Abstract
The Saccharomyces cerevisiae INO1 gene encodes the structural enzyme inositol-3-phosphate synthase for the synthesis de novo of inositol and inositol-containing phospholipids. The transcription of INO1 is completely derepressed in the absence of inositol and choline (I− C−). Derepression requires the binding of the Ino2p-Ino4p basic helix-loop-helix (bHLH) heterodimer to the UASINO promoter element. We report here the requirement of a third bHLH protein, centromere-binding factor 1 (Cbf1p), for the complete derepression of INO1 transcription. We found that Cbf1p regulates INO1 transcription by binding to sites distal to the INO1 promoter and encompassing the upstream SNA3 open reading frame (ORF) and promoter. The binding of Cbf1p requires Ino2p-Ino4p binding to the UASINO sites in the INO1 promoter and vice versa, suggesting a cooperative mechanism. Furthermore, Cbf1p binding to the upstream sites was required for the binding of the ISW2 chromatin-remodeling complex to the Ino2p-Ino4p-binding sites on the INO1 promoter. Consistent with this, ISW2 was also required for the complete derepression of INO1 transcription.
The Saccharomyces cerevisiae INO1 (inositol-3-phosphate synthase) gene is required for the de novo synthesis of phosphatidylinositol (PI) from glucose-6-phosphate (19, 54). The regulation of INO1 transcription has been studied for 35 years as a model for understanding the regulation of phospholipid biosynthesis. Its transcription is repressed by inositol and choline (I+ C+) and completely derepressed in their absence (I− C−) by an intricate cascade of cis DNA elements and trans factors (11, 15, 32, 39). Most significantly, investigations into its regulation have been driven by the fact that altered INO1 expression is a hallmark of general defects in transcription (35). This study is significant because it reports novel findings regarding new regulators of INO1 transcription.
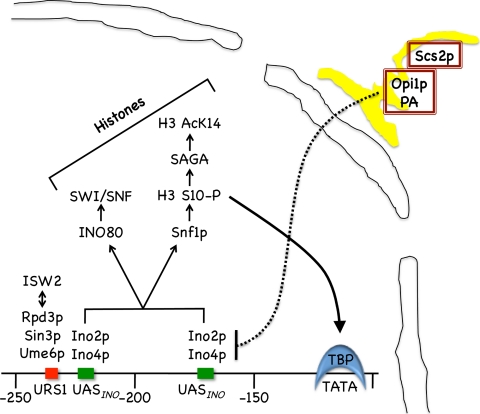
The mechanism for the derepression of INO1 transcription has been extensively studied. Derepression requires the basic helix-loop-helix (bHLH) transcription factors Ino2p and Ino4p that bind as a heterodimer to two cis-regulatory (UASINO) elements in the INO1 promoter (Fig. 1) (2, 6, 55, 67, 73). The Ino2p activation domain then recruits the Snf1p histone kinase to the INO1 promoter, which phosphorylates Ser10 of histone H3 (62). Phosphorylated histone H3 serves as a docking site for the SAGA acetyltransferase, which acetylates Lys14 on histone H3, but also recruits the TATA-binding protein (TBP) (Fig. 1) (61, 62). Ino2p was also shown recently to cause an increase in H3 and H4 acetylation across the INO1 gene under derepressing conditions (22).
Fig. 1.
Model for regulation of INO1 transcription. Shown is a schematic of the INO1 promoter containing two UASINO elements (shown in green) and a repressor site (URS1) (shown in red). Under derepressing conditions (I− C−), the Ino2p-Ino4p heterodimer binds to two UASINO elements and recruits INO80 and Snf1p. INO80 is a chromatin-remodeling complex that recruits another remodeling complex, SWI/SNF. Snf1p is a kinase that phosphorylates serine 10 (S10) on histone H3, which in turn recruits SAGA, which acetylates lysine 14 of histone H3. Histone 3 S10-P promotes the interaction of the TATA-binding protein (TBP) with the INO1 TATA sequence. Under derepressing conditions, the Opi1p repressor is complexed with phosphatidic acid (PA) and retained in the endoplasmic reticulum (ER) (shown in yellow) by association with Scs2p. Under repressing conditions (I+ C+), PA levels drop, and Opi1p is released from the ER, translocates to the nucleus, and associates with Ino2p, repressing transcription. URS1 is a binding site for the general repressor Ume6p, which recruits the Sin3p corepressor and the Rpd3p histone deacetylase (HDAC) complex.
The regulation of INO1 expression also requires several chromatin remodelers. For example, Ino2p recruits INO80 to the INO1 promoter, which then recruits SWI/SNF, which leads to chromatin remodeling at the INO1 promoter (74, 75). Both ISW1 and ISW2 complexes have been reported to play regulatory roles in INO1 transcription (70). Isw2p has been shown to remodel INO1 chromatin (53), and the ISW2 complex, containing Isw2p and Itc1p, has been shown to be required for the complete repression of INO1 through the Ume6p/Sin3p/Rpd3 histone deacetylase (HDAC) (Fig. 1) (24, 31, 53, 82).
INO1 transcription is also affected by the physical location of the gene. Upon derepression, the INO1 locus is recruited to the nuclear periphery (8, 9). It was shown recently that the localization of INO1 to the periphery is dependent on sequences, called DNA zip codes, within the INO1 promoter and upstream region and that these zip codes are required for complete derepression (1).
Under repressing conditions (I+ C+), INO1 gene expression is reduced by the Opi1p repressor protein (9, 36, 44, 47, 48, 50, 63, 65). Opi1p senses phosphatidic acid (PA) levels, which are elevated under derepressing conditions (I− C−) (Fig. 1) (63). Opi1p bound to PA is tethered to the endoplasmic reticulum (ER) via Scs2p, an ER integral membrane protein (Fig. 1) (63–65). Under I+ C+ conditions, PA levels decrease, releasing Opi1p, which translocates to the nucleus and prevents INO1 transcription by interacting with Ino2p (Fig. 1) (40, 48, 63). It has been suggested that Opi1p recruits the Sin3p/Rpd3p HDAC complex to the UASINO elements (87). It is more strongly supported that Sin3p/Rpd3p is recruited by Ume6p to an URS1 element on the INO1 promoter and leads to the general repression of INO1 transcription (Fig. 1) (21, 31, 49, 79).
The bHLH transcription factors function by dimerization to regulate transcription (5, 7, 34, 38, 46, 58, 59, 68, 77, 78). Previous studies in our laboratory have shown that multiple yeast bHLH proteins can regulate a single gene (14, 16). This study began with the goal of determining if bHLH proteins other than Ino2p and Ino4p are also involved in the regulation of INO1. We found that Cbf1p is also required for the complete derepression of INO1 transcription. Our results show that Cbf1p and Ino2p-Ino4p bindings are interdependent. Cbf1p plays an important role in chromatin remodeling (25, 52, 70, 71). Consistent with this role, we show that Cbf1p is required for the binding of the ISW2 chromatin-remodeling complex to INO1 upstream sequences bound by the Ino2p-Ino4p heterodimer.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The S. cerevisiae strains used in this study were BY4742 (MATα his3-1 leu2-0 lys2-0 ura3-0); isogenic strains containing the ino2Δ, ino4Δ, pho4Δ, cbf1Δ, sgc1Δ, rtg1Δ, rtg3Δ, hms1Δ, ygr290wΔ, isw2Δ, itc1Δ, isw1Δ, ioc2Δ, ioc3Δ, and ioc4Δ alleles (30, 88); and a strain harboring an INO1 promoter deletion (INO1-100 or OPI5) (66, 83). Strains with tandem affinity purification (TAP)-tagged INO2, INO4, CBF1, and ISW2 were purchased from Open Biosystems (Huntsville, AL) (29).
A CBF1-TAP-tagged strain harboring an ino2Δ mutant allele was generated by transformation with a 2,000-bp fragment amplified from the ino2Δ strains using primers KANino2ΔF and KANino2ΔR (Table 1) and wild-type (WT) genomic DNA. Genomic DNA was extracted by using a Zymo yeast DNA extraction kit (Zymo Research, Orange, CA). The 2,000-bp fragment contained the KanMX cassette (86) flanked by 200 bp of DNA upstream and 200 bp of DNA downstream of the INO2 open reading frame (ORF). The ino4Δ CBF1-TAP and cbf1Δ ISW2-TAP strains were created in a similar manner by using the KAN primer pairs listed in Table 1. INO2-TAP- and INO4-TAP-tagged strains harboring a cbf1Δ mutant allele were similarly created by replacing the CBF1 ORF with the URA3 gene. This was accomplished by amplifying the URA3 gene flanked by 200 bp of promoter sequences from plasmid YEp357R (61), using the cbf1orf URA3 primer pair (Table 1). The amplified fragment containing the URA3 gene flanked by 45 bp of sequence homologous to the CBF1 ORF was transformed into the INO2-TAP and INO4-TAP strains. Transformants were selected on plates lacking Ura (Ura−), and the cbf1Δ::URA3 allele was confirmed by PCR and sequencing. All integration-based transformations were carried out with a Yeast Maker transformation kit (Clontech, Mountain View, CA).
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| KANino2ΔF | 5′-TTTTCTATCTCCCTCCGTCAT-3′ |
| KANino2ΔR | 5′-ATGAAGATACTGGTAATTCTT-3′ |
| KANino4ΔF | 5′-TCTTTGTTATAAATAGATTAG-3′ |
| KANino4ΔR | 5′-TATAGTAAGTTGAACACTAAA-3′ |
| KANcbf1ΔF | 5′-ACGAGAAAAGTATTGGGCAAA-3′ |
| KANcbf1ΔR | 5′-TAACGTACAAAGACATATTTG-3′ |
| cbf1orf URA3 F | 5′-TCAAGTGCTTAAAATATAATACGGTTTTCTACACTTTTATTAACGTAGCTTTTCAATTCAATTCA-3′ |
| cbf1orf URA3 R | 5′-TACATAGGGAGACTCGAAATACATTTAGCTATCTATTTTTAACTCGTTTTGCTGGCCGCATCTT-3′ |
| ISW2 URA3 F | 5′-ATGACGACCCAGCAAGAGGAGCAACGAAGTGATACCAAGAATAGCTAGCTTTTCAATTCAATTCA-3′ |
| ISW2 URA3 R | 5′-TCATGCTTCTTGATCAATTTTGGTTCTTTTATCAACATGATCGTTGTTTTGCTGGCCGCATCTTC-3′ |
| INO1-Ebox1F | 5′-CCCAGAATATTGAACTTATTTAATTGAGCTCGAGCAGAGAAAGCGCACCTCTGCGTTGG-3′ |
| INO1-Ebox1R | 5′-CCAACGCAGAGGTGCGCTTTCTCTGCTCGAGCTCAATTAAATAAGTTCAATATTCTGGG-3′ |
| INO1-Ebox2F | 5′-CCAAGTATGCGCTTCGGCGGCTAAATGCGGTCTAGAAAAAGTATTGTCTATTTTATCTTCATCC-3′ |
| INO1-Ebox2R | 5′-GGATGAAGATAAAATAGACAATACTTTTTCTAGACCGCATTTAGCCGCCGAAGCGCATACTTGG-3′ |
| Cbf1-XbaI F | 5′-TCTAGAACATGTCATCCGTGAGCG-3′ |
| Cbf1-Sal1R | 5′-GTCGACGCAGATACATAGGGAGACT-3′ |
| INO1-100 KpnI F | 5′-GGTACCAAAACAAGTAGAGGAAAAG-3′ |
| INO1-100 EcoRI R | 5′-GAATTCATTGTTACTTCTTTTTCACTG-3′ |
| SNA3-500F (F) | 5′-TCCTCTTTGTGTGGGACGAT-3′ |
| SNA3-500R (F) | 5′-TCAATGCAACGCTTTACTGC-3′ |
| SNA3-200F (E) | 5′-ACGTGATGAAGGCTCGTTTT-3′ |
| SNA3-200R (E) | 5′-TGGTTGTTTGCTTTCTGCTG-3′ |
| INO1-849F (D) | 5′-TAATTTAGAAATGGACAGAGACCA-3′ |
| INO1-849R (D) | 5′-GTATCCCTGTTGAACATACCCTTA-3′ |
| INO1-549F (C) | 5′-CCCTGCAGAGGAATCTCAAG-3′ |
| INO1-549R (C) | 5′-CACTAAGTACGGCCGGAAGA-3′ |
| INO1-383F (B) | 5′-ATTGCCTTTTTCTTCGTTCC-3′ |
| INO1-383R (B) | 5′-CATTCAACACTTTCGATTCC-3′ |
| INO1E1F (A) | 5′-CTTCATCCTTCTTTCCCAGAATATTGAAC-3′ |
| INO1E1R (A) | 5′-GACGAAAGCTCCAATTTATATACGTCTC-3′ |
| INO1-ORF1 F | 5′-CAAACTACTTCGGCTCCATGAC-3′ |
| INO1-ORF1 R | 5′-CTTGACTTCTCTGCATAGCTTCG-3′ |
| INO1-ORF2 F | 5′-GTATTAAACCGGTCTCCATTGC-3′ |
| INO1-ORF2 R | 5′-CCGACGGGCTTCATATATTTG-3′ |
| TCM1-ORF F | 5′-CCAGAGCTGGTCAAAGAGGT-3′ |
| TCM1-ORF R | 5′-ACCGTAGTGGACGAAACCAC-3′ |
Yeast cultures were grown at 30°C in a complete synthetic medium lacking inositol, choline, KH2PO4 (16, 51, 84), and uracil and/or leucine (in the case of reporter plasmids). Where indicated, 75 μM inositol (I+) and/or 1 mM choline (C+) was added. Low-Pi medium contained 0.22 mM KH2PO4 and 20 mM KCl, and high-Pi medium contained 11 mM KH2PO4. The high-Pi medium was used in all figures where the [Pi] is not indicated. Plasmids containing Escherichia coli DH5α cells (Invitrogen, Carlsbad, CA) were grown at 37°C in Luria-Bertani broth containing 50 μg/ml ampicillin.
Plasmid construction.
Plasmid pJH330 was described previously (21) and contains 543 bp upstream of the INO1 ORF and 132 codons of the INO1 ORF fused in frame to the lacZ reporter in YEp357R (72). The upstream sequences include 439 bp of the SNA3-INO1 intergenic region and 104 bp of the 3′ end of the SNA3 ORF. Two E boxes in the INO1 promoter (positions −173 to −178 and −238 to −243) were mutagenized by using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). A pGEM-T-INO1 derivative, which contains the INO1 fragment from pJH330, was used for this mutagenesis. INO1 E-box primers (Table 1) were used to create the single E-box mutants. One of the single mutants was used to create the double mutant. The mutant fragments were digested with EcoRI and KpnI and cloned into YEp357R to yield the INO1 E1 boxΔ, E2 boxΔ, and E1-E2 boxΔ versions of pJH330. A lithium acetate-based one-step method was used for plasmid transformations (13).
A plasmid to complement the cbf1Δ allele was constructed by cloning 500 bp of the CBF1 promoter and the CBF1 ORF into pRS315 (80). Plasmid pRS315-CBF1 was constructed by amplifying a 1,563-bp fragment from BY4742 genomic DNA, using primers Cbf1-XbaI F (position −500) and Cbf1-SalI R (position +1051) (Table 1). The fragment was cloned into pGEM-T, sequenced, excised with XbaI and SalI, and ligated into pRS315.
A strain with an INO1 promoter mutant lacking both E boxes and a URS1 element (Ume6p-binding site) (239-bp deletion) was previously denoted INO1-100 (OPI5) (66, 83). Two INO1-lacZ reporter plasmids that contained either 1,239 bp of INO1 5′-flanking sequences from a wild-type strain or 1,000 bp from the INO1-100 mutant strain were constructed. Wild-type and INO1-100 genomic sequences were amplified by using primers INO1-100 KpnI F (position −1239) and INO-100 EcoRI R (position −1) (Table 1). The resulting fragments (1,240 bp for the wild type and 1,000 bp for INO1-100) were cloned into pGEM-T, sequenced, excised with KpnI and EcoRI, and ligated into YEp357R (72) to yield lacZ fusions called INO1-1200-lacZ and INO1-100-lacZ, respectively.
Enzyme assays.
β-Galactosidase assays were performed with microtiter plates as described previously (16). Units of β-galactosidase activity were calculated as the optical density at 420 nm (OD420)/min/mg total protein × 1,000.
RNA extraction and quantitative real-time PCR (QRT-PCR) analysis.
RNA was extracted by using a hot-acid phenol method (17), subjected to DNase digestion using Qiagen (Valencia, CA) DNase, and purified by using a Qiagen (Valencia, CA) RNeasy RNA extraction kit. RNA (1 μg) was used to synthesize cDNA using either Superscript II or Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). For quantification, cDNA was diluted 1:10, and quantitative PCR (QPCR) was performed as previously described, with either 300 nM or 500 nM primer concentrations (43). INO1 and TCM1 transcripts were quantified by using the INO1-ORF2 and TCM1-ORF primer pairs (Table 1).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (3), with some modifications. Cells were fixed with formaldehyde for 15 min for the INO2-TAP and INO4-TAP strains and 2 h for the CBF1-TAP, ino2Δ CBF1-TAP, ino4Δ CBF1-TAP, ISW2-TAP, and cbf1Δ ISW2-TAP strains (52). Lysis was performed with a Multivortexer using glass beads. The cell extract was sonicated by using a model 100 Sonic Dismembrator with a Branson 250 Microtip sonicator (Fisher Scientific, Pittsburgh, PA) at a 50% duty cycle with a power of 6. Sonication was performed 20 times for 20 s with at least 1 min on ice between pulses to fractionate DNA to ∼300 bp. Immunoprecipitations were performed by incubating 800 μl chromatin with 40 μl IgG Sepharose beads for 1 h at room temperature. Beads were washed twice each with FA lysis buffer (3), FA lysis buffer containing 500 mM NaCl, and ChIP wash buffer followed by a wash with Tris-EDTA (TE) buffer. Protein-DNA complexes were eluted from the beads by incubating the beads in ChIP elution buffer for 10 min at 65°C followed by TE buffer. The supernatants from the two steps were combined, treated with 25 μg RNase A (Invitrogen, Carlsbad, CA), and incubated for 15 min at 37°C. DNA was eluted by incubating the supernatant at 65°C overnight with 5 μg proteinase K (Invitrogen, Carlsbad, CA) and 0.1% SDS. DNA was purified by using ChIP DNA Clean and Concentrator kits (Zymo Research, Orange, CA). For QPCR analysis, ChIP DNA and input DNA were diluted 1:10 and 1:100, respectively. QPCR analysis was performed as previously described (43). Primers used for QPCR analysis are listed in Table 1.
RESULTS
Transcription of INO1 is regulated by Cbf1p.
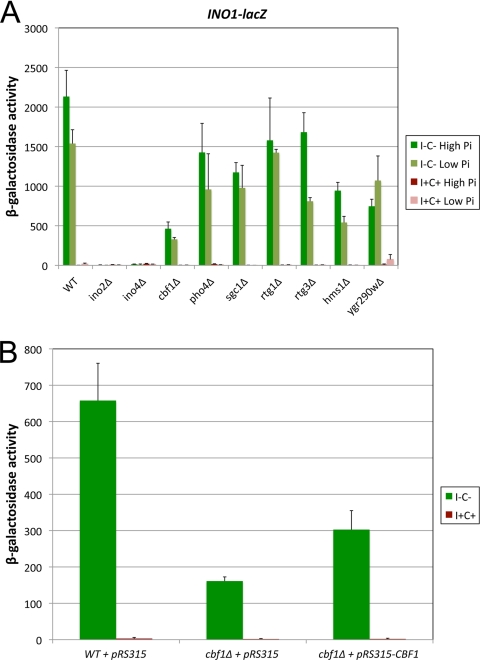
The Ino2p and Ino4p bHLH proteins are known regulators of INO1 transcription (2, 67, 73). We recently reported that the expression of two well-studied bHLH target genes, CIT2 and ENO1, is regulated by multiple bHLH factors (14, 16). Thus, we tested whether other bHLH proteins, besides Ino2p and Ino4p, also regulate INO1 transcription. To this end, we transformed WT and isogenic bHLH knockout strains with an INO1-lacZ reporter (pJH330) that contains 543 bp of the INO1 promoter. Inositol (I) and choline (C) regulate Ino2p and Ino4p function (11, 15, 32, 39), and phosphate (Pi) concentrations regulate Pho4p function (45, 81); hence, transformants were grown in four different media that varied these components (I− C− high Pi, I− C− low Pi, I+ C+ high Pi, and I+ C+ low Pi). In the case of ino2Δ and ino4Δ mutant strains, the I− C− media contained 10 μM inositol, which allows growth but still derepresses the expression of Ino2p-Ino4p target genes (4, 20).
As expected, Ino2p and Ino4p were required for INO1-lacZ expression under both derepressing conditions (I− C−) (Fig. 2A). In addition, the data showed that mutations in all bHLH proteins affected transcription from the INO1 promoter, although in most cases the effect was modest (see rtg1Δ, rtg3Δ, and pho4Δ strains in Fig. 2A). The most dramatic defect, besides those seen with the ino2Δ and ino4Δ strains, was observed with the cbf1Δ strain. Activity was reduced to 21% of that seen for the WT under both derepressing conditions (I− C−) (Fig. 2A). For the remainder of this study, we focused on Cbf1p because it was found to exhibit the most dramatic effect on INO1 derepression. While we conducted most experiments under the four growth conditions described above, for the reminder of this report we show only data for the high-Pi conditions to facilitate presentation and because the low-Pi results recapitulated the high-Pi results.
Fig. 2.
CBF1 regulates INO1-lacZ expression. (A) WT and isogenic bHLH knockout strains were transformed with an INO1-lacZ plasmid (pJH330). Transformants were grown in four different media: I− C− high Pi, I− C− low Pi, I+ C+ high Pi, and I+ C+ low Pi. Green and red bars indicate derepressing and repressing conditions, whereas dark and light indicate high and low Pi, respectively. In the case of ino2Δ and ino4Δ, the I− C− media had 10 μM inositol to allow the growth of these auxotrophic strains. Cells were harvested in mid-log phase and assayed for β-galactosidase activity. (B) Complementation test of the cbf1Δ INO1-lacZ phenotype. The cbf1Δ strain was cotransformed with the INO1-lacZ plasmid and either a pRS315-CBF1 plasmid or pRS315, and an isogenic WT strain was cotransformed with INO1-lacZ and plasmid pRS315. These transformants were assayed for β-galactosidase activity. The data represent means and standard errors of the means from at least three different experiments.
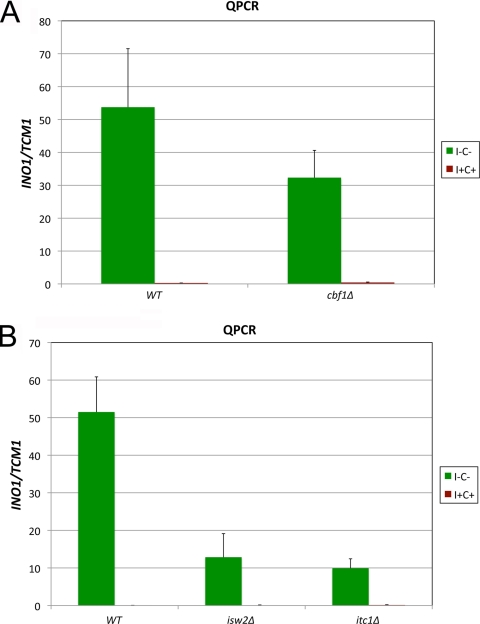
Two different experiments were carried out to confirm the effect of the cbf1Δ allele on INO1 expression. A complementation test was performed to confirm that the phenotype of the cbf1Δ strain was due to the knockout allele (Fig. 2B). The cbf1Δ mutant strain was transformed with either a pRS315 plasmid or a pRS315 derivative carrying the CBF1 gene driven by its own promoter, and an isogenic wild-type strain was transformed with the empty plasmid pRS315. The cbf1Δ strain carrying plasmid pRS315-CBF1 yielded INO1-lacZ expression that was 2-fold higher than that of the pRS315 transformant, confirming that the phenotype was due to the cbf1Δ mutation. However, plasmid pRS315-CBF1 did not completely restore activity to wild-type levels (Fig. 2B). This is not an unusual observation, since the expression of the plasmid-based CBF1 may not completely recapitulate the native state. We also directly tested the effect of cbf1Δ on the transcription of the INO1 gene by QRT-PCR analysis. The data showed that INO1 transcript levels were reduced by ∼2-fold in a cbf1Δ strain compared to a WT strain under derepressing conditions (I− C−) (Fig. 3A). Thus, both the QRT-PCR analysis and the lacZ reporter assay showed that Cbf1p is required for the complete derepression of INO1 transcription and that this occurs through the INO1 promoter.
Fig. 3.
Quantification of INO1 transcript levels in WT and cbf1Δ strains. (A) Isogenic WT and cbf1Δ strains were grown to mid-log phase under derepressing (I− C−) and repressing (I+ C+) conditions, and INO1 transcript levels were quantified by QRT-PCR. (B) Similarly, isogenic WT, isw2Δ, and itc1Δ strains were grown as described above. INO1 transcript levels were normalized to TCM1 transcript levels. The data represent means and standard errors of the means from at least three different experiments.
Cbf1p binds distal sites in the INO1 promoter.
bHLH transcription factors regulate transcription by binding DNA (42, 77). As stated above, Ino2p and Ino4p interact with the INO1 promoter at two E boxes (UASINO elements) (2, 67, 73), and Cbf1p activates the expression of MET genes by binding their promoters (52). Since Cbf1p regulates INO1 transcription, ChIP was used to determine if Cbf1p binds to the INO1 promoter.
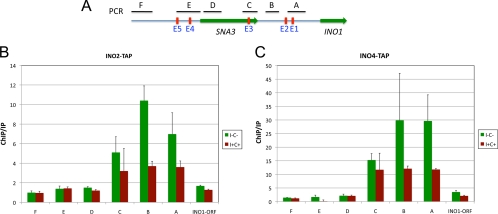
We probed for the binding of Ino2p, Ino4p, and Cbf1p on the INO1 promoter and regions further upstream (Fig. 4A) under derepressing and repressing conditions. The experiments with Ino2p and Ino4p served as controls, since they are known to bind the two E boxes in the INO1 promoter, but in addition, we also tested binding to regions much further upstream (60, 76). We used C-terminal TAP-tagged bHLH strains and quantitative PCR with primers to six different regions spanning ∼1.6 kb upstream of the INO1 ORF (Fig. 4A). As expected, Ino2p-TAP and Ino4p-TAP were enriched on two regions of the INO1 promoter that contain E boxes (E1 and E2) under derepressing conditions (Fig. 4B and C). The B set of primers (Fig. 4A) did not overlap the E2 element but were close enough (46 bp) to identify binding at this site. We did make repeated attempts to identify a primer set that overlapped the E2 element, but every combination tested yielded multiple PCR products or other artifacts. The enrichment of Ino2p-TAP was 7- to 10-fold higher at the INO1 promoter than at TCM1 (normalizer) or the INO1 ORF (Fig. 4B) under derepressing conditions. Ino4p-TAP was enriched ∼30-fold at the INO1 promoter. There was also enrichment at a third region that includes the 3′ end of the adjacent SNA3 ORF (primer set C in Fig. 4). These results are in good agreement with data from previous studies, which showed that the Ino2p-Ino4p heterodimer binds the INO1 promoter and activates INO1 transcription (2, 9, 67, 73).
Fig. 4.
ChIP analysis of Ino2p-Ino4p binding to the INO1 promoter and upstream regions. (A) Schematic showing primer positions (A to E) and E boxes (E1 to E5) relative to the INO1 and SNA3 ORFs. (B and C) Ino2p-TAP and Ino4p-TAP bind to the INO1 promoter. ChIP analysis was performed by using TAP-tagged strains grown under derepressing (I− C−) and repressing (I+ C+) conditions. Enrichment on the INO1 promoter and upstream regions was quantified by using QRT-PCR. ChIP/immunoprecipitation (IP) ratios were normalized by using TCM1. The INO1 ORF primers cover a region within the INO1 coding sequence and serve as a negative control. The data represent means and standard errors of the means from at least three different experiments.
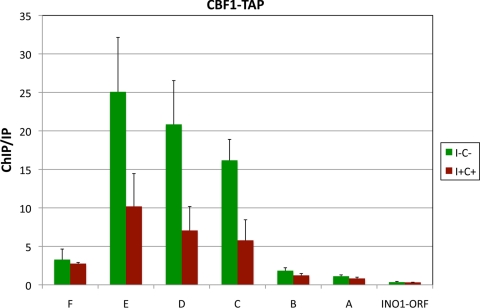
Surprisingly, we did not observe any significant enrichment of Cbf1p-TAP in the same region bound by Ino2p-TAP and Ino4p-TAP in the INO1 promoter (compare Fig. 4 and 5). Instead, Cbf1p was enriched in the region from −439 bp upstream to −1,019 bp upstream of INO1 covered by three primer pairs (Fig. 4A and 5). Notably, two of the three PCR primer sets that identified Cbf1p binding include three potential E boxes. Binding was enhanced 2-fold under derepressing conditions, suggesting that it might depend on Ino2p-Ino4p binding. Thus, Cbf1p regulates INO1 transcription by binding regions upstream of the canonical promoter that include the upstream SNA3 ORF and its promoter.
Fig. 5.
ChIP analysis of Cbf1p binding to the INO1 promoter and upstream regions. Cbf1p-TAP binds to regions upstream of the INO1 promoter within and upstream of the SNA3 ORF. ChIP analysis was performed by using a CBF1-TAP-tagged strain grown under derepressing (I− C−) and repressing (I+ C+) conditions. Enrichment on the INO1 promoter and upstream regions was quantified by using QRT-PCR. ChIP/IP ratios were normalized by using TCM1. The data represent means and standard errors of the means from at least three different experiments.
Cbf1p regulation of INO1 expression is dependent on an Ino2p-Ino4p-binding site.
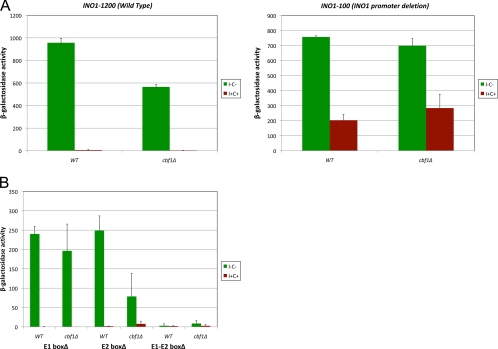
The results described above raised the possibility that Cbf1p regulation of INO1 might depend on Ino2p-Ino4p binding to the INO1 promoter. We tested this possibility in two ways. First, we used a construct carrying a deletion of the INO1 promoter (previously called INO1-100 or OPI5) fused to lacZ. The INO1-100 mutation has been described to be a dominant allele of INO1 lacking 239 bp of the INO1 promoter, including the two UASINO elements (E1 and E2 in Fig. 4); however, it demonstrates incomplete dominance with respect to INO1 transcription (83). The INO1-100 allele exhibits a nearly constitutive expression of INO1, which is independent of Ino2p and Ino4p (83). The repressive region required for the INO1-100 phenotype was mapped to a 20-bp region adjacent to the URS1 element and overlapping the distal UASINO element (E2 box) (Fig. 1) (83). It is not definitively known why the INO1 gene is expressed in the absence of the two UASINO elements. For the purposes of our studies, the INO1-100 allele was used simply to determine if Cbf1p regulation of INO1 expression required the region, including Ino2p-Ino4p-binding sites. As a control, we transformed a construct containing 1,200 bp of DNA upstream of the INO1 ORF fused to the lacZ reporter into WT and cbf1Δ strains and assayed for β-galactosidase activity. This full-length construct recapitulated the results observed with the cbf1Δ allele in the INO1-lacZ reporter described above (pJH330, which contained only 543 bp upstream of INO1), namely, decreased expression in the cbf1Δ strain (compare Fig. 6A and 2). However, expression from the INO1-100 promoter was unaffected by the cbf1Δ mutation (Fig. 6A). Also, in accordance with previously reported results (83), the INO1-100 construct yielded only 50% repression in inositol and choline (Fig. 6A) compared to the wild-type INO1 promoter construct (Fig. 6A). These results support the model whereby the Cbf1p-mediated regulation of INO1 requires the Ino2p-Ino4p heterodimer bound to the INO1 regulatory region.
Fig. 6.
Cbf1p regulation of INO1 expression depends on Ino2p-Ino4p-binding sites in the INO1 promoter. (A) WT and isogenic cbf1Δ knockout strains were transformed with an INO1-1200-lacZ plasmid (wild-type INO1 promoter) or an INO1-100-lacZ plasmid (UASINO-deleted INO1 promoter). (B) WT and isogenic cbf1Δ knockout strains were transformed with pJH330 containing mutations of the E1 box, the E2 box, or both the E1 and E2 boxes. These transformants were assayed for β-galactosidase activity as described above. The data represent means and standard errors of the means from at least three different experiments.
The results described above did not preclude that Cbf1p regulation of INO1 might be due to other factors that bind the 239-bp region deleted in INO1-100. To more precisely define if Ino2p-Ino4p binding to the INO1 promoter is required for Cbf1p binding, we created specific E-box point mutations in the pJH330 construct. The E1-boxΔ, E2-boxΔ, and E1-E2-boxΔ mutants were transformed into WT and cbf1Δ mutant strains and assayed for lacZ expression. Deleting the two E boxes eliminated the expression of the reporter (Fig. 6B). However, the data also show that deleting the E1 box effectively eliminated the cbf1Δ phenotype (Fig. 6B). This epistatic effect suggests that the cbf1Δ phenotype is dependent on Ino2p-Ino4p binding to the E1 box. The results also show that eliminating the E2 box did not affect the cbf1Δ phenotype. It is curious that the E box that is more distal to the Cbf1p-binding sites appears to be required for Cbf1p activity. A model to explain this result is discussed below.
Ino2p-Ino4p and Cbf1p bindings are interdependent.
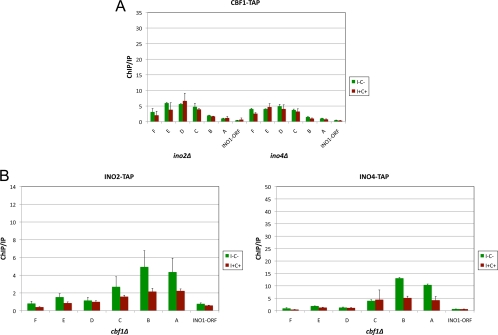
The results described above led us to determine the sequence of events of Cbf1p and Ino2p-Ino4p binding to the INO1 promoter and upstream sequences. Given that the effect of Cbf1p on the derepression of INO1 depended on the Ino2p-Ino4p-binding sites in the INO1 promoter (Fig. 6), we reasoned that the Ino2p-Ino4p heterodimer might recruit Cbf1p to the upstream regions. To test this, we performed a ChIP assay using ino2Δ CBF1-TAP and ino4Δ CBF1-TAP strains, where the INO2 and INO4 genes have been replaced with a KanMX cassette. The data show that deleting INO2 and INO4 severely decreased (∼80% decrease) Cbf1p-TAP binding to regions upstream of INO1 (compare Fig. 7A and 5). These results indicate that Cbf1p recruitment to the region from positions −439 to −1019 upstream from the INO1 gene is dependent on Ino2p-Ino4p binding to downstream regions. Likewise, we performed ChIP using cbf1Δ INO2-TAP and INO4-TAP strains (Fig. 7B). The data show that Cbf1p is required for the complete binding of the Ino2p-Ino4p heterodimer. There was a ∼60% drop in Ino2p and Ino4p binding in the cbf1Δ strain relative to the wild-type strain (compare Fig. 4B and C and 7B). Note that the ChIP experiments were normalized by using binding to TCM1, which allows comparisons between data sets.
Fig. 7.
Cbf1p and Ino2p-Ino4p bindings to the INO1 promoter are interdependent. (A) Cbf1p binding requires the Ino2p-Ino4p heterodimer. ChIP analysis was performed by using CBF1-TAP-tagged ino2Δ and ino4Δ strains under derepressing (I− C−) and repressing (I+ C+) conditions. (B) Ino2p-Ino4p binding requires Cbf1p. ChIP analysis was performed by using INO2- and INO4-TAP-tagged cbf1Δ strains as described above. Enrichment on the INO1 promoter and upstream regions was quantified by using QRT-PCR and the primer pairs described in the legend of Fig. 4A. ChIP/IP ratios were normalized by using TCM1. The data represent means and standard errors of the means from at least three different experiments.
The ISW2 chromatin-remodeling complex binds to the INO1 promoter and is required for complete derepression of INO1 expression.
Cbf1p has been shown to regulate transcription by recruiting chromatin remodelers of the imitation switch (ISWI) class family, composed of ISW1a, ISW1b, and ISW2 (53, 70, 71). ISWI family proteins were shown previously to be involved in the repression of INO1 expression (70, 82). We reasoned that Cbf1p might recruit ISWI complexes to regulate INO1 transcription under derepressing conditions. We first tested the effect of ISWI complex mutants on INO1-lacZ expression (pJH330). We found that Isw2p and Itc1p were required for the complete derepression of the INO1 promoter (see Fig. S1 in the supplemental material). These two proteins are members of the ISW2 complex (70). We also tested the effect of ISW2 mutants on the transcription of INO1 by QRT-PCR. Both Isw2p and Itc1p were required for the complete derepression of INO1 transcription (Fig. 3B). INO1 transcript levels were reduced more than 60% in isw2Δ and itc1Δ mutant strains compared to WT levels.
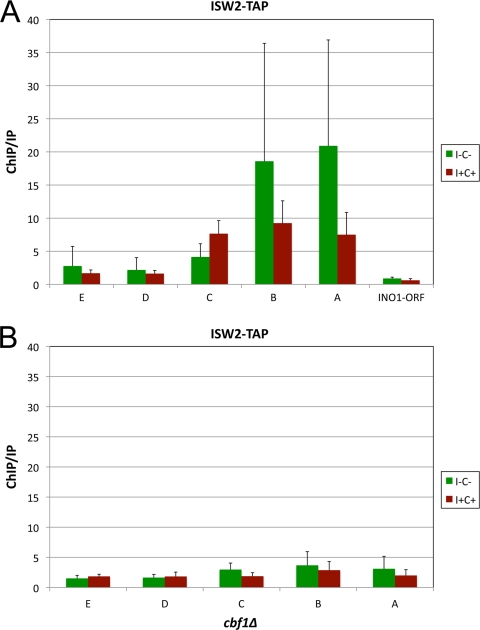
The results described above led us to determine if ISW2 components are associated with the INO1 promoter. We performed a ChIP analysis using TAP-tagged Isw2p under activating and repressing conditions using the same primer sets described above in order to compare ISW2 binding to Ino2p-Ino4p and Cbf1p binding patterns. We found that Isw2p-TAP binding was enriched in the INO1 promoter (Fig. 8A). The pattern of ISW2 binding was similar to that seen for Ino2p-TAP and Ino4p-TAP (compare Fig. 6B to 4B and C). Note that the large variability in binding for Isw2p was previously documented by others in the field. However, our results are consistent with genomic ChIP analysis using a catalytically inactive Isw2p that was found to bind the INO1 promoter (28).
Fig. 8.
Isw2p recruitment to the INO1 promoter requires Cbf1p. ChIP analysis was performed by using ISW2-TAP (A) and ISW2-TAP cbf1Δ strains (B) under derepressing (I− C−) and repressing (I+ C+) conditions. Enrichment on the INO1 promoter was quantified by using QRT-PCR and the primer pairs described in the legend of Fig. 4A. ChIP/IP ratios were normalized by using TCM1. The data represent means and standard errors of the means from at least three different experiments.
To further elucidate the role of Cbf1p in INO1 transcription, we explored the possibility that Cbf1p plays a role in Isw2p binding to the INO1 promoter. We performed a ChIP assay using a cbf1Δ ISW2-TAP strain. The data showed that the binding of Isw2p-TAP was substantially decreased in the cbf1Δ strain (Fig. 8B).
DISCUSSION
The expression of the S. cerevisiae INO1 gene is regulated by a variety of environmental cues such as inositol, nitrogen starvation, and the unfolded-protein response (9, 11, 12, 15, 33, 39). These responses require intricate regulatory cascades involving the concerted action of at least 16 activators, repressors, general transcription factors, histone modifiers, and chromatin remodelers (15, 26, 27). For more than 3 decades, the INO1 gene has been a model for studies of transcription regulation (15). In spite of the wealth of information available, novel mechanisms of transcription regulation are still being uncovered from studies of INO1. For instance, just this year it was reported that the INO1 promoter and regions further upstream harbor sequences called DNA zip codes (1). These sequences are important for the recruitment of INO1 to the nuclear periphery and are required for optimal transcription. The current study underscores the fact that there is still more to be learned from analyzing the regulation of INO1 expression.
In recent years we have reported that multiple bHLH proteins regulate the transcription of single genes in yeast (e.g., CIT2 and ENO1) (14, 16). We can now add INO1 to this target list, since we showed here that Cbf1p acted in concert with the two known activators of INO1 transcription, Ino2p and Ino4p (Fig. 2 and 3). Cbf1p plays an important role in chromosome segregation and the regulation of methionine biosynthesis (10, 57). Strains bearing cbf1Δ alleles display several phenotypes, such as increased chromosome loss, sensitivity to microtubule-disrupting drugs (thiabendazole and benomyl), and methionine auxotrophy (69, 77). In addition, genomic studies have unearthed a myriad of new physical (protein-protein) and genetic interactions, suggesting that Cbf1p has other important biological functions in the cell (18, 56, 85). Collectively, these observations suggest that PI or, more generally, phospholipid synthesis may be coordinated with all of these processes.
Our results showed that Cbf1p was required for the full derepression of INO1 transcription under derepressing (I− C−) conditions (Fig. 2 and 3). We also found that the Cbf1p-mediated regulation of INO1 required one of the two Ino2p-Ino4p-binding sites (Fig. 6B). Surprisingly, we found that Cbf1p did not bind the INO1 promoter, or at least not the SNA3-INO1 intergenic region (bound by the Ino2p-Ino4p heterodimer) (Fig. 5). Instead, it bound to sites distal to the INO1 promoter within the SNA3 ORF and promoter sequences. Genome-wide transcription factor binding analyses done previously did not identify a binding of Cbf1p to these regions (37). This could be due to a number of reasons such as growth conditions or cross-linking conditions. It is important that while data from the ChIP studies suggest that Cbf1p may bind multiple locations, they do not conclusively prove that it does bind multiple locations. In fact, the pJH330 construct that was used for most experiments in this study contains 439 bp of the SNA3-INO1 intergenic region and 104 bp of the 3′ end of the SNA3 ORF. The latter 104 bp includes the E3 box (Fig. 4). Thus, maybe this is the only E box required for regulation by Cbf1p. Alternatively, multiple sites may be involved in vivo, but the E3 box is enough to yield a phenotype on a reporter plasmid.
Our results also showed that Cbf1p was required for maximal Ino2p-Ino4p binding to the INO1 promoter (Fig. 4 and 7). This finding suggests that the bindings of Cbf1p and the Ino2p-Ino4p heterodimer are cooperative. Our results showed that Cbf1p was more dependent on the Ino2p-Ino4p heterodimer than vice versa. This finding is consistent with the effect of mutations in each of the factors on the expression of INO1. An interesting observation was that the E1 box was required for a cbf1Δ phenotype (Fig. 6B). One possible explanation for this observation may lie in the fact that the E1 and E2 boxes lie in different orientations (2, 6, 55, 67, 73). Thus, Cbf1p may interact with either Ino4p or Ino2p but not both. Another possible explanation may have to do with the phasing of the binding sites.
Our results also showed that the ISW2 complex was required for complete INO1 derepression (Fig. 3). Isw2p appeared to regulate INO1 transcription through the same pathway as that of Cbf1p since INO1 transcript levels were nearly identical in the isw2Δ and isw2Δ cbf1Δ strains (see Fig. S2 in the supplemental material). Furthermore, Cbf1p was required for the recruitment of Isw2p onto the INO1 promoter (Fig. 8). This is consistent with the current model for ISW2 activity, which includes a requirement for a DNA-binding factor to recruit the ISW2 complex to promoters (23, 28). It seemed likely Cbf1p was the target-specific DNA-binding factor required for the ISW2 chromatin-remodeling activity on INO1. However, our experiments suggest that ISW2 binding is likely through the Ino2p-Ino4p heterodimer (or something recruited by these bHLH proteins) and that the requirement for Cbf1p is indirect, since Cbf1p is required for maximal Ino2p-Ino4p binding.
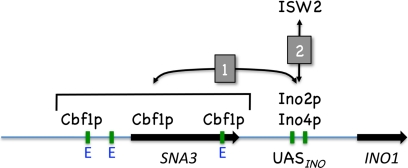
Based on our results, we propose the following model for Cbf1p-mediated INO1 derepression (Fig. 9). Under activating conditions (I− C−), the Ino2p-Ino4p heterodimer bound to the INO1 promoter enhances the binding of Cbf1p to more-distal sites. Likewise, Cbf1p binding enhances Ino2p-Ino4p binding. Cbf1p binds across a region, which includes the upstream SNA3 ORF and its promoter. Cbf1p is required for the interaction of the ISW2 complex with the Ino2p-Ino4p heterodimer. ISW2 remodels chromatin in the INO1 promoter, facilitating transcription under derepressing conditions. It is important that ISW2 has been associated with the repression of INO1 (I+ C+ conditions) (70, 82). This phenotype was not as obvious in the bar graphs presented here, because they included derepressed levels of expression. However, it is evident that the isw2Δ and itc1Δ mutants yielded elevated expression levels of INO1-lacZ under derepressing conditions (see Fig. S1 in the supplemental material).
Fig. 9.
Model for regulation of INO1 transcription by Ino2p-Ino4p, Cbf1p, and ISW2. Black arrows indicate the positions of genes, and green bars indicate the positions of UASINO elements and other potential E boxes. Numbered arrows indicate the sequence of events. Refer to Discussion for a complete description of the model.
Our results raise several questions. Where does Cbf1p bind specifically, and how does the Ino2p-Ino4p heterodimer mediate the recruitment of Cbf1p to distal sites? Genomic regulator localization studies showed that ∼83% of yeast intergenic regions that contain the palindromic E-box sequence CACGTG are likely to bind Cbf1p (52, 60). It is therefore plausible that Cbf1p binds to E box 5 (CACGTG) upstream of the SNA3 ORF (Fig. 4A). However, our ChIP results indicated that Cbf1p is likely to interact at multiple sites in the SNA3 promoter and ORF (Fig. 5). This region contains three potential E boxes that could serve as binding sites for Cbf1p. The fact that Cbf1p enrichment at the INO1 flanking sites is lost in the absence of Ino2p and Ino4p (Fig. 7) shows that Cbf1p binding is consequential for INO1 transcription and not merely an artifact of the ChIP assay. One possible explanation for our results is that one or more of these E boxes are weak binding sites for Cbf1p and that the proximal binding of the Ino2p-Ino4p heterodimer cooperatively enhances the binding of Cbf1p to the distal sites.
Another interesting question is whether Cbf1p regulates SNA3 gene expression. It has been known for some time that SNA3 transcription is repressed by inositol and choline (similarly to INO1), but in contrast to INO1, SNA3 expression is upregulated in the absence of INO2 and INO4 (41). Since the absence of the Ino2p-Ino4p heterodimer results in the loss of Cbf1p binding at distal sites (Fig. 7), it is possible that the binding of Cbf1p at the SNA3 promoter regulates the transcription of both INO1 and SNA3.
Yet another question is whether ISW2-mediated chromatin remodeling at the INO1 locus occurs under depressing conditions. ISW1 and ISW2 complexes have both been reported to be required for INO1 repression (28, 70). So how is ISW2 involved in derepression? INO1 transcript levels were reported previously to be higher in an isw2Δ background than in a wild-type background, suggesting that ISW2 is a repressor of INO1 transcription (53, 82). However, those studies were done only under repressing conditions (I+ C+). Our data clearly showed that under activating (I− C−) conditions, isw2Δ strains show reduced INO1 transcript levels (Fig. 3), indicating that ISW2p is also required for complete derepression. Previous reports showed that nucleosome profiles on the INO1 promoter are similar in isw2 and ume6 strains, suggesting that they act in the same pathway (53). However, the removal of the Ume6p-binding site has no effect on Isw2p enrichment on the INO1 promoter (28), suggesting that ISW2 is recruited independent of Ume6p. These results, combined with our data, suggest that the Ino2p-Ino4p heterodimer may be the target-specific DNA-binding factor that interacts with ISW2 on the INO1 promoter under derepressing conditions but that this interaction is driven by Cbf1p.
Finally, it will also be necessary to determine how Cbf1p regulation is coordinated with other cascades that regulate INO1 expression. For example, how is it coordinated with the INO80 and Snf1p pathways that are also driven by Ino2p-Ino4p binding to the UASINO elements (Fig. 1)? Moreover, how are all of these regulatory cascades coordinated with the recruitment of the INO1 gene to the nuclear periphery by DNA zip codes present within the INO1 regulatory region and distal sites within SNA3 (1)?
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Heather Geisler for technical assistance with the INO1-100 experiments, and we thank Ying He, Christina Paul, and Maybelline Panta for discussions and comments on the manuscript.
This work was supported by a National Science Foundation grant (MCB-0718608) to J.M.L.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Ahmed S., Brickner D. G., Light W. H., Cajigas I., McDonough M., Froyshteter A. B., Volpe T., Brickner J. H. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambroziak J., Henry S. A. 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269:15344–15349 [PubMed] [Google Scholar]
- 3.Aparicio O., Geisberg J. V., Struhl K. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. 17:17.17. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner B. P., Lopes J. M. 1995. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol. Cell. Biol. 15:1709–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atchley W. R., Fitch W. M. 1997. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U. S. A. 94:5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachhawat N., Ouyang Q., Henry S. A. 1995. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 270:25087–25095 [DOI] [PubMed] [Google Scholar]
- 7.Bailey P. C., Martin C., Toledo-Ortiz G., Quail P. H., Huq E., Heim M. A., Jakoby M., Werber M., Weisshaar B. 2003. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15:2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., Widom J., Brickner J. H. 2007. H2A. Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brickner J. H., Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai M., Davis R. W. 1990. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61:437–446 [DOI] [PubMed] [Google Scholar]
- 11.Carman G. M., Han G. S. 2009. Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50(Suppl.):S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H. J., Jones E. W., Henry S. A. 2002. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 162:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D. C., Yang B. C., Kuo T. T. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83–84 [DOI] [PubMed] [Google Scholar]
- 14.Chen L., Lopes J. M. 2010. Multiple bHLH proteins regulate CIT2 expression in Saccharomyces cerevisiae. Yeast 27:345–359 [DOI] [PubMed] [Google Scholar]
- 15.Chen M., Hancock L. C., Lopes J. M. 2007. Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim. Biophys. Acta 1771:310–321 [DOI] [PubMed] [Google Scholar]
- 16.Chen M., Lopes J. M. 2007. Multiple basic helix-loop-helix proteins regulate expression of the ENO1 gene of Saccharomyces cerevisiae. Eukaryot. Cell 6:786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collart M. A., Oliviero S. 2001. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 13:13.12. [DOI] [PubMed] [Google Scholar]
- 18.Costanzo M. C., Hogan J. D., Cusick M. E., Davis B. P., Fancher A. M., Hodges P. E., Kondu P., Lengieza C., Lew-Smith J. E., Lingner C., Roberg-Perez K. J., Tillberg M., Brooks J. E., Garrels J. I. 2000. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 28:73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue T. F., Henry S. A. 1981. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J. Biol. Chem. 256:7077–7085 [PubMed] [Google Scholar]
- 20.Eiznhamer D. A., Ashburner B. P., Jackson J. C., Gardenour K. R., Lopes J. M. 2001. Expression of the INO2 regulatory gene of Saccharomyces cerevisiae is controlled by positive and negative promoter elements and an upstream open reading frame. Mol. Microbiol. 39:1395–1405 [PubMed] [Google Scholar]
- 21.Elkhaimi M., Kaadige M. R., Kamath D., Jackson J. C., Biliran H., Jr., Lopes J. M. 2000. Combinatorial regulation of phospholipid biosynthetic gene expression by the UME6, SIN3 and RPD3 genes. Nucleic Acids Res. 28:3160–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esposito M., Konarzewska P., Odeyale O., Shen C. H. 2010. Gene-wide histone acetylation at the yeast INO1 requires the transcriptional activator Ino2p. Biochem. Biophys. Res. Commun. 391:1285–1290 [DOI] [PubMed] [Google Scholar]
- 23.Fazzio T. G., Gelbart M. E., Tsukiyama T. 2005. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 25:9165–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fazzio T. G., Kooperberg C., Goldmark J. P., Neal C., Basom R., Delrow J., Tsukiyama T. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreiro J. A., Powell N. G., Karabetsou N., Kent N. A., Mellor J., Waters R. 2004. Cbf1p modulates chromatin structure, transcription and repair at the Saccharomyces cerevisiae MET16 locus. Nucleic Acids Res. 32:1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ford J., Odeyale O., Eskandar A., Kouba N., Shen C. H. 2007. A SWI/SNF- and INO80-dependent nucleosome movement at the INO1 promoter. Biochem. Biophys. Res. Commun. 361:974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ford J., Odeyale O., Shen C. H. 2008. Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochem. Biophys. Res. Commun. 373:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelbart M. E., Bachman N., Delrow J., Boeke J. D., Tsukiyama T. 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19:942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 30.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Guldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kotter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 31.Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433 [DOI] [PubMed] [Google Scholar]
- 32.Greenberg M. L., Lopes J. M. 1996. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 60:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griac P. 1997. Regulation of yeast phospholipid biosynthetic genes in phosphatidylserine decarboxylase mutants. J. Bacteriol. 179:5843–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove C. A., De Masi F., Barrasa M. I., Newburger D. E., Alkema M. J., Bulyk M. L., Walhout A. J. 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138:314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampsey M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13:1099–1133 [DOI] [PubMed] [Google Scholar]
- 36.Hancock L. C., Behta R. P., Lopes J. M. 2006. Genomic analysis of the Opi− phenotype. Genetics 173:621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., Danford T. W., Hannett N. M., Tagne J. B., Reynolds D. B., Yoo J., Jennings E. G., Zeitlinger J., Pokholok D. K., Kellis M., Rolfe P. A., Takusagawa K. T., Lander E. S., Gifford D. K., Fraenkel E., Young R. A. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heim M. A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P. C. 2003. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20:735–747 [DOI] [PubMed] [Google Scholar]
- 39.Henry S. A., Patton-Vogt J. L. 1998. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acid Res. Mol. Biol. 61:133–179 [DOI] [PubMed] [Google Scholar]
- 40.Heyken W. T., Repenning A., Kumme J., Schuller H. J. 2005. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol. Microbiol. 56:696–707 [DOI] [PubMed] [Google Scholar]
- 41.Hirsch J. P., Henry S. A. 1986. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol. Cell. Biol. 6:3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooker C. W., Hurlin P. J. 2006. Of Myc and Mnt. J. Cell Sci. 119:208–216 [DOI] [PubMed] [Google Scholar]
- 43.Jani N. M., Lopes J. M. 2008. Transcription regulation of the Saccharomyces cerevisiae PIS1 gene by inositol and the pleiotropic regulator, Ume6p. Mol. Microbiol. 70:1529–1539 [DOI] [PubMed] [Google Scholar]
- 44.Jiranek V., Graves J. A., Henry S. A. 1998. Pleiotropic effects of the opi1 regulatory mutation of yeast: its effects on growth and on phospholipid and inositol metabolism. Microbiology 144(Pt. 10):2739–2748 [DOI] [PubMed] [Google Scholar]
- 45.Johnston M., Carlson M. 1992. Regulation of phosphate and carbon utilization, p. 193–281InJones E. W., Pringle J. R., Broach J. R. (ed.), The molecular biology of the yeast Saccharomyces cerevisiae: gene expression, vol. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46.Jones S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biol. 5:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaadige M. R., Lopes J. M. 2003. Opi1p, Ume6p and Sin3p control expression from the promoter of the INO2 regulatory gene via a novel regulatory cascade. Mol. Microbiol. 48:823–832 [DOI] [PubMed] [Google Scholar]
- 48.Kaadige M. R., Lopes J. M. 2006. Analysis of Opi1p repressor mutants. Curr. Genet. 49:30–38 [DOI] [PubMed] [Google Scholar]
- 49.Kadosh D., Struhl K. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365–371 [DOI] [PubMed] [Google Scholar]
- 50.Kagiwada S., Hashimoto M. 2007. The yeast VAP homolog Scs2p has a phosphoinositide-binding ability that is correlated with its activity. Biochem. Biophys. Res. Commun. 364:870–876 [DOI] [PubMed] [Google Scholar]
- 51.Kelly B. L., Greenberg M. L. 1990. Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1046:144–150 [DOI] [PubMed] [Google Scholar]
- 52.Kent N. A., Eibert S. M., Mellor J. 2004. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J. Biol. Chem. 279:27116–27123 [DOI] [PubMed] [Google Scholar]
- 53.Kent N. A., Karabetsou N., Politis P. K., Mellor J. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klig L. S., Henry S. A. 1984. Isolation of the yeast INO1 gene: located on an autonomously replicating plasmid, the gene is fully regulated. Proc. Natl. Acad. Sci. U. S. A. 81:3816–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koipally J., Ashburner B. P., Bachhawat N., Gill T., Hung G., Henry S. A., Lopes J. M. 1996. Functional characterization of the repeated UASINO element in the promoters of the INO1 and CHO2 genes of yeast. Yeast 12:653–665 [DOI] [PubMed] [Google Scholar]
- 56.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St. Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643 [DOI] [PubMed] [Google Scholar]
- 57.Kuras L., Barbey R., Thomas D. 1997. Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 16:2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ledent V., Paquet O., Vervoort M. 2002. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 3:RESEARCH0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ledent V., Vervoort M. 2001. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11:754–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., Gerber G. K., Hannett N. M., Harbison C. T., Thompson C. M., Simon I., Zeitlinger J., Jennings E. G., Murray H. L., Gordon D. B., Ren B., Wyrick J. J., Tagne J. B., Volkert T. L., Fraenkel E., Gifford D. K., Young R. A. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799–804 [DOI] [PubMed] [Google Scholar]
- 61.Lo W. S., Duggan L., Emre N. C., Belotserkovskya R., Lane W. S., Shiekhattar R., Berger S. L. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142–1146 [DOI] [PubMed] [Google Scholar]
- 62.Lo W. S., Gamache E. R., Henry K. W., Yang D., Pillus L., Berger S. L. 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24:997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304:1644–1647 [DOI] [PubMed] [Google Scholar]
- 64.Loewen C. J., Levine T. P. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280:14097–14104 [DOI] [PubMed] [Google Scholar]
- 65.Loewen C. J., Roy A., Levine T. P. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loewy B. S. 1985. Ph.D. dissertation. Albert Einstein College of Medicine, New York, NY [Google Scholar]
- 67.Lopes J. M., Henry S. A. 1991. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 19:3987–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Massari M. E., Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mellor J., Jiang W., Funk M., Rathjen J., Barnes C. A., Hinz T., Hegemann J. H., Philippsen P. 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9:4017–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mellor J., Morillon A. 2004. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1677:100–112 [DOI] [PubMed] [Google Scholar]
- 71.Moreau J. L., Lee M., Mahachi N., Vary J., Mellor J., Tsukiyama T., Goding C. R. 2003. Regulated displacement of TBP from the PHO8 promoter in vivo requires Cbf1 and the Isw1 chromatin remodeling complex. Mol. Cell 11:1609–1620 [DOI] [PubMed] [Google Scholar]
- 72.Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299–310 [DOI] [PubMed] [Google Scholar]
- 73.Nikoloff D. M., Henry S. A. 1994. Functional characterization of the INO2 gene of Saccharomyces cerevisiae. A positive regulator of phospholipid biosynthesis. J. Biol. Chem. 269:7402–7411 [PubMed] [Google Scholar]
- 74.Peterson C. L., Kruger W., Herskowitz I. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135–1143 [DOI] [PubMed] [Google Scholar]
- 75.Pollard K. J., Peterson C. L. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Volkert T. L., Wilson C. J., Bell S. P., Young R. A. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306–2309 [DOI] [PubMed] [Google Scholar]
- 77.Robinson K. A., Lopes J. M. 2000. Survey and summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28:1499–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ross S. E., Greenberg M. E., Stiles C. D. 2003. Basic helix-loop-helix factors in cortical development. Neuron 39:13–25 [DOI] [PubMed] [Google Scholar]
- 79.Rundlett S. E., Carmen A. A., Suka N., Turner B. M., Grunstein M. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831–835 [DOI] [PubMed] [Google Scholar]
- 80.Sikorski R. S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Springer M., Wykoff D. D., Miller N., O'Shea E. K. 2003. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1:E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sugiyama M., Nikawa J. 2001. The Saccharomyces cerevisiae Isw2p-Itc1p complex represses INO1 expression and maintains cell morphology. J. Bacteriol. 183:4985–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swift S., McGraw P. 1995. INO1-100: an allele of the Saccharomyces cerevisiae INO1 gene that is transcribed without the action of the positive factors encoded by the INO2, INO4, SWI1, SWI2 and SWI3 genes. Nucleic Acids Res. 23:1426–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toh-E A., Ueda Y., Kakimoto S. I., Oshima Y. 1973. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J. Bacteriol. 113:727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Menard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A. M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., Boone C. 2004. Global mapping of the yeast genetic interaction network. Science 303:808–813 [DOI] [PubMed] [Google Scholar]
- 86.Wach A., Brachat A., Pohlmann R., Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808 [DOI] [PubMed] [Google Scholar]
- 87.Wagner C., Dietz M., Wittmann J., Albrecht A., Schuller H. J. 2001. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41:155–166 [DOI] [PubMed] [Google Scholar]
- 88.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Veronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.