DE gels were apparently not cold regulated, which seems to confirm the psychrotrophic behavior of the strain. In mesophilic and psychrotrophic bacteria, cold shock proteins (Csp) are overexpressed for a limited time after cold shock. They assist in overcoming the immediate effects of temperature downshifts. In psychrotrophic bacteria during prolonged growth at low temperatures, cold acclimation proteins (Cap) are overexpressed and allow growth at chilled temperatures (17, 22). Cold-induced proteins (Csp and Cap proteins) play roles in a variety of cellular processes such as fatty acid metabolism, chromosome structuring, transcription, translation, general metabolism, energetic metabolism, and the stress response (19, 49, 51). In contrast to what has been observed for several species, including E. coli, B. subtilis, and L. lactis, after cold shock, this report reveals the absence of a major cold-induced protein of the 7-kDa Csp family. However, one non-cold-inducible 7-kDa Csp-E was identified, which is probably involved in cryoprotection in L. lactis (50). The role of Csp-E at optimal and chilled temperatures remains to be evaluated, but the constitutive expression of this 7-kDa Csp could be deeply implicated in the efficient cold adaptation of this strain.
To maintain membrane fluidity, the synthesis of normal unsaturated fatty acids is required, involving enzymes of the Fab (16) family, some of which are cold regulated. FabF is cold activated in E. coli (32) and controls the temperature-dependent regulation of the branched-chain fatty acid pathway in L. monocytogenes (53). In L. piscium CNCM I-4031, the cold regulation of FabF, FabG, and FabH indicates a global increase in fatty acid metabolism correlated with low temperatures. In order to overcome the efficient decrease in transcription and translation, proteins involved in these cellular processes have been identified in several species after cold shock (18) and particularly in our study.
The decreased enzyme-catalyzed reaction rates due to cold temperatures can be offset by an induction of some glycolytic enzymes to compensate for an overall lower energetic capacity. For example, the maximal glycolytic activity measured at 30°C in L. lactis increased approximately 2.5-fold following a cold shock (48). Cold induction has been observed for glycolytic enzymes, including GAPDH in B. subtilis and GAPDH, fructose-bisphosphate aldolase (Fba), and phosphoglycerate kinase (PGK) in L. piscium CNCM I-4031. A role in the control of glycolysis in L. lactis has been assigned to GAPDH, which was shown to be a rate-limiting enzyme in the glycolytic pathway during starvation (41). Transcriptional regulation of glycolysis has been strongly linked to HPr phosphorylation (10). Moreover, HPr is involved in the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS) implicated in the concomitant uptake and phosphorylation of carbohydrates. The cold induction of this protein has been observed for B. subtilis, Lactobacillus sakei (18, 20), and L. piscium CNCM I-4031. In our study, two very close spot occurrences were upregulated, first after cold shock and second during acclimation. We suggest that these two spots are related to different phosphorylated forms and that HPr phosphorylation could be a key to glycolysis regulation in cold adaptation. In addition, alternative fermentative pathways have been described for lactic acid bacteria under nutritional stress conditions (9), and in our study, AdhE was strongly cold upregulated, suggesting the induction of possible mixed-acid fermentation.
Cold stress provokes protein misfolding and oxidative compounds as well as inducing the expression of general stress proteins, including antioxidants and protein chaperones. DnaK is described as the major heat shock protein (HSP) molecular chaperone in E. coli. HSPs are commonly induced by multiple stresses and play an important role in withstanding and surviving harmful conditions (44). In L. lactis, the expression of DnaK is induced by heat, saline, and acid stresses (4, 14, 21, 25, 46), and it was reported previously that the in vitro overexpression of DnaK conferred freeze tolerance to E. coli (8). Until now, cold induction of DnaK has been described only for Leuconostoc mesenteroides (43). We speculate that this chaperone is also involved in the cold shock response of L. piscium CNCM I-4031.
Previous studies of Shewanella oneidensis and L. sakei reported the cold induction of Usp universal stress proteins (15, 33), as shown in this report during cold acclimation. The expression of this small cytoplasmic protein is enhanced in E. coli when cellular viability is challenged with any unfavorable agents, such as heat shock, nutrient starvation, DNA-damaging agents, and stress agents that inhibit cell growth (26). The oxidative stress protein Ahp is required by B. subtilis upon oxidative heat and salt stresses (1, 23), by Staphylococcus aureus upon osmotic stress (3), by LAB in aerotolerance (24), and by Shewanella putrefaciens upon osmotic stress and during cold acclimation (28). In L. piscium CNCM I-4031, three oxidative stress proteins were cold induced, Ahp, OsmC, and thioredoxin (Trx). In L. lactis, Trx is implicated in aerobic environment life, and it was assumed to trigger the induction of several mechanisms acting at the membrane and of metabolic levels, including GAPDH activation (45). It is noteworthy that GAPDH is also cold induced in L. piscium CNCM I-4031. On the other hand, OsmC is known to belong to the general stress regulons of E. coli and B. subtilis, enhanced by the stress sigma factor. In L. lactis, despite the absence of a sigma factor, the OsmC homologue was upregulated upon cold shock, and in L. piscium CNCM I-4031, a putative sigma 54 modulation protein was 4-fold upregulated during cold acclimation.
In L. piscium CNCM I-4031, comparative analyses of 2-DE gel profiles highlighted specific responses to cold shock and cold acclimation versus responses under standard conditions. The proteome was not analyzed kinetically, but it was assumed that cold shock is a transient state before cold acclimation. Among other things, specific stress proteins were observed for cold shock and cold acclimation. This suggests dynamic protein regulation during cold adaptation, with Csp proteins (DnaK, OsmC, and Ahp) involved in stress responses being time relayed by Cap proteins (UspA and thioredoxin) involved in the same pathways.
In conclusion, we suggest that L. piscium CNCM I-4031 adapts quickly to low temperatures by a constitutive expression of the potentially cryoprotective CspE and by an increase in glycolysis with potential HPr phosphorylation regulation. In a biopreservation strategy, the protective strains are generally cultivated at their optimum temperatures, eventually freeze-dried, and then directly inoculated into products that are stored at chilled temperatures. Cold acclimation thus constitutes a real advantage in bacterial competition against spoiling and pathogenic psychrotrophic bacteria in terms of food preservation. It would enable cells to multiply at optimal temperatures and be inoculated into chilled foodstuffs without delaying the food preservative effect.
Acknowledgments
We thank Frederique Chevalier from IFREMER for technical support with bacterial cultures and Carol Robins for English corrections.
This work was partially supported by grants from the European Union commission under Integrated Project (IP) SEAFOODplus contract no. FOOD-CT-2004-506359.
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Antelmann, H., S. Engelmann, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki, T. 1991. The effect of temperature shifts on protein synthesis by the psychrophilic bacterium Vibrio sp. strain ANT-300. J. Gen. Microbiol. 137:817-826. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong-Buisseret, L., M. B. Cole, and G. S. A. B. Stewart. 1995. A homolog to the Escherichia coli alkyl hydroperoxide reductase AhpC is induced by osmotic upshock in Staphylococcus aureus. Microbiology 141:1655-1661. [DOI] [PubMed] [Google Scholar]
- 4.Arnau, J., K. I. Sorensen, K. F. Appel, F. K. Vogensen, and K. Hammer. 1996. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology 142:1685-1691. [DOI] [PubMed] [Google Scholar]
- 5.Bayles, D. O., B. A. Annous, and B. J. Wilkinson. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshock and growth at low temperatures. Appl. Environ. Microbiol. 62:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, F., N. Morellet, F. Menu, and P. Potier. 1996. Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 178:2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champomier-Vergès, M.-C., E. Maguin, M.-Y. Mistou, P. Anglade, and J.-F. Chich. 2002. Lactic acid bacteria and proteomics: current knowledge and perspectives. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 771:329-342. [DOI] [PubMed] [Google Scholar]
- 8.Chow, K. C., and W. L. Tung. 1998. Overexpression of dnaK/dnaJ and groEL centers freeze tolerance to Escherichia coli. Biochem. Biophys. Res. Commun. 253:502-505. [DOI] [PubMed] [Google Scholar]
- 9.Cocaign-Bousquet, M., C. Garrigues, P. Loubiere, and N. D. Lindley. 1996. Physiology of pyruvate metabolism in Lactococcus lactis. Antonie Van Leeuwenhoek 70:253-267. [DOI] [PubMed] [Google Scholar]
- 10.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliker, P. R., A. W. Anderson, and G. Hannesson. 1956. An agar culture medium for lactic acid streptococci and lactobacilli. J. Dairy Sci. 39:1611-1612. [Google Scholar]
- 12.Fall, P. A., F. Leroi, M. Cardinal, F. Chevalier, and M. F. Pilet. Inhibition of Brochothrix thermosphacta and sensory improvement of tropical peeled cooked shrimp by Lactococcus piscium CNCM I-4031. Lett. Appl. Microbiol. 50:357-361. [DOI] [PubMed]
- 13.Fall, P. A., F. Leroi, F. Chevalier, C. Guerin, and M. F. Pilet. Protective effect of a non-bacteriocinogenic Lactococcus piscium CNCM I-4031 strain against Listeria monocytogenes in sterilized tropical cooked peeled shrimp. J. Aquat. Food Prod. Technol. 19:84-92.
- 14.Frees, D., F. K. Vogensen, and H. Ingmer. 2003. Identification of proteins induced at low pH in Lactococcus lactis. Int. J. Food Microbiol. 87:293-300. [DOI] [PubMed] [Google Scholar]
- 15.Gao, H. C., Z. M. K. Yang, L. Y. Wu, D. K. Thompson, and J. Z. Zhou. 2006. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 188:4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garwin, J. L., A. L. Klages, and J. E. Cronan. 1980. Beta-ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J. Biol. Chem. 255:3263-3265. [PubMed] [Google Scholar]
- 17.Graumann, P., and M. A. Marahiel. 1996. Some like it cold: response of microorganisms to cold shock. Arch. Microbiol. 166:293-300. [DOI] [PubMed] [Google Scholar]
- 18.Graumann, P., K. Schroder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graumann, P., T. M. Wendrich, M. H. W. Weber, K. Schroder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 20.Graumann, P. L., and M. A. Marahiel. 1999. Cold shock response in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 1:203-209. [PubMed] [Google Scholar]
- 21.Hartke, A., S. Bouche, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr. Microbiol. 33:194-199. [DOI] [PubMed] [Google Scholar]
- 22.Hébraud, M., and P. Potier. 1999. Cold shock response and low temperature adaptation in psychrotrophic bacteria. J. Mol. Microbiol. Biotechnol. 1:211-219. [PubMed] [Google Scholar]
- 23.Hecker, M., C. Heim, U. Volker, and L. Wolfel. 1988. Induction of stress proteins by sodium chloride treatment in Bacillus subtilis. Arch. Microbiol. 150:564-566. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, R. R., B. R. Riebel, and A. S. Bommarius. 2005. Comparison of alkyl hydroperoxide reductase (AhpR) and water-forming NADH oxidase from Lactococcus lactis ATCC 19435. Adv. Synth. Catal. 347:1139-1146. [Google Scholar]
- 25.Kilstrup, M., S. Jacobsen, K. Hammer, and F. K. Vogensen. 1997. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 27.Lai, C. Y., and J. E. Cronan. 2003. Beta-ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 278:51494-51503. [DOI] [PubMed] [Google Scholar]
- 28.Leblanc, L., F. Leroi, A. Hartke, and Y. Auffray. 2000. Do stresses encountered during the smoked salmon process influence the survival of the spoiling bacterium Shewanella putrefaciens? Lett. Appl. Microbiol. 30:437-442. [DOI] [PubMed] [Google Scholar]
- 29.Leroi, F., J. J. Joffraud, J. C. Arboleya, F. Amarita, Z. Cruz, E. Izurieta, A. Lasagabaster, I. Martinez de Maranon, I. Miranda, M. Nuin, I. Olabarrieta, H. L. Lauzon, G. Laurentzen, I. Bjorkevoll, R. Olsen, M. F. Pilet, F. Prevost, X. Dousset, S. Matamoros, and T. Skjerdal. 2008. Hurdle technology to ensure the safety of seafood products, p. 399-425. In T. Borresen (ed.), Improving seafood products for the consumer. Woodhead Publishing Ltd., Cambridge, United Kingdom.
- 30.Lottering, E. A., and U. N. Streips. 1995. Induction of cold shock proteins in Bacillus subtilis. Curr. Microbiol. 30:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Mackey, A. J., T. A. J. Haystead, and W. R. Pearson. 2002. Getting more from less—algorithms for rapid protein identification with multiple short peptide sequences. Mol. Cell. Proteomics 1:139-147. [DOI] [PubMed] [Google Scholar]
- 32.Mansilla, M. C., L. E. Cybulski, D. Albanesi, and D. de Mendoza. 2004. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 186:6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marceau, A., M. Zagorec, S. Chaillou, T. Mera, and M. C. Champomier-Verges. 2004. Evidence for involvement of at least six proteins in adaptation of Lactobacillus sakei to cold temperatures and addition of NaCl. Appl. Environ. Microbiol. 70:7260-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matamoros, S., F. Leroi, M. Cardinal, F. Gigout, F. K. Chadli, J. Cornet, H. Prevost, and M. F. Pilet. 2009. Psychrotrophic lactic acid bacteria used to improve the safety and quality of vacuum-packaged cooked and peeled tropical shrimp and cold-smoked salmon. J. Food Prot. 72:365-374. [DOI] [PubMed] [Google Scholar]
- 35.Matamoros, S., M. F. Pilet, F. Gigout, H. Prevost, and F. Leroi. 2009. Selection and evaluation of seafood-borne psychrotrophic lactic acid bacteria as inhibitors of pathogenic and spoilage bacteria. Food Microbiol. 26:638-644. [DOI] [PubMed] [Google Scholar]
- 36.McGovern, V. P., and J. D. Oliver. 1995. Induction of cold-responsive proteins in Vibrio vulnificus. J. Bacteriol. 177:4131-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel, V., I. Lehoux, G. Depret, P. Anglade, J. Labadie, and M. Hebraud. 1997. The cold shock response of the psychrotrophic bacterium Pseudomonas fragi involves four low-molecular-mass nucleic acid binding proteins. J. Bacteriol. 179:7331-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita, R. Y. 1975. Psychrophilic bacteria. Bacteriol. Rev. 39:144-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Farrell, P. H. 1975. High-resolution 2-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 40.Panoff, J. M., S. Legrand, B. Thammavongs, and P. Boutibonnes. 1994. The cold shock response in Lactococcus lactis subsp. lactis. Curr. Microbiol. 29:213-216. [Google Scholar]
- 41.Poolman, B., B. Bosman, J. Kiers, and W. N. Konings. 1987. Control of glycolysis by glyceraldehyde-3-phosphate dehydrogenase in Streptococcus cremoris and Streptococcus lactis. J. Bacteriol. 169:5887-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodgers, S. 2001. Preserving non-fermented refrigerated foods with microbial cultures—a review. Trends Food Sci. Technol. 12:276-284. [Google Scholar]
- 43.Salotra, P., D. K. Singh, K. P. Seal, N. Krishna, H. Jaffe, and R. Bhatnagar. 1995. Expression of DnaK and GroEL homologs in Leuconostoc esenteroides [mesenteroides] in response to heat shock, cold shock or chemical stress. FEMS Microbiol. Lett. 131:57-62. [DOI] [PubMed] [Google Scholar]
- 44.Sugimoto, S., A.-Al-Mahin, and K. Sonomoto. 2008. Molecular chaperones in lactic acid bacteria: physiological consequences and biochemical properties. J. Biosci. Bioeng. 106:324-336. [DOI] [PubMed] [Google Scholar]
- 45.Vido, K., N. Diemer, A. Van Dorsselaer, E. Leize, V. Juillard, A. Gruss, and P. Gaudu. 2005. Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J. Bacteriol. 187:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitaker, R. D., and C. A. Batt. 1991. Characterization of the heat shock response in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 57:1408-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wouters, J. A., B. Jeynov, F. M. Rombouts, W. M. de Vos, O. P. Kuipers, and T. Abee. 1999. Analysis of the role of 7 kDa cold-shock proteins of Lactococcus lactis MG1363 in cryoprotection. Microbiology 145:3185-3194. [DOI] [PubMed] [Google Scholar]
- 48.Wouters, J. A., H. H. Kamphuis, J. Hugenholtz, O. P. Kuipers, W. M. De Vos, and T. Abee. 2000. Changes in glycolytic activity of Lactococcus lactis induced by low temperature. Appl. Environ. Microbiol. 66:3686-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wouters, J. A., F. M. Rombouts, O. P. Kuipers, W. M. de Vos, and T. Abee. 2000. The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst. Appl. Microbiol. 23:165-173. [DOI] [PubMed] [Google Scholar]
- 50.Wouters, J. A., J. W. Sanders, J. Kok, W. M. de Vos, O. P. Kuipers, and T. Abee. 1998. Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology 144:2885-2893. [DOI] [PubMed] [Google Scholar]
- 51.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-255. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y. M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222-233. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, K., X. Ding, M. Julotok, and B. J. Wilkinson. 2005. Exogenous isoleucine and fatty acid shortening ensure the high content of anteiso-C15:0 fatty acid required for low-temperature growth of Listeria monocytogenes. Appl. Environ. Microbiol. 71:8002-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. Vantriet. 1990. Modeling of the bacterial-growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]