Abstract
Using a combination of cultivation-dependent and -independent methods, this study aimed to elucidate the diversity of microorganisms involved in iron cycling and to resolve their in situ functional links in sediments of an acidic lignite mine lake. Using six different media with pH values ranging from 2.5 to 4.3, 117 isolates were obtained that grouped into 38 different strains, including 27 putative new species with respect to the closest characterized strains. Among the isolated strains, 22 strains were able to oxidize Fe(II), 34 were able to reduce Fe(III) in schwertmannite, the dominant iron oxide in this lake, and 21 could do both. All isolates falling into the Gammaproteobacteria (an unknown Dyella-like genus and Acidithiobacillus-related strains) were obtained from the top acidic sediment zones (pH 2.8). Firmicutes strains (related to Bacillus and Alicyclobacillus) were only isolated from deep, moderately acidic sediment zones (pH 4 to 5). Of the Alphaproteobacteria, Acidocella-related strains were only isolated from acidic zones, whereas Acidiphilium-related strains were isolated from all sediment depths. Bacterial clone libraries generally supported and complemented these patterns. Geobacter-related clone sequences were only obtained from deep sediment zones, and Geobacter-specific quantitative PCR yielded 8 × 105 gene copy numbers. Isolates related to the Acidobacterium, Acidocella, and Alicyclobacillus genera and to the unknown Dyella-like genus showed a broad pH tolerance, ranging from 2.5 to 5.0, and preferred schwertmannite to goethite for Fe(III) reduction. This study highlighted the variety of acidophilic microorganisms that are responsible for iron cycling in acidic environments, extending the results of recent laboratory-based studies that showed this trait to be widespread among acidophiles.
Biologically mediated iron (Fe) cycling is performed by a number of prokaryotes that are capable of either ferrous iron [Fe(II)] oxidation or/and of dissimilatory ferric iron [Fe(III)] reduction, including respiratory growth with Fe(III) as the sole electron acceptor (43, 55, 56, 68). While Fe(III) exists predominantly in the solid phase as oxyhydroxide minerals at circumneutral pH, Fe(III) tends to be more soluble under extremely acidic conditions. This, coupled with the high redox potential of the redox pair Fe3+/Fe2+ (+770 mV) at pH 1 compared with that of equivalent couples at neutral pH [ranging from +100 mV to −300 mV depending on the form of Fe(III) mineral (68)] makes ferric iron an attractive alternative to molecular oxygen as an electron acceptor. Therefore, prokaryotes involved in the microbial reduction of Fe(III) under acidic and pH-neutral conditions appear to be dissimilar.
The potential for Fe(III) reduction seems to be widespread among acidophilic heterotrophs (15), including bacteria of the genera Acidiphilium (15, 38, 51) and Acidocella (15), Acidicaldus organivorans (44) and Acidimicrobium ferrooxidans (10), Ferrimicrobium acidiphilum (36), Ferrithrix thermotolerans (36), and Acidobacterium spp., including the type strain Acidobacterium capsulatum DSM 11244T (7, 15). Some, such as Ferrimicrobium acidiphilum and Ferrithrix thermotolerans, even have the capacity to oxidize Fe(II) with oxygen as the electron acceptor, a trait shared with the two chemolithoautotrophs Acidithiobacillus ferrooxidans (47) and Acidithiobacillus ferrivorans (26). Other reported non-Fe(III)-reducing Fe(II) oxidizers are Acidianus brierleyi (66), Alicyclobacillus ferrooxydans (35), Leptospirillum ferrooxidans (30), Leptospirillum ferriphilum (13), Sulfobacillus acidophilus (58), Sulfolobus metallicus (33), Thiomonas spp. (39), and Ferroplasma spp. (21). Most Fe(III) reducers cultured to date have no or only a minor capacity to reduce Fe(III) over a pH range from 3 to 6. Acidiphilium cryptum (ATCC 33463) and Acidobacterium capsulatum can reduce small amounts of Fe(III) up to pH 5 (6, 7), and the acidotolerant Serratia-related Mam Tor isolate reduces chelated Fe(III) at similar rates from pH 3 to 6 (1). However, due to the lack of a common functional gene marker and the lack of studies in the moderately acidic pH range (pH 4 to 5), the diversity of prokaryotes which are able to reduce Fe(III) is probably underestimated.
A number of acidic locations have been studied in the last 10 years with respect to Fe cycling, ranging from extremely acidic mine drainage within the Richmond Mine system, Iron Mountain, CA (20), to less acidic mine tailings (25, 70), acidic rivers (23), acidic lakes (7, 49, 61), and moderately acidic subsurface sediments and peatlands (50, 60). While proteomic analyses have greatly extended our knowledge about the ecophysiology of microbes inhabiting an extreme site with low diversity (5, 17), laboratory isolates are still paramount for a detailed analysis of their unique biological traits. We have studied Fe cycling in sediments of lignite mine lakes that are characterized by high concentrations of Fe(II), sulfate, and protons due to the oxidation of pyrite in the surrounding mine tailings (49). Microbial Fe(II) oxidation in the water column yields poorly crystalline Fe(III) oxides that precipitate to the sediment, where they can be utilized as electron acceptors (51). Within 20 cm of the sediment surface, the pH in the sediment increases from 2.8 to 6, and previous molecular-based research has revealed a pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms (7). However, the key drivers of iron reduction could not be identified in those studies. Here, we used a combination of cultivation-dependent and -independent approaches to elucidate the diversity of microorganisms involved in Fe cycling in the same lignite mine lake. With this approach, we extend the previous work to get a concrete handle on those microbes responsible for the reductive half of the cycle. The study of individual isolates can reveal the ecophysiology of potential drivers of Fe(III) reduction at low pH.
MATERIALS AND METHODS
Field site description and sampling.
The acidic lignite mine lake (Lake 77) is located in Lower Lusatia near Lauchhammer in east-central Germany, a historical region which was shaped by the lignite industry and extensive open-pit mining. It is a dimictic lake with an anoxic hypolimnion during summer stratification. During each sampling period, pH, dissolved oxygen, and temperature were measured over the depth at the deep central part of the lake with a U-10 multiparameter water quality meter (Horiba, Japan). Sediment cores were obtained on the north shore line in 3 m of water (North) and in the deep central basin in 7 m of water (Deep) using a gravity corer equipped with Plexiglas tubes (60 cm in length) (see Fig. S1 in the supplemental material). The Deep site has been studied intensively over the last 12 years (7, 51, 59). A total of 3 cores were obtained during each sampling period (July 2008, November 2008, and December 2008) from each site, transported to the laboratory at 4°C, sectioned under anoxic conditions according to visual differences in stratification, and characterized geochemically (Table 1). Northern cores were used for DNA extraction (July 2008) and for potential Fe(III) reduction rate measurements (November 2008). Cores obtained from both sites after the lake became mixed were used for oxygen profile measurements (November 2008) with a microelectrode (tip diameter, 100 μm; Unisense, Aarhus, Denmark) connected to a picoammeter (PA2000; Unisense, Aarhus, Denmark) and for isolation studies (December 2008). The North cores contained an orange, fluffy upper sediment zone (North I) (dry weight, 13.3%) that was enriched with reactive iron, mainly schwertmannite [Fe8O8(OH)x(SO4)y] (64). The second sediment zone (North II) was yellowish brown (see Fig. S1 in the supplemental material). The third sediment zone (North III) had dark stripes, and the remaining part of the sediment cores (North IV) was brownish to black and had a higher density (dry weight, 28.4%), with goethite (α-FeOOH) as the dominant Fe solid phase (59). In general, Deep core sediments (Deep I to IV) had a yellowish-brown color with fewer dark stripes (see Fig. S1 in the supplemental material).
TABLE 1.
Geochemistry of the north and deep central sediment cores obtained from lignite mine Lake 77
| Sediment zone | Depth (cm) | pH | Amt (mM) of: |
|||
|---|---|---|---|---|---|---|
| Fe(II) |
Fe(III) |
|||||
| Dissolveda | HCl extractedb | Dissolved | HCl extracted | |||
| North I | 0-1 | 2.8 | 4.5 | 17.4 | NDc | 211.7 |
| North II | 1-4 | 2.9 | 7.3 | 13.1 | ND | 87.5 |
| North III | 4-10.5 | 3.7 | 15.1 | 11.9 | ND | ND |
| North IV | 10.5-20.5 | 5.3 | 12.5 | 30.6 | ND | ND |
| Deep I | 0∼1 | 2.7 | 1.5 | 7.0 | ND | 81.0 |
| Deep II | 1∼4 | 2.9 | 4.6 | 11.9 | ND | 129.7 |
| Deep III | 4∼10.5 | 3.2 | 5.9 | 13.8 | ND | 10.9 |
| Deep IV | 10.5∼20.5 | 4.1 | 4.2 | 16.3 | ND | ND |
Fe measured in the supernatant after centrifugation.
Fe measured after preextraction of sediment in 0.5 M HCl.
ND, not detected.
Sediment Fe(III) reduction measurement.
To study potential Fe(III) reduction, 20 g (fresh weight) of homogenized sediment were mixed under a continuous flow of sterile nitrogen with 10 ml of an anoxic 0.7% NaCl solution and placed into a sterile 180-ml incubation flask (Mueller & Krempel, Buelach, Switzerland). Flasks were closed with rubber stoppers and screw caps and incubated in the dark at 15°C with an initial overpressure of ∼100 mbar in three replicates. The formation of Fe(II) during 9 days of incubation was determined by measuring the amount of Fe(II) after acid extraction (7). Fe(III) reduction rates were calculated from the linear increase of Fe(II) formation. HCl-extractable Fe(III) was calculated from the increase in Fe(II) concentration after the addition of ascorbic acid.
DNA extraction and PCR amplification.
Samples from North I and North IV of the July 2008 core were pelleted by centrifugation, and amounts of 0.25 to 0.3 g of sediment (wet weight) were used for DNA extraction with a PowerSoil DNA kit according to the manufacturer's instructions (Mo Bio Laboratories, Solana Beach, CA). Aliquots of purified DNA were PCR amplified (Primus 96 advanced; Peqlab, Germany) using either the bacterial small-subunit rRNA gene-specific forward primer 27F (53) or the archaeal forward primer ARC363F (63) together with the universal reverse primer 1492R (57) for clone library construction. In addition, phylogenetic-group-specific primers for Anaeromyxobacter, Acidiphilium, and Geothrix “bioleaching-associated bacteria”(7) and for Shewanella (71) were used. The primer set specific for acidophilic bioleaching-associated bacteria detects seven bacterial phylotypes (Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans, Acidithiobacillus caldus, Sulfobacillus thermosulfidooxidans, Leptospirillum ferrooxidans, Acidiphilium cryptum, and Acidiphilium organovorum) which are involved in the bioleaching of mineral ores. Bacterial isolates were picked and resuspended in 100 μl of 5% (wt/vol) Chelex-100 (Bio-Rad, United States) solution in 0.5-ml tubes. The tubes were then boiled in a water bath for 10 min, followed by 2 min of centrifugation at maximum speed. For the few isolates which failed to produce any PCR amplicon, a lysis solution (0.05 M NaOH and 0.25% SDS) was used following the method given by Johnson et al. (43). One or 2 μl of the supernatant served as template for bacterial 16S rRNA gene PCR with the universal primers 27F and 1492R.
Quantitative PCR for detecting Geobacter isolates was performed using Geobacteraceae-specific 16S rRNA gene primers 561F (69) and Geo858R (5′-CTAGGTGTTGCGGGTATTGA-3′) and 10 ng of purified environmental DNA as the template in the reaction mixture SensiMix LowRef (Quantace, distributed by Bioline GmbH, Luckenwalde, Germany). Thermocycling was performed with the following temperature program: 10 min at 95°C and 50 cycles of 15 s at 95°C, 30 s at 61°C, 30 s at 72°C, and 10 s at 78°C. Fluorescence measurements were made at 61, 72, and 78°C for each cycle, followed by a dissociation curve analysis with 1 min at 95°C, 30 s at 55°C, and heating to 95°C at a rate of 0.01°C s−1. Genomic DNA from a pure culture of Geobacter sulfurreducens (DSM 12127) was used as the standard. Reactions that yielded cycle threshold (CT) values with less than a three-cycle difference from those of nontemplate controls were considered to be within the error of measurement.
16S rRNA gene library construction and statistical analysis.
For bacterial clone libraries, purified PCR products were ligated into the pGEM-T vector (Promega, United States) and transformed into Escherichia coli cells according to the manufacturer's instructions. Amplified rRNA gene restriction analysis (ARDRA) with restriction enzymes HhaI and BsuRI (HaeIII) (Fermentas, Canada) was applied, and clones were subsequently grouped into operational taxonomic units (OTUs) according to their ARDRA banding patterns. Representative clones of the respective OTUs were sequenced at Macrogen, Inc. (South Korea). For archaeal libraries, purified PCR products were ligated into the pCR4-TOPO vector (Invitrogen, United States) and sent to the Genome Center at Washington University, St. Louis, MO, for downstream processing. All sequences were pooled, assembled, end trimmed, and revised using Geneious Pro software (Biomatters Ltd., New Zealand). The sequences were checked for chimeric properties using the Bellerophon chimera detection program (34) and Chimera Check from Ribosomal Database Project II (11). The sequences were then aligned using the greengenes Align tool (18) and dereplicated using FastGroupII (76) with the percentage sequence identity method with gaps of 97% similarity. The phylogenetic affiliations were assigned using the greengenes Compare tool (18). Rarefaction curves and percent coverage were calculated using Analytic Rarefaction 1.3 (http://www.uga.edu/strata/software/anRareReadme.html) (32). EstimateS (12) was used to calculate the abundance-based coverage estimator (ACE), incidence-based coverage estimator (ICE), and Chao1 values, as well as the Shannon and Simpson's diversity indices.
Isolation of acidophilic microorganisms.
One-gram subsamples of sediment from North core zones I and IV and from Deep core zones I and IV were diluted with 4 ml of filter-sterilized lake water (pH ∼ 3) and homogenized by shaking for 30 min at ∼1,000 rpm (Vortex-Genie2 shaker; Scientific Industries, United States). Samples were serially diluted (100 to 10−2) with filter-sterilized lake water, and 100 μl of each dilution was plated in triplicate on one of several selective solid overlay media specific for different groups of autotrophic and heterotrophic acidophilic microorganisms (40). These were (i) ferrous iron overlay medium (Feo, final pH 2.5) with tryptone soya broth (TSB; Oxoid, United Kingdom), (ii) ferrous sulfate and potassium tetrathionate overlay medium with TSB (FeSo, final pH 2.5), (iii) inorganic ferrous iron overlay medium without TSB (iFeo, final pH 2.5), (iv) ferrous iron and sodium thiosulfate (0.01 M) overlay medium with TSB (FeTo, final pH 4.3), and yeast extract overlay medium at (v) pH 3 (YE3) or (vi) pH 4 (YE4). Cycloheximide (Carl Roth, Germany) was added into the top-layer medium at 50 μg ml−1 to inhibit fungal growth. All plates were incubated at room temperature (∼20°C) in the dark and checked frequently. The picked colonies were transferred at least 5 times, and purity was controlled both by colony morphology assessment and microscopic examination (Axiovert 25; Carl Zeiss, Germany). Fe(II) oxidation capacities were determined by the appearance of an orange Fe rust color of the colonies due to the microbial oxidation of ferrous sulfate contained in the media. Heterotrophic bacteria obtained from yeast extract media were transferred to other ferrous sulfate-containing media to determine their Fe(II) oxidation capacities. Purified colonies were screened by ARDRA, and representative OTUs were chosen for sequencing. Isolates were grouped if the 16S rRNA gene sequences and colony morphologies were identical. The cultivated strains and type strains closest to these isolates were determined through the online BLAST database (45) and EzTaxon server (9), respectively. Strain designations were assigned as follows, by combining information about the medium, source, and isolate number separated by hyphens. The first part designates the type of medium, and the second part designates the origin of the sediment, e.g., D1 or D4 means the Deep site of the lake (D) and either the upper sediment zone I (1) or the deeper sediment zone IV (4). N1 or N4 means the North shore site of the lake (N) and either the upper sediment zone I (1) or the deeper sediment zone IV (4). The next number is the sequential number of the isolate, and CH designates the presence of cycloheximide if it was added.
Fe(III) reduction assays of isolates.
All the isolates obtained were first inoculated into 5 ml of liquid medium (same media as described above but without agarose) with fructose as the electron donor and incubated at room temperature on a rotary shaker. The Fe(III)-reducing capacities of the grown isolates were determined under microoxic and anoxic conditions in the dark. Microoxic conditions were established in sterile 150-ml conical flasks filled with 100 ml of sterile liquid medium (15, 38), while anoxic conditions were established using 20 ml of medium flushed with nitrogen (N2) gas in 30-ml serum bottles (Wheaton, United States) in an anaerobic chamber (100% N2 gas phase). The bottles were sealed with rubber stoppers and aluminum caps. After the medium was autoclaved, filter-sterilized fructose (∼10 mM final concentration) and 100 mg schwertmannite (equivalent to 10 mM) were added as an electron donor and acceptor, respectively. Reduction assays started after the addition of 1 or 2 ml of active cultures into the flasks and bottles at room temperature. Uninoculated flasks or bottles were used as controls. Selected isolates (YE3-D1-10-CH [a strain of an unknown Dyella-like genus], FeSo-D4-20-CH and YE4-D4-4i-CH [Alicyclobacillus-related strains], YE3-D1-20 [an Acidobacterium-related strain], and YE4-N1-5-CH [an Acidocella-related strain]) were studied in more detail for their Fe(III)-reducing capacities by using 5 mM fructose and (i) chelated Fe(III) citrate (50 mM) and Fe(III) pyrophosphate (40 mM) under strict anoxic conditions and (ii) either artificially synthesized schwertmannite or goethite (10 mM) (14, 64) under microanoxic conditions. Potential microbial contamination in synthesized schwertmannite, which could not be autoclaved, was checked by spreading 0.5 ml of the suspension onto different overlay plates as described above and, also, on nutrient-rich Luria-Bertani agar plates. No growth was observed. Samples (1 ml) were taken from the flasks or bottles at intervals and used for measuring pH (Semi-Micro pH meter; Mettler Toledo, Switzerland) and Fe(II) concentration.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences determined in this study have been deposited in the EMBL database under accession numbers FN870189 to FN870350.
RESULTS
Biogeochemical characteristics of sediment cores.
Sediments obtained from the North site contained higher concentrations of dissolved and HCl-extractable Fe(II) than those from the deep part of the lake (Table 1). Similarly, the sulfate concentrations were higher in North (19.7 to 27.8 mM) samples than in Deep sediments (8.1 to 11.9 mM), suggesting a higher input from the surrounding mine tailings. Nonetheless, the pH was highest in North IV, reaching 5.3 (Table 1). The pH of the Deep sediment cores did not increase beyond 4.1 with depth, which was in contrast to the results of earlier studies (7, 49, 59). After seasonal mixing of the lake water in autumn, the dissolved oxygen concentration of the water in the center (Deep) of the lake was stable at 88% saturation near the surface and 83% above the sediment. However, oxygen microprofiles measured in the North and Deep sediment cores declined sharply, to levels below the detection limit for O2 (314.9 μmol liter−1), from the water sediment interface to within 2 and 10 mm deep, respectively (see Fig. S1 in the supplemental material). Anoxic Fe(III) reduction rates approximated 1.37 and 2.08 μmol g−1 (dry weight) day−1 in North I and North II samples, respectively, but were not detectable in North III or IV samples. Deep core sediment Fe(III) reduction rates approximated 6.19 and 1.2 μmol g−1 (dry weight) day−1 when sediments from depths of 0 to 6 and 6 to 10.5 cm, respectively, were incubated.
16S rRNA gene-based analyses of North sediments.
As had been done in previous work with Deep sediments (7), we tried to identify microorganisms potentially involved in Fe cycling in the North I and IV sediment zones by using primer sets for known phylogenetic groups of bacteria and general primer sets for Bacteria and Archaea. PCR products were obtained from North I for the acidophilic genus Acidiphilium and bioleaching-associated bacteria but not for Fe(III)-reducing, neutrophilic bacteria of the genera Anaeromyxobacter, Geothrix, Geobacter, and Shewanella. However, with the newly designed primer set for quantitative PCR, Geobacter genes could be amplified, and copy numbers approximated 8.34 × 105 g−1 (fresh weight sediment) in North IV but only 4.89 × 103 g−1 (fresh weight sediment) in North I.
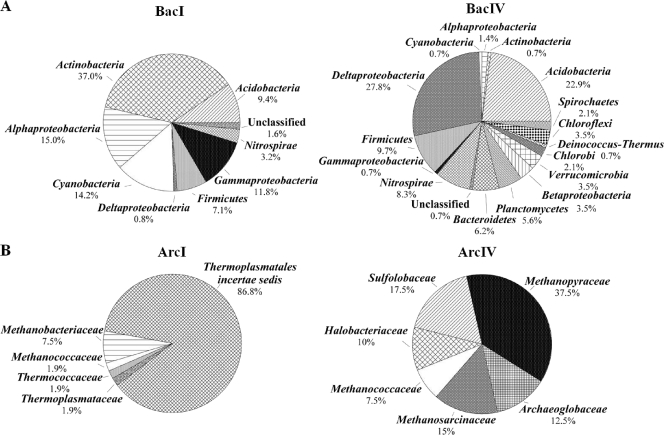
Totals of 127 and 144 clones of bacterial 16S rRNA genes from North I (BacI) and North IV (BacIV) were screened, and ARDRA revealed 36 and 65 distinct phylotypes, respectively (Table 2). There were no identical phylotypes present in either library based on FastGroupII results. The rarefaction curves did not indicate saturation (data not shown), and coverage was 81.4% for the BacI and 61.7% for the BacIV library. The ACE, ICE, and Chao1 values and the Shannon and Simpson's diversity indices indicated a higher diversity in the BacIV than in the BacI library (Table 2). The most frequently detected lineage in the BacI library was the Actinobacteria (37% of all clones), with members of the Alphaproteobacteria and Cyanobacteria each comprising 15.0% and 14.2% of the total clones, followed by Gammaproteobacteria (11.8%), Acidobacteria (9.4%), and Firmicutes (7.1%) (Fig. 1). The most frequently detected lineages in the BacIV library were Deltaproteobacteria (27.8%), Acidobacteria (22.9%), Firmicutes (9.7%), and Nitrospirae (8.3%). More lineages were detected in zone IV than in zone I (Fig. 1).
TABLE 2.
Statistical analysis of 16S rRNA gene clone libraries using ecological and molecular estimates of phylotype diversity
| Sediment samplea | No. of clones | No. of phylotypes | % Coverage | ACEb | ICEc | Chao1 ± SD | Diversity index |
|
|---|---|---|---|---|---|---|---|---|
| Shannon | Simpson's (1/D) | |||||||
| BacI | 127 | 36 | 81.4 | 42.5 | 36 | 43.7 ± 5.6 | 3.13 | 17.39 |
| BacIV | 144 | 65 | 61.7 | 119.1 | 65 | 126.3 ± 29.4 | 3.85 | 40.54 |
| ArcI | 53 | 8 | 79.3 | 9.5 | 8 | 12.5 ± 7.2 | 1.27 | 2.34 |
| ArcIV | 40 | 25 | 38.2 | 38.1 | 25 | 41.1 ± 11.0 | 3.08 | 33.91 |
BacI and BacIV designate the bacterial 16S rRNA gene clone libraries obtained from the northern shore of the lake from either the upper sediment zone I or the deeper sediment zone IV. ArcI and ArcIV designate the archaeal 16S rRNA gene clone libraries obtained from the northern shore of the lake from either the upper sediment zone I or the deeper sediment zone IV.
ACE, abundance-based coverage estimator.
ICE, incidence-based coverage estimator.
FIG. 1.
Frequencies of bacterial (A) and archaeal (B) phylogenetic lineages detected in 16S rRNA gene clone libraries derived from sediment zones North I (BacI and ArcI) and North IV (BacIV and ArcIV) of a core obtained from the northern basin. Calculations were based on the total number of clones associated with phylotypes of sequenced representatives at the phylum or class level for Proteobacteria for Bacteria libraries and the family level for Archaea libraries.
A total of 53 and 40 archaeal clones from the North I (ArcI) and North IV (ArcIV) libraries were screened, yielding 8 and 25 distinct phylotypes, respectively, based on ARDRA (Table 2). Again, identical phylotypes were not detected in both libraries, and indices indicated higher diversity in the ArcIV library than in the ArcI library (Table 2). The percent coverage was 79.3% for the ArcI and 38.2% for the ArcIV library. Only Crenarchaeota were detected in the ArcI library (17.5%), whereas members of the Euryarchaeota comprised 100 and 82.5% of the clones in zones I and IV, respectively (Fig. 1). The Sulfolobaceae family was the only family detected in the phylum Crenarchaeota, whereas the phylum Euryarchaeota consisted of various families each representing 1.9 to 86.8% of the total clones in the ArcI and 7.5 to 37.5% in the ArcIV libraries (Fig. 1B; also see Table S3 in the supplemental material).
Isolation of acidophilic or acidotolerant microorganisms and their Fe(II)-oxidizing capacities.
Media were chosen with a pH range to cover the pH range of the sediment in order to isolate acidophilic or acidotolerant Fe(II) oxidizers, aerobic heterotrophs, and potential Fe(III) reducers. Colonies appeared from 5 to 14 days after inoculation with North and Deep zone I and IV sediment dilutions. Based on the colony morphologies and the types of media, more than 240 single colonies were picked and isolated. From these, 117 isolates were grouped according to ARDRA fragment patterns (16, 13, 22, 21, 18, and 27 patterns from Feo, FeSo, FeTo, iFeo, YE3, and YE4 selective medium, respectively). The 16S rRNA genes of 38 representatives were sequenced (2, 8, 7, 2, 7, and 12 from Feo, FeSo, FeTo, iFeo, YE3, and YE4 medium, respectively), yielding a total of 28 representative strains (Table 3). According to ARDRA patterns, the occurrence of some strains was restricted to the upper acidic sediment zone from the North and Deep cores, whereas others could be obtained from both sediment zones (see Table S2 in the supplemental material). At the genus level, 36% of the isolates were related to 16S rRNA genes detected in bacterial clone libraries obtained from North I and North IV (see Table S2 in the supplemental material).
TABLE 3.
Bacterial isolates from acidic lignite mine lake sediments using 6 different agarose overlay plates, with taxonomic status, identity values based on analysis of 16S rRNA gene sequences, and the results of microoxic and anoxic Fe(III) reduction experiments and Fe(II) oxidation capacities observed on Fe(II)-containing agarose overlay plates
| Representative straina | No. of isolatesb | Closest type strain (GenBank sequence accession no., % identity) | Fe-cycling capacity |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Reported (reference[s])c | Oxidationd | Microoxic reductionf | Anoxic reductionf | ||||||
| YE3-D1-20 | 1 | Edaphobacter modestus Jbg-1T (DQ528760, 93.9) | UO, R (7, 25) | − (Feo) | + | + | |||
| YE4-D4-2-CH | 2 | Streptomyces ferralitis SFOp68T (AY262826, 98.4) | UO, UR | NAe | + | + | |||
| FeSo-D1-6-CH | 1 | Ferrimicrobium acidiphilum T23T (AF251436, 99.4) | O, R (36) | + | + | + | |||
| YE4-D4-16-CH | 1 | Actinocorallia aurea IFO 14752T (AB006177, 94.2) | UO, UR | − (FeTo) | + | (−) | |||
| YE4-D4-1-CH | 1 | Bacillus acidicola 105-2T (AF547209, 99.8) | UO, UR | NA | + | (−) | |||
| YE4-D4-4-CH | 1 | Bacillus pocheonensis Gsoil 420T (AB245377, 99) | UO, UR | − (FeTo) | + | (+) | |||
| YE3-D4-3i-CH | 2 | Alicyclobacillus contaminans 3-A191T (AB264026, 89.6) | O (37), UR | + (iFeo) | + | + | |||
| iFeo-D4-31-CH | 1 | Alicyclobacillus contaminans 3-A191T (AB264026, 90.6) | O, R (42) | + | − | + | |||
| YE3-D4-31i-CH | 3 | Alicyclobacillus ferrooxydans TC-34T (EU137838, 98.6) | O (72), UR | + (FeTo) | + | + | |||
| Feo-D4-16-CH | 1 | Alicyclobacillus ferrooxydans TC-34T (EU137838, 91.4) | O, R (42) | + | + | + | |||
| FeSo-D4-20-CH | 1 | Alicyclobacillus pomorum 3AT (AB089840, 91) | O (19), UR | + | + | (−) | |||
| FeSo-N4-1-CH | 2 | Alicyclobacillus tolerans K1T (AF137502, 95.1) | O (72), UR | + | + | + | |||
| YE4-D4-4i-CH | 2 | Alicyclobacillus tolerans K1T (AF137502, 94.9) | O (72), UR | + (FeTo) | + | + | |||
| Feo-N4-15-CH | 1 | Alicyclobacillus tolerans K1T (AF137502, 100) | O, R (46) | + | + | + | |||
| FeSo-N4-2-CH | 1 | Sulfobacillus acidophilus DSM 10332T (AB089842, 94.2) | O (58, 72), UR | − | + | + | |||
| FeSo-N4-3-CH | 1 | Sulfobacillus thermotolerans Kr1T (DQ124681, 99.1) | O, R (42) | + | + | + | |||
| FeSo-D1-15-CH | 1 | Acidiphilium acidophilum ATCC 27807T (D86511, 99.8) | UO, R (38) | − | + | + | |||
| YE3-D4-63i-CH | 1 | Acidiphilium acidophilum ATCC 27807T (D86511, 98.2) | UO, R (38) | NA | + | + | |||
| YE3-D1-35 | 1 | Acidiphilium cryptum ATCC 33463T (D30773, 100) | UO, R (6, 51) | NA | + | + | |||
| FeTo-D1-1-CH | 1 | Acidisphaera rubrifaciens HS-AP3T (D86512, 99.7) | UO, NR (15) | − | − | (−) | |||
| YE4-N1-5-CH | 2 | Acidocella facilis ATCC 35904T (D30774, 97) | UO, R (15) | − (FeTo) | + | + | |||
| FeTo-D4-17-CH | 1 | Methylocella tundrae T4T (AJ555244, 99.9) | UO, UR | + | − | (−) | |||
| YE3-D4-2i-CH | 1 | Methylocapsa acidiphila DSM 13967T (AJ278726, 98.6) | UO, UR | NA | (−) | (−) | |||
| YE4-D1-1-CH | 1 | Thiomonas cuprina NBRC 102094T (AB331953, 98.8) | UO, UR | − (FeTo) | + | − | |||
| YE3-D1-10-CH | 4 | Dyella yeojuensis R2A16-10T (DQ181549, 94.3) | O, R (15, 72) | + (FeTo) | + | + | |||
| FeSo-D1-9-CH | 1 | Acidithiobacillus ferrivorans NO-37T (AF376020, 99.4) | O, R (26) | + | + | (−) | |||
| FeSo-N1-1-CH | 1 | Acidithiobacillus ferrooxidans ATCC 23270T (AF465604, 99.3) | O, R (26) | + | + | (−) | |||
| FeTo-N1-3-CH | 1 | Acidithiobacillus ferrooxidans ATCC 23270T (AF465604, 99.5) | O, R (26) | − | + | (−) | |||
Strain designation was constructed as follows, by combining information about the medium, source, and isolate number separated by hyphens. The first part designates the medium type, the second part designates the origin of the sediment, e.g., D1 or D4 means deep site of the lake (D) and either upper sediment zone I (1) or deeper sediment zone IV (4). N1 or N4 means north shore site of the lake (N) and either upper sediment zone I (1) or deeper sediment zone IV (4). The next number is the sequential number of the isolate, and CH designates the presence of cycloheximide (CH) if it was added. The pHs of the media Feo, iFeo, FeSo, FeTo, YE3, and YE4 were 2.5, 2.5, 2.5, 4.3, 3.0, and 4.0, respectively. Strains which appear to be putative new species are presented in boldface.
A number greater than one indicates that isolates were grouped based on identical 16S rRNA gene sequences and colony morphologies on the plates.
Known Fe redox capacities of the closest cultivated strains based on the literature: O means Fe(II) oxidation; R means Fe(III) reduction; NO means no Fe(II) oxidation; NR means no Fe(III) reduction; UO means unknown for Fe(II) oxidation function; and UR means unknown for Fe(III) reduction.
Colonies from yeast extract media were checked for Fe(II) oxidation capacities on iron-rich media. +, positive for Fe(II) oxidation; −, negative for Fe(II) oxidation. The medium code in brackets indicates the iron-rich medium on which the colonies were able to grow.
NA, not applicable. Colonies from yeast extract media were tested for Fe(II) oxidation capacities on iron-rich media, but strains could not grow on the indicated medium.
+, Fe(II) formation in the liquid medium; −, no Fe(III) formation compared to uninoculated controls with cells growing in the medium; (−), tested, but no cell growth or Fe(III) formation was observed; (+), Fe(II) formation detected but not confirmed.
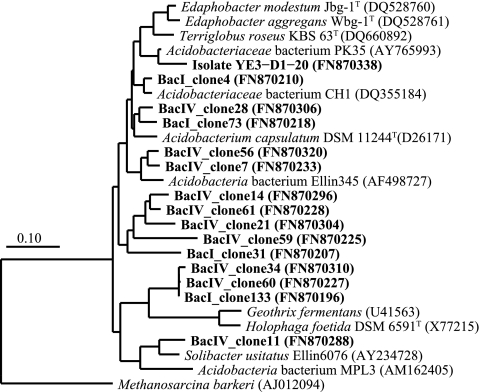
One strain belonging to the Acidobacteria family Acidobacteriaceae was obtained from a YE3 plate (YE3-D1-20) (Table 3 and Fig. 2). Strains related to the phylum Actinobacteria were YE4-D4-2-CH (Streptomycetaceae related), FeSo-D1-6-CH (Acidimicrobiaceae related), and YE4-D4-16i-CH (Thermomonosporaceae related). Isolate FeSo-D1-6-CH produced rusty, orange-brown precipitates on the plate (see Table S1 in the supplemental material) and was identical to a heterotrophic, iron-oxidizing, extremely acidophilic Actinobacterium able to reduce Fe(III) (36). Seventeen Firmicutes strains were obtained from all six media and were affiliated with the families Bacillaceae and Alicyclobacillaceae. Strain YE4-D4-1-CH was most closely related to Bacillus acidicola 105-2T, which grows at pH 3.5 to 7 (2). Thirteen of the 15 Alicyclobacillaceae strains were affiliated with the genus Alicyclobacillus and showed “fried-egg”-shaped rusty orange-brown colonies. Three appeared to be new species, based on <97% sequence similarity (see Table S1 in the supplemental material). Two other putative novel strains were affiliated with the genus Sulfobacillus. Isolate FeSo-N4-2-CH did not show an Fe(II)-oxidizing capacity, in contrast to Sulfobacillus acidophilus DSM 10332T (94%) (58). In contrast, strain FeSo-N4-3-CH oxidized Fe(II) and was highly related (99.1%) to Sulfobacillus thermotolerans Kr1T, a mixotrophic Fe(II) oxidizer with a pH range of 1.2 to 2.4 (8).
FIG. 2.
Phylogenetic tree of Acidobacteriaceae-related sequences showing the close relationship of 16S rRNA gene clones obtained from sediment zones I and IV of a core obtained from the northern basin and of a strain isolated from sediment zone I of a core obtained from the deep central basin. GenBank sequence accession numbers are shown; sequences from this study are shown in boldface. The Archaea Methanosarcina barkeri (AJ012094) was used as an out-group. Scale bar shows 0.1 change per nucleotide position.
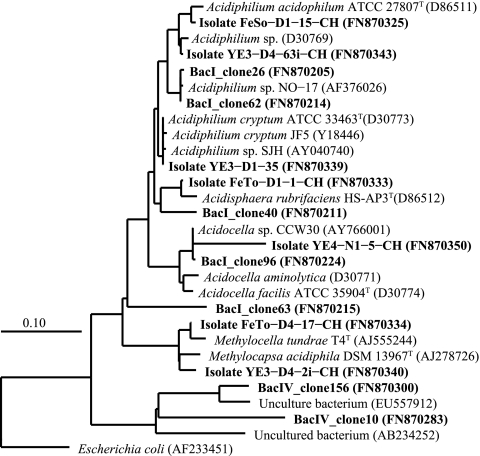
Within the Alphaproteobacteria, representative strains FeSo-D1-15-CH, YE3-D4-63i-CH, YE3-D1-35, and YE4-N1-5-CH were related to the Acetobacteraceae family (Fig. 3 and Table 3). One strain was related to Acidisphaera rubrifaciens HS-AP3T (31), an aerobe that can grow in a pH range of 3.5 to 6.0, and strain YE4-N1-5-CH was identical to Acidocella sp. CCW30 (AY766001) isolated from acidic, metal-rich mine waters (25). None of our Acetobacteraceae-related isolates oxidized Fe(II) (Table 3). Of two Beijerinckiaceae strains (FeTo-D4-17-CH and YE3-D4-2i-CH) related to methane-oxidizing acidophiles (16), only one (FeTo-D4-17-CH) could oxidize Fe(II).
FIG. 3.
Phylogenetic tree of Alphaproteobacteria-related sequences showing the close relationship of 16S rRNA gene clones obtained from sediment zones I and IV of a core obtained from the northern basin and of the strains isolated from sediment zones I and IV of cores obtained from the northern and deep central basin. GenBank sequence accession numbers are shown; sequences from this study are shown in boldface. The Gammaproteobacteria Escherichia coli (AF233451) was used as an out-group. Scale bar shows 0.1 change per nucleotide position.
The only Betaproteobacteria strain isolated (YE4-D1-1-CH) could not oxidize Fe(II). A total of 7 strains were isolated from the Gammaproteobacteria. Strain YE4-D1-10-CH (within the Xanthomonadaceae) was identical to strain WJ2 from acidic mine drainage waters (27) and produced orange precipitates. Acidithiobacillaceae-related strains FeSo-D1-9-CH and FeSo-N1-1-CH were related to known Fe(II)-oxidizing strains (26), but no oxides were produced by FeTo-N1-3-CH (see Table S1 in the supplemental material).
Fe(III)-reducing capacities of isolates and potential Fe cycling.
Thirty-six of the 38 tested representatives showed the capacity to form Fe(II) under microoxic and/or anoxic conditions when incubated with schwertmannite, the dominant Fe(III) mineral of Lake 77 (Table 3). The exceptions were strain FeTo-D1-1-CH that is related to Acidisphaera rubrifaciens HS-AP3T, which is unable to reduce Fe(III) (15), and strain YE3-D4-2i-CH. The highest levels of Fe(II) formation were observed for FeSo-N4-1-CH, FeSo-D1-15-CH, FeSo-D1-9-CH, Feo-N4-15-CH, FeSo-N4-3-CH, and FeSo-D1-6-CH (data not shown). Specific rates of reduction could not be determined because conditions might vary within a flask, from anoxic in the bottom of the flasks to oxic at the medium-air interface, leading to a partial reoxidation of Fe(II). In general, Fe(II) formation did not increase under strictly anoxic conditions. After another 4 weeks, we observed evidence of Fe(II) reoxidation, with some isolates forming a rusty orange solid layer (YE4-D4-4-CH, final medium pH 5.8) or a light yellowish soft layer (YE3-D4-3i-CH, final medium pH 3.3) on the top of the medium. Another isolate (FeSo-N4-1-CH, final medium pH 3.7) had a rusty orange crust attached to the inner flask surface at the medium-air interface. Raman spectroscopy revealed that the solid iron that reformed at the medium-air interface consisted of schwertmannite (YE3-D4-3i-CH and FeSo-N4-1-CH) or ferrihydrite (YE4-D4-4-CH) (Valerian Ciobota, personal communication).
The following five isolates were selected for further experiments (Table 4): YE3-D1-20 due to the high relative abundance of Acidobacteria-related sequences in clone libraries; two new Alicyclobacillus species with high Fe(III)-reducing capacities; YE4-N1-5-CH because little is known about Fe(III) reduction by Acidocella; and YE3-D1-10-CH. Most of these strains showed a broad pH range for growth, ranging from 1.5 to 6.0, and the pH optimum for most strains was 3.5 (Table 4). While all 5 of these isolates could reduce iron in schwertmannite, chelated forms of Fe(III), e.g., ferric citrate and ferric pyrophosphate, were not reduced during 14 days. However, after this lag phase, two isolates (FeSo-D4-20-CH and YE3-D1-10-CH) formed up to 7.5 mM Fe(II) (data not shown). Only very small amounts of synthetic goethite were reduced (Table 4).
TABLE 4.
Growth pH range and Fe(II) formation assay results for five selected isolates
| Genus, strain | Growth pH range (optimal growth pH)a | Schwertmanniteb |
Goethiteb |
||||
|---|---|---|---|---|---|---|---|
| pH on day: |
Amt of Fe(II) (mM) on day 22 | pH on day: |
Amt of Fe(II) (mM) on day 22 | ||||
| 0 | 22 | 0 | 22 | ||||
| Acidobacteria sp. | |||||||
| YE3-D1-20 | 2.5-5.0 (3.5) | 3.0 ± 0.0 | 4.0 ± 0.2 | 3.1 ± 0.3 | 3.2 ± 0.2 | 3.2 ± 0.1 | 0.6 ± 0.3 |
| Alicyclobacillus spp. | |||||||
| FeSo-D4-20-CH | 1.5-6.0 (3.5) | 3.0 ± 0.0 | 3.6 ± 0.6 | 2.4 ± 1.6 | 3.1 ± 0.2 | 2.9 ± 0.2 | 0.1 ± 0.1 |
| YE4-D4-4i-CH | 2.5-5.0 (3.5) | 3.0 ± 0.1 | 3.9 ± 0.1 | 3.6 ± 0.0 | 3.0 ± 0.0 | 3.0 ± 0.1 | 0.9 ± 0.2 |
| Acidocella sp. | |||||||
| YE4-N1-5-CH | 2.5-4 (2.5) | 3.0 ± 0.0 | 3.6 ± 0.4 | 2.6 ± 0.8 | 3.2 ± 0.2 | 3.1 ± 0.2 | 0.7 ± 0.2 |
| Unknown Dyella-like genus sp. | |||||||
| YE3-D1-10-CH | 2.0-5.0 (3.5) | 3.0 ± 0.0 | 4.0 ± 0.1 | 3.6 ± 0.6 | 3.3 ± 0.1 | 3.3 ± 0.2 | 0.8 ± 0.1 |
| Control | 3.1 ± 0.0 | 2.8 ± 0.0 | 0.1 ± 0.1 | 3.1 ± 0.0 | 3.0 ± 0.0 | 0.1 ± 0.1 | |
Tested in the YE media under oxic conditions without the presence of Fe(III) minerals.
Tested in the YE media under microoxic conditions.
DISCUSSION
The diversity of microorganisms involved in Fe cycling in acidic sediments was elucidated by cultivation-based and molecular methods. This approach extended previous knowledge by providing information on those microbes responsible for the reductive half of the cycle at low pHs. From 117 selected isolates, a total of 28 different 16S rRNA gene sequences were obtained. Despite identical 16S rRNA genes, some isolates showed different morphologies or differed in their Fe-cycling capabilities. Sequence analysis suggests that 18 isolates are putative new species when compared to fully characterized relatives. However, the number decreases to 5 new species if we make our comparison to isolates which have been obtained over the last several years but have not been fully characterized. For example, strains YE3-D4-3i-CH and iFeo-D4-31-CH shared only 90 and 91% identity, respectively, with the type strain Alicyclobacillus contaminans 3-A191T (24) but more than 99 and 94%, respectively, with the closest cultivated strains SLC66 (iron-oxidizing acidophile, GenBank sequence accession number AY040739) (37) and Y0010 (GenBank sequence accession number AY140235) (42). Not surprisingly, many of these closest cultivated relatives were isolated from acidic hot springs, gold mine soils, porphyry copper tailings, or sulfide ores (19, 42, 72, 75). Thus, the use of “overlay” plates has greatly improved the isolation success of a diversity of autotrophic and heterotrophic acidophiles, which tend to grow poorly on normal solid media (40). The greater diversity of isolates beyond the well-known acidophiles Acidithiobacillus ferrooxidans and Acidiphilium cryptum isolated in other studies (70) might be due to the broad pH range of the different media used in this study. Nonetheless, 15 and 33 isolates in our study were related to those two species, respectively.
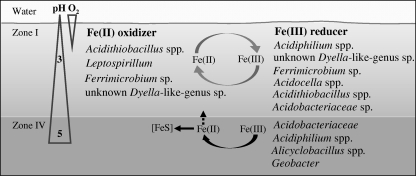
Several phylogenetic groups involved in Fe(II) oxidation were restricted to the top acidic sediment zones of Lake 77 as revealed by the combination of our methods (Fig. 4). All isolates of the genus Acidithiobacillus were obtained from the acidic top zones of the Deep and North basins of the lake, but Acidithiobacillus spp.-related sequences were not detected in the clone library. From the other Gammaproteobacteria family Xanthomonadaceae, 8 isolates and 15 clones were obtained only from the top sediment zones. These 15 clones were closely related to those obtained from the river Rio Tinto, Spain (GenBank sequence accession number DQ303270) (22), from acid mine drainage (GenBank sequence accession numbers DQ480485 and AY096032) (15, 29), and also from the top sediment zone of a manipulated lignite mine lake (KacFeR1-3, GenBank sequence accession number EF544680) (61). Isolate YE3-D1-10-CH showed Fe(II)-oxidizing and Fe(III)-reducing capacities, a trait shared with the closely related strain WJ2 (27). Additional clones were related to the well-known Fe(II) oxidizer Leptospirillum from the Nitrospiraceae family.
FIG. 4.
Schematic of the key microbial players involved in Fe cycling in the acidic lignite mine Lake 77 ecosystem. Microbial players detected by isolation and 16S rRNA gene cloning included Fe(II) oxidizers and Fe(III) reducers. The oxygen content declined sharply to 0 at 2 to 10 mm below the water-sediment interface, and the pH increased from 3 in the water and the top sediment zone I to ca. 5 in zone IV.
Groups of Fe(III)-reducing bacteria varied in their distribution over sediment zones (Fig. 4). All of the 13 Acidocella isolates were obtained from the top acidic zones, matching the pH optimum of the more thoroughly investigated isolate YE4-N1-5-CH. This isolate could reduce schwertmannite similarly to the other four isolates tested (15) but did not oxidize Fe(II). Clones related to the known Fe(III)-reducing genus Acidiphilium (15, 51) were only detected in the BacI library (Fig. 3), but isolates of this genus, with high capacities to reduce schwertmannite under strictly anoxic conditions, could be obtained from all sediment depths and sites. Since at least one Acidiphilium isolate can reduce Fe(III) at a pH of up to 5 (6), bacteria of the genus Acidiphilium might be important drivers for Fe(III) respiration in acidic sediments.
Acidobacteria occurred at higher relative abundances in the deeper sediment zones, and the majority of clones were affiliated with the Acidobacteria subdivision 1, which contains known Fe(III) reducers. Despite the dominance of subdivision 1 Acidobacteria in various habitats, including uranium-contaminated subsurface sediments (4), mine tailings (74), the soil surrounding iron-manganese nodules (28), extremely acidic, metal-rich streams (65), and acidic lignite mine lakes (7, 48), their ecological roles remain poorly documented. Acidobacterium capsulatum DSM 11244T and other related isolates (PK35, RIT23, KP3, and WJ7) are capable of dissimilatory Fe(III) reduction (7, 15, 27) under strict anoxic or microoxic conditions, similar to our isolate YE3-D1-20 that was closely related to strain PK35. The genome study of Acidobacteria strains, including an A. capsulatum strain, strain Ellin345 (subdivision 1), and strain Ellin6076 (subdivision 3), provides some evidence for a probable Fe(III) reduction pathway (73).
All of the 20 Alicyclobacillus-related isolates were obtained from deep, slightly acidic sediment zones despite their Fe(II)-oxidizing capacities. Characterization of these acidophilic iron- and sulfur-oxidizing Firmicutes shows that they are aerobes or facultative anaerobes with considerable metabolic versatility (46, 75). Thirteen of our tested isolates could oxidize Fe(II) and reduce schwertmannite, but the extent of Fe(II) formation was not enhanced under strictly anoxic conditions. Several Alicyclobacillus strains can utilize Fe(III), and one Alicyclobacillus isolate, GSM, is able to couple Fe(III) reduction with cell growth under anoxic conditions (75). Isolate FeSo-D4-20-CH, which probably represents a new species, could grow within a pH range of 1.5 to 6, suggesting a high adaptability to geochemical gradients. Surprisingly, none of the molecular techniques detected members of the Alicyclobacillaceae family, although these organisms were cultivated in abundance. This missing overlap might be due to the inherent biases of molecular techniques and/or to their low in situ abundance.
Geobacter-related sequences were obtained from the deep but not from the top acidic sediment zone, a result which was supported by Geobacter-specific quantitative PCR. Sixteen of 17 clones were closely related to Geobacter sp. G02 or Geobacter chapellei strain 172 (54), suggesting that some Geobacteraceae species may be more acid tolerant than previously thought. However, attempts to isolate Fe(III) reducers at pH 5 from zone IV under strictly anoxic conditions failed. Sulfate reducer sequences from the families Desulfuromonadaceae, Desulfobulbaceae, Desulfobacteraceae, and Desulfovibrionaceae were not closely related to strains known for Fe(III) reduction.
In contrast to previous work (7), archaeal sequences were obtained, which might be due to the different primer set used in this study. Unfortunately, no archaeal strain could be isolated. Both libraries were dominated by Euryarchaeota, including known methanogens. In addition, one clone was affiliated with the family Archaeoglobaceae, which contains strains capable of sulfur reduction (67), a process that appears to be restricted to the deeper, moderately acidic sediment zone of this lignite mine lake (59). Picrophilaceae and Thermoplasmatales-like phylotypes (but not Ferroplasma related) were detected which were related to sequences from an extremely acidic, metal-rich stream in Spain (65). This result is similar to those from other acid mine drainage sites where Thermoplasmatales represent a dominant group (3, 20, 22, 62) but contrasts with the results for other sites where Ferroplasma spp. appear to be responsible for pyrite leaching (20, 21).
Collectively, these results demonstrate that cultivation revealed a much broader diversity of microorganisms with the capacity to reduce Fe(III) under both acidic and moderately acidic conditions (Fig. 4). These findings broaden our previous understanding of the lignite mine Lake 77 ecosystem obtained from a molecular-based study (7). In this work, molecular methods showed that 13 of 101 distinctive phylotypes obtained in the bacterial clone libraries were closely related to our isolated strains. The presence of Fe(II) oxidizers appeared to be restricted to the top acidic sediment zone, where oxygen is present after lake water mixing (Fig. 4). Fe(III)-reducing bacteria included many genera, such as Acidobacterium, Acidocella, Alicyclobacillus, and an unknown Dyella-like genus, which are not commonly found in moderately acidic habitats. The isolates studied in more detail showed an average pH range for growth from 2.5 to 5 with a maximum range of 1.5 to 6, suggesting that they can inhabit a wide ecological niche. In addition to this broad pH tolerance, the isolates obtained reduced Fe(III) under ecosystem-relevant conditions. We found that the isolates preferred microoxic conditions to reduce Fe(III) and were unable to reduce chelated forms of Fe(III), which is in contrast to the results for the commonly studied neutrophilic Fe(III) reducers. Since other acidophilic Fe(III) reducers have been shown to reduce Fe(III) in the presence of oxygen (41) or even to corespire Fe(III) and oxygen (52), the presence of low oxygen concentrations will not inhibit the reduction of Fe(III) in acidic sediments. Thus, the order of redox processes might differ at the sediment-water interface of acidic ecosystems compared to the order of these processes in neutral-pH sediments, with pH acting as a major driving force in shaping biogeochemical redox processes.
Supplementary Material
Acknowledgments
This work was supported by the Graduate School of Excellence Jena School for Microbial Communication (JSMC) and grant KU 1367/8-1 funded by the German Research Foundation (Deutsche Forschungsgemeinschaft [DFG]).
We thank Peter Bouwma for proofreading the manuscript and Susanne Grube for technical assistance.
Footnotes
Published ahead of print on 22 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, L. K., C. Boothman, and J. R. Lloyd. 2007. Identification and characterization of a novel acidotolerant Fe(III)-reducing bacterium from a 3000-year-old acidic rock drainage site. FEMS Microbiol. Lett. 268:151-157. [DOI] [PubMed] [Google Scholar]
- 2.Albert, R. A., J. Archambault, R. Rossello-Mora, B. J. Tindall, and M. Matheny. 2005. Bacillus acidicola sp. nov., a novel mesophilic, acidophilic species isolated from acidic sphagnum peat bogs in Wisconsin. Int. J. Syst. Evol. Microbiol. 55:2125-2130. [DOI] [PubMed] [Google Scholar]
- 3.Almeida, W. I., R. P. Vieira, A. M. Cardoso, C. B. Silveira, R. G. Costa, A. M. Gonzalez, R. Paranhos, J. A. Medeiros, F. A. Freitas, R. M. Albano, and O. B. Martins. 2009. Archaeal and bacterial communities of heavy metal contaminated acidic waters from zinc mine residues in Sepetiba Bay. Extremophiles 13:263-271. [DOI] [PubMed] [Google Scholar]
- 4.Barns, S. M., E. C. Cain, L. Sommerville, and C. R. Kuske. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belnap, C. P., C. Pan, N. C. VerBerkmoes, M. E. Power, N. F. Samatova, R. L. Carver, R. L. Hettich, and J. F. Banfield. 2010. Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 4:520-530. [DOI] [PubMed] [Google Scholar]
- 6.Bilgin, A. A., J. Silverstein, and J. D. Jenkins. 2004. Iron respiration by Acidiphilium cryptum at pH 5. FEMS Microbiol. Ecol. 49:137-143. [DOI] [PubMed] [Google Scholar]
- 7.Blöthe, M., D. M. Akob, J. E. Kostka, K. Goschel, H. L. Drake, and K. Küsel. 2008. pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments. Appl. Environ. Microbiol. 74:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogdanova, T. I., I. A. Tsaphna, T. F. Kondra'eva, V. I. Duda, N. E. Suzina, V. S. Melamud, T. P. Tourova, and G. I. Karavaiko. 2006. Sulfobacillus thermotolerans sp. nov., a thermotolerant, chemolithotrophic bacterium. Int. J. Syst. Evol. Microbiol. 56:1039-1042. [DOI] [PubMed] [Google Scholar]
- 9.Chun, J., J. H. Lee, Y. Jung, M. Kim, S. Kim, B. K. Kim, and Y. W. Lim. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2259-2261. [DOI] [PubMed] [Google Scholar]
- 10.Clark, D. A., and P. R. Norris. 1996. Acidimicrobium ferrooxidans gen. nov., sp. nov.: mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology (Reading, England) 142:785-790. [DOI] [PubMed] [Google Scholar]
- 11.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colwell, R. K., C. X. Mao, and J. Chang. 2004. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology 85:2717-2727. [Google Scholar]
- 13.Coram, N. J., and D. E. Rawlings. 2002. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptosphillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40°C. Appl. Environ. Microbiol. 68:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornell, R. M., and U. Schwertmann. 2003. The iron oxides: structure, properties, reactions, occurrences and uses, 2nd ed. Wiley-VCH Verlagsgesellschaft, Weinheim, Germany.
- 15.Coupland, K., and D. B. Johnson. 2008. Evidence that the potential for dissimilatory ferric iron reduction is widespread among acidophilic heterotrophic bacteria. FEMS Microbiol. Lett. 279:30-35. [DOI] [PubMed] [Google Scholar]
- 16.Dedysh, S. N., Y. Y. Berestovskaya, L. V. Vasylieva, S. E. Belova, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, W. Liesack, and G. A. Zavarzin. 2004. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54:151-156. [DOI] [PubMed] [Google Scholar]
- 17.Denef, V. J., R. S. Mueller, and J. F. Banfield. 2010. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 4:599-610. [DOI] [PubMed] [Google Scholar]
- 18.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaby, N., B. Dold, H. R. Pfeifer, C. Holliger, D. B. Johnson, and K. B. Hallberg. 2007. Microbial communities in a porphyry copper tailings impoundment and their impact on the geochemical dynamics of the mine waste. Environ. Microbiol. 9:298-307. [DOI] [PubMed] [Google Scholar]
- 20.Druschel, G. K., B. J. Baker, T. M. Gihring, and J. F. Banfield. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, K. J., P. L. Bond, T. M. Gihring, and J. F. Banfield. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796-1799. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Moyano, A., E. Gonzalez-Toril, A. Aguilera, and R. Amils. 2007. Prokaryotic community composition and ecology of floating macroscopic filaments from an extreme acidic environment, Rio Tinto (SW, Spain). Syst. Appl. Microbiol. 30:601-614. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Toril, E., E. Llobet-Brossa, E. O. Casamayor, R. Amann, and R. Amils. 2003. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69:4853-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto, K., K. Mochida, Y. Kato, M. Asahara, R. Fujita, S. Y. An, H. Kasai, and A. Yokota. 2007. Proposal of six species of moderately thermophilic, acidophilic, endospore-forming bacteria: Alicyclobacillus contaminans sp. nov., Alicyclobacillus fastidiosus sp. nov., Alicyclobacillus kakegawensis sp. nov., Alicyclobacillus macrosporangiidus sp. nov., Alicyclobacillus sacchari sp. nov and Alicyclobacillus shlzuokensis sp. nov. Int. J. Syst. Evol. Microbiol. 57:1276-1285. [DOI] [PubMed] [Google Scholar]
- 25.Hallberg, K. B., K. Coupland, S. Kimura, and D. B. Johnson. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in north Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72:2022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallberg, K. B., E. Gonzalez-Toril, and D. B. Johnson. 2010. Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 14:9-19. [DOI] [PubMed] [Google Scholar]
- 27.Hallberg, K. B., and D. B. Johnson. 2003. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 71:139-148. [Google Scholar]
- 28.He, J. Z., L. M. Zhang, S. S. Jin, Y. G. Zhu, and F. Liu. 2008. Bacterial communities inside and surrounding soil iron-manganese nodules. Geomicrobiol. J. 25:14-24. [Google Scholar]
- 29.He, Z. G., S. M. Xiao, X. H. Xie, H. Zhong, Y. H. Hu, Q. H. Li, F. L. Gao, G. Y. Li, J. S. Liu, and G. Z. Qiu. 2007. Molecular diversity of microbial community in acid mine drainages of Yunfu sulfide mine. Extremophiles 11:305-314. [DOI] [PubMed] [Google Scholar]
- 30.Hippe, H. 2000. Leptospirillum gen. nov. (ex Markosyan 1972), nom. rev., including Leptospirillum ferrooxidans sp. nov. (ex Markosyan 1972), nom. rev., and Leptospirillum thermoferrooxidans sp. nov. (Golovacheva et al). Int. J. Syst. Evol. Microbiol. 50:501-503. [DOI] [PubMed] [Google Scholar]
- 31.Hiraishi, A., Y. Matsuzawa, T. Kanbe, and N. Wakao. 2000. Acidisphaera rubrifaciens gen. nov., sp. nov., an aerobic bacteriochlorophyll-containing bacterium isolated from acidic environments. Int. J. Syst. Evol. Microbiol. 50:1539-1546. [DOI] [PubMed] [Google Scholar]
- 32.Holland, S. M. 2003. Analytic Rarefaction 1.3 user's guide. University of Georgia, Athens, GA.
- 33.Huber, G., and K. O. Stetter. 1991. Sulfolobus metallicus, sp. nov., a novel strictly chemolithoautotrophic thermophilic archaeal species of metal-mobilizers. Syst. Appl. Microbiol. 14:372-378. [Google Scholar]
- 34.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 35.Jiang, C. Y., Y. Liu, Y. Y. Liu, X. Y. You, X. Guo, and S. J. Liu. 2008. Alicyclobacillus ferrooxydans sp. nov., a ferrous-oxidizing bacterium from solfataric soil. Int. J. Syst. Evol. Microbiol. 58:2898-2903. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, D. B., P. Bacelar-Nicolau, N. Okibe, A. Thomas, and K. B. Hallberg. 2009. Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic actinobacteria. Int. J. Syst. Evol. Microbiol. 59:1082-1089. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, D. B., P. Bacelar-Nicolau, N. Okibe, A. Yahya, and K. B. Hallberg. 2001. Role of pure and mixed cultures of gram-positive eubacteria in mineral leaching, p. 461-470. In V. S. T. Ciminelli and O. J. Garcia (ed.), Biohydrometallurgy: fundamentals, technology and sustainable development, 1st ed., vol. 11A. Elsevier Science, Amsterdam, Netherlands. [Google Scholar]
- 38.Johnson, D. B., and T. A. M. Bridge. 2002. Reduction of ferric iron by acidophilic heterotrophic bacteria: evidence for constitutive and inducible enzyme systems in Acidiphilium spp. J. Appl. Microbiol. 92:315-321. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, D. B., and K. B. Hallberg. 2005. Biogeochemistry of the compost bioreactor components of a composite acid mine drainage passive remediation system. Sci. Total Environ. 338:81-93. [DOI] [PubMed] [Google Scholar]
- 40.Johnson, D. B., and K. B. Hallberg. 2007. Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms, p. 237-261. In D. E. Rawlings and D. B. Johnson (ed.), Biomining. Springer-Verlag, Berlin, Germany.
- 41.Johnson, D. B., and S. McGinness. 1991. Ferric iron reduction by acidophilic heterotrophic bacteria. Appl. Environ. Microbiol. 57:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson, D. B., N. Okibe, and F. F. Roberto. 2003. Novel thermo-acidophilic bacteria isolated from geothermal sites in Yellowstone National Park: physiological and phylogenetic characteristics. Arch. Microbiol. 180:60-68. [DOI] [PubMed] [Google Scholar]
- 43.Johnson, D. B., S. Rolfe, K. B. Hallberg, and E. Iversen. 2001. Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ. Microbiol. 3:630-637. [DOI] [PubMed] [Google Scholar]
- 44.Johnson, D. B., B. Stallwood, S. Kimura, and K. B. Hallberg. 2006. Isolation and characterization of Acidicaldus organivorus, gen. nov., sp. nov.: a novel sulfur-oxidizing, ferric iron-reducing thermo-acidophilic heterotrophic Proteobacterium. Arch. Microbiol. 185:212-221. [DOI] [PubMed] [Google Scholar]
- 45.Johnson, M., I. Zaretskaya, Y. Raytselis, Y. Merezhuk, S. McGinnis, and T. L. Madden. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karavaiko, G. I., T. I. Bogdanova, T. P. Tourova, T. F. Kondrat'eva, I. A. Tsaplina, M. A. Egorova, E. N. Krasil'nikova, and L. M. Zakharchuk. 2005. Reclassification of “Sulfobacillus thermosulfidooxidans subsp thermotolerans” strain K1 as Alicyclobacillus tolerans sp. nov. and Sulfobacillus disulfidooxidans Dufresne et al. 1996. as Alicyclobacillus disulfidooxidans comb. nov., and emended description of the genus Alicyclobacillus. Int. J. Syst. Evol. Microbiol. 55:941-947. [DOI] [PubMed] [Google Scholar]
- 47.Karavaiko, G. I., T. P. Turova, T. F. Kondrat'eva, A. M. Lysenko, T. V. Kolganova, S. N. Ageeva, L. N. Muntyan, and T. A. Pivovarova. 2003. Phylogenetic heterogeneity of the species Acidithiobacillus ferrooxidans. Int. J. Syst. Evol. Microbiol. 53:113-119. [DOI] [PubMed] [Google Scholar]
- 48.Kleinsteuber, S., F. D. Müller, A. Chatzinotas, K. Wendt-Potthoff, and H. Harms. 2008. Diversity and in situ quantification of Acidobacteria subdivision 1 in an acidic mining lake. FEMS Microbiol. Ecol. 63:107-117. [DOI] [PubMed] [Google Scholar]
- 49.Küsel, K. 2003. Microbial cycling of iron and sulfur in acidic coal mining lake sediments. Water Air Soil Pollut. 3:67-90. [Google Scholar]
- 50.Küsel, K., M. Blöthe, D. Schulz, M. Reiche, and H. L. Drake. 2008. Microbial reduction of iron and porewater biogeochemistry in acidic peatlands. Biogeosciences 5:1537-1549. [Google Scholar]
- 51.Küsel, K., T. Dorsch, G. Acker, and E. Stackebrandt. 1999. Microbial reduction of Fe(III) in acidic sediments: isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. Appl. Environ. Microbiol. 65:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Küsel, K., U. Roth, and H. L. Drake. 2002. Microbial reduction of Fe(III) in the presence of oxygen under low pH conditions. Environ. Microbiol. 4:414-421. [DOI] [PubMed] [Google Scholar]
- 53.Lane, D. J. 1991. 16S/23S rRNA sequencing in E. coli, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 54.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. P. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lovley, D. R. 2006. Dissimilatory Fe (III)- and Mn (IV)-reducing prokaryotes, p. 635-658. In M. Dworkin (ed.), The prokaryotes, 3rd ed., vol. 2. Springer, New York, NY. [Google Scholar]
- 56.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction, p. 219-286. In R. K. Poole, Advances in microbial physiology, vol. 49. Academic Press Ltd., London, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 57.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norris, P. R., D. A. Clark, J. P. Owen, and S. Waterhouse. 1996. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiology (Reading, England) 142:775-783. [DOI] [PubMed] [Google Scholar]
- 59.Peine, A., A. Tritschler, K. Küsel, and S. Peiffer. 2000. Electron flow in an iron-rich acidic sediment—evidence for an acidity-driven iron cycle. Limnol. Oceanogr. 45:1077-1087. [Google Scholar]
- 60.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porsch, K., J. Meier, S. Kleinsteuber, and K. Wendt-Potthoff. 2009. Importance of different physiological groups of iron reducing microorganisms in an acidic mining lake remediation experiment. Microb. Ecol. 57:701-717. [DOI] [PubMed] [Google Scholar]
- 62.Qiu, G. Z., M. X. Wan, L. Qian, Z. Y. Huang, K. Liu, X. D. Liu, W. Y. Shi, and Y. Yang. 2008. Archaeal diversity in acid mine drainage from Dabaoshan Mine, China. J. Basic Microbiol. 48:401-409. [DOI] [PubMed] [Google Scholar]
- 63.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Regenspurg, S., A. Brand, and S. Peiffer. 2004. Formation and stability of schwertmannite in acidic mining lakes. Geochim. Cosmochim. Acta 68:1185-1197. [Google Scholar]
- 65.Rowe, O. F., J. Sanchez-Espana, K. B. Hallberg, and D. B. Johnson. 2007. Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ. Microbiol. 9:1761-1771. [DOI] [PubMed] [Google Scholar]
- 66.Segerer, A., A. Neuner, J. K. Kristjansson, and K. O. Stetter. 1986. Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi comb. nov.: facultatively aerobic, extremely acidophilic thermophilic sulfur-metabolizing archaebacteria. Int. J. Syst. Bacteriol. 36:559-564. [Google Scholar]
- 67.Speich, N., C. Dahl, P. Heisig, A. Klein, F. Lottspeich, K. O. Stetter, and H. G. Truper. 1994. Adenylylsulphate reductase from the sulphate-reducing archaeon Archaeoglobus fulgidus: cloning and characterization of the genes and comparison of the enzyme with other iron-sulphur flavoproteins. Microbiology (Reading, England) 140:1273-1284. [DOI] [PubMed] [Google Scholar]
- 68.Straub, K. L., M. Benz, and B. Schink. 2001. Iron metabolism in anoxic environments at near neutral pH. FEMS Microbiol. Ecol. 34:181-186. [DOI] [PubMed] [Google Scholar]
- 69.Stults, J. R., O. Snoeyenbos-West, B. Methe, D. R. Lovley, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan, G. L., W. S. Shu, K. B. Hallberg, F. Li, C. Y. Lan, and L. N. Huang. 2007. Cultivation-dependent and cultivation-independent characterization of the microbial community in acid mine drainage associated with acidic Pb/Zn mine tailings at Lechang, Guangdong, China. FEMS Microbiol. Ecol. 59:118-126. [DOI] [PubMed] [Google Scholar]
- 71.Todorova, S. G., and A. M. Costello. 2006. Design of Shewanella-specific 16S rRNA primers and application to analysis of Shewanella in a minerotrophic wetland. Environ. Microbiol. 8:426-432. [DOI] [PubMed] [Google Scholar]
- 72.Wakeman, K., H. Auvinen, and D. B. Johnson. 2008. Microbiological and geochemical dynamics in simulated-heap leaching of a polymetallic sulfide ore. Biotechnol. Bioeng. 101:739-750. [DOI] [PubMed] [Google Scholar]
- 73.Ward, N. L., J. F. Challacombe, P. H. Janssen, B. Henrissat, P. M. Coutinho, M. Wu, G. Xie, D. H. Haft, M. Sait, J. Badger, R. D. Barabote, B. Bradley, T. S. Brettin, L. M. Brinkac, D. Bruce, T. Creasy, S. C. Daugherty, T. M. Davidsen, R. T. Deboy, J. C. Detter, R. J. Dodson, A. S. Durkin, A. Ganapathy, M. Gwinn-Giglio, C. S. Han, H. Khouri, H. Kiss, S. P. Kothari, R. Madupu, K. E. Nelson, W. C. Nelson, I. Paulsen, K. Penn, Q. H. Ren, M. J. Rosovitz, J. D. Selengut, S. Shrivastava, S. A. Sullivan, R. Tapia, L. S. Thompson, K. L. Watkins, Q. Yang, C. H. Yu, N. Zafar, L. W. Zhou, and C. R. Kuske. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winch, S., H. J. Mills, J. E. Kostka, D. Fortin, and D. R. S. Lean. 2009. Identification of sulfate-reducing bacteria in methylmercury-contaminated mine tailings by analysis of SSU rRNA genes. FEMS Microbiol. Ecol. 68:94-107. [DOI] [PubMed] [Google Scholar]
- 75.Yahya, A., K. B. Hallberg, and D. B. Johnson. 2008. Iron and carbon metabolism by a mineral-oxidizing Alicyclobacillus-like bacterium. Arch. Microbiol. 189:305-312. [DOI] [PubMed] [Google Scholar]
- 76.Yu, Y. N., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinform. 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.