Abstract
Calmodulin in Schizosaccharomyces pombe is encoded by the cam1+ gene, which is indispensable for both vegetative growth and sporulation. Here, we report how Cam1 functions in spore formation. We found that Cam1 preferentially localized to the spindle pole body (SPB) during meiosis and sporulation. Formation of the forespore membrane, a precursor of the plasma membrane in spores, was blocked in a missense cam1 mutant, which was viable but unable to sporulate. Three SPB proteins necessary for the onset of forespore membrane formation, Spo2, Spo13, and Spo15, were unable to localize to the SPB in the cam1 mutant although five core SPB components that were tested were present. Recruitment of Spo2 and Spo13 is known to require the presence of Spo15 in the SPB. Notably, Spo15 was unstable in the cam1 mutant, and as a result, SPB localization of Spo2 and Spo13 was lost. Overexpression of Spo15 partially alleviated the sporulation defect in the cam1 mutant. These results indicate that calmodulin plays an essential role in forespore membrane formation by stably maintaining Spo15, and thus Spo2 and Spo13, at the SPB in meiotic cells.
Calmodulin is a calcium-binding protein that is ubiquitously distributed and highly conserved among eukaryotes. It contains four EF-hand Ca2+-binding sites, which are required for function. Calmodulin controls a variety of cellular processes mostly related to calcium signaling. When bound to calcium, calmodulin undergoes a characteristic conformational change to an active configuration. Activated calmodulin then binds effector proteins and transmits the signal to downstream regulators.
Yeast is a genetically tractable model organism suitable for studying the biological function of calmodulin, using conditional-lethal calmodulin mutants (4). In the budding yeast Saccharomyces cerevisiae, calmodulin is encoded by the CMD1 gene (5). Cmd1p is implicated in a wide variety of cellular processes, including initiation of budding and mitotic spindle formation (24). The fission yeast Schizosaccharomyces pombe has a typical calmodulin encoded by the cam1+ gene, which plays an indispensable role in cell proliferation, dependent on its Ca2+-binding activity (18, 19, 30). A green fluorescent protein (GFP)-Cam1 fusion protein localizes to sites of polarized cell growth and to the spindle pole body (SPB) in vegetative cells (19). Thus, an essential role of Cam1 might be its regulatory function in chromosome segregation (19). The role of calmodulin in the sexual cycle has been documented to a lesser extent in previous studies. A missense mutant, cam1-117, in which the Arg117 codon is changed to a Phe codon, exhibits reduced sporulation efficacy (29), suggesting that calmodulin plays a role in sporulation in fission yeast.
Spore formation in fission yeast initiates with assembly of the forespore membrane (FSM), composed of double-unit membranes within the cytoplasm of a diploid zygote cell (10, 27, 28, 34). The FSM expands to encapsulate each haploid nucleus generated by meiosis and then forms a nucleated prespore. The inner bilayer of the FSM subsequently becomes the plasma membrane of the newborn spores. During meiosis II, the SPB undergoes morphological alteration from a compact single plaque to a multilayered expanded structure (10). Such modification of the SPB is a prerequisite for FSM assembly, which occurs close to the outermost layer of the modified SPB (9, 10, 20, 21).
Three SPB component proteins, Spo2, Spo13, and Spo15, have been identified as essential for SPB modification and formation of the FSM (11, 23). Spo15, a large coiled-coil protein, is associated with the SPB throughout the life cycle and is indispensable for recruitment of Spo2 and Spo13 to the cytoplasmic surface of the meiotic SPB. The latter two proteins are produced only during meiosis (23). These observations imply that the SPB serves as a platform for assembly of the FSM. Cam1 has been reported to localize to the SPB during vegetative growth (19), raising the intriguing possibility that fission yeast calmodulin is involved in sporulation through proper construction of a modified meiotic SPB. To test this possibility, we report herein a detailed analysis of Cam1 localization during meiosis and the consequence of a missense mutation of cam1 on SPB modification and FSM formation.
MATERIALS AND METHODS
Yeast strains, media, and culture conditions.
The S. pombe strains used in this study are listed in Table 1. The complete medium YEA (yeast extract agar) was used for growth. Malt extract agar (MEA) medium and synthetic sporulation media (MM-N and SSA) were used for mating and sporulation. These media have been described in Moreno et al. (17). S. pombe cells were grown and sporulated at 28°C.
Table 1.
S. pombe strains used in this study
| Strain (accession no.)a | Genotypeb | Source or reference |
|---|---|---|
| AI22 (FY19908) | h90cam1-22,117≪ura4+ura4-D18 leu1-32 | This study |
| AI52 (FY19909) | h90cam1-22,117≪ura4+ura4-D18 ade6≪gfp-psy1 | This study |
| AI154 (FY19910) | h90spo15-gfp≪LEU2 leu1-32 | This study |
| AI156 (FY19911) | h90cam1-22,117≪ura4+ura4-D18 spo15-GFP≪LEU2 leu1-32 | This study |
| AI190 (FY19912) | h90cam1-22,117≪ura4+ura4-D18 sid4-gfp::Knrleu1-32 | This study |
| AI197 (FY19913) | h90cam1-22,117≪ura4+ura4-D18 cut12-gfp::ura4+ | This study |
| AI200 (FY19914) | h90cut12-gfp≪ura4+ura4-D18 | This study |
| AI207 (FY19915) | h90cam1-22,117≪ura4+ura4-D18 alp4-gfp::Knr | This study |
| AI210 (FY19916) | h90spo15::ura4+ade6≪gfp-cam1 ura4-D18 | This study |
| AI248 (FY19917) | h90ade6≪gfp-cam1 | This study |
| AI258 (FY19918) | h90leu1≪spo13-gfp | This study |
| AI259 (FY19919) | h90leu1≪spo2-gfp | This study |
| AI262 (FY19920) | h90cam1-22,117≪ura4+ura4-D18 leu1≪spo2-gfp | This study |
| AI263 (FY19921) | h90cam1-22,117≪ura4+ura4-D18 leu1≪spo13-gfp | This study |
| AI350 (FY19922) | h90cam1-22,117≪ura4+ura4-D18 leu1≪pcp1-gfp | This study |
| AI351 (FY19923) | h90leu1≪pcp1-gfp | This study |
| AI509 (FY19924) | h90/h90cam1::ura4+/cam1+ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| AI510 (FY19925) | h90cam1::ura4+leu1≪cam1+ura4-D18 leu1-32 ade6-M216 | This study |
| AI512 (FY19926) | h90cam1::ura4+leu1≪cam1-22 ura4-D18 leu1-32 ade6-M216 | This study |
| AI518 (FY19927) | h90cam1::ura4+leu1≪cam1-22,117 ura4-D18 leu1-32 ade6-M216 | This study |
| AI520 (FY19928) | h90cam1::ura4+leu1≪cam1-EF1 ura4-D18 leu1-32 ade6-M216 | This study |
| AI522 (FY19929) | h90cam1::ura4+leu1≪cam1-EF2 ura4-D18 leu1-32 ade6-M216 | This study |
| AI524 (FY19930) | h90cam1::ura4+leu1≪cam1-EF3 ura4-D18 leu1-32 ade6-M216 | This study |
| AI526 (FY19931) | h90cam1::ura4+leu1≪cam1-EF4 ura4-D18 leu1-32 ade6-M216 | This study |
| AI16 (FY12806) | h90ura4-D18 ade6-M210 leu1-32 | A. Itadani |
| AI2030 (FY19932) | h90leu1≪gfp-cam1+ura4-D18 ade6-M210 leu1-32 | This study |
| AI2031 (FY19933) | h90leu1≪gfp-cam1-22,117 ura4-D18 ade6-M210 leu1-32 | This study |
| AI2032 (FY19934) | h90leu1≪gfp-cam1-22 ura4-D18 ade6-M210 leu1-32 | This study |
| AI2033 (FY19935) | h90leu1≪gfp-cam1-EF1 ura4-D18 ade6-M210 leu1-32 | This study |
| AI2034 (FY19936) | h90leu1≪gfp-cam1-EF2 ura4-D18 ade6-M210 leu1-32 | This study |
| AI2035 (FY19937) | h90leu1≪gfp-cam1-EF3 ura4-D18 ade6-M210 leu1-32 | This study |
| AI2036 (FY19938) | h90leu1≪gfp-cam1-EF4 ura4-D18 ade6-M210 leu1-32 | This study |
| AI3001 (FY19939) | h90cam1::ura4+leu1≪cam1-22 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)] | This study |
| AI3002 (FY19940) | h90cam1::ura4+leu1≪cam1-22 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1+] | This study |
| AI3003 (FY19941) | h90cam1::ura4+leu1≪cam1-22 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1-22] | This study |
| AI3004 (FY19942) | h90cam1::ura4+leu1≪cam1-22,117 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)] | This study |
| AI3005 (FY19943) | h90cam1::ura4+leu1≪cam1-22,117 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1+] | This study |
| AI3006 (FY19944) | h90cam1::ura4+leu1≪cam1-22,117 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1-22,117] | This study |
| AI3007 (FY19945) | h90cam1::ura4+leu1≪cam1-EF2 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)] | This study |
| AI3008 (FY19946) | h90cam1::ura4+leu1≪cam1-EF2 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1+] | This study |
| AI3009 (FY19947) | h90cam1::ura4+leu1≪cam1-EF2 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1-EF2] | This study |
| AI3010 (FY19948) | h90cam1::ura4+leu1≪cam1-EF3 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)] | This study |
| AI3011 (FY19949) | h90cam1::ura4+leu1≪cam1-EF3 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1+] | This study |
| AI3012 (FY19950) | h90cam1::ura4+leu1≪cam1-EF3 ura4-D18 leu1-32 ade6-M216 [pREP1(ade6)cam1-EF3] | This study |
| MFP7 (FY19951) | h−gfp::cam1 ade1-D25 ade6-M210 leu1-32 ura4-D18 | 19 |
| S60-3 (FY12208) | h90sid4-gfp::Knrleu1-32 | K. Okazaki |
| TN8 (FY7132) | h90leu1-32 | 20 |
| YN24 (FY12250) | h90ade6-M210 | Y. Nakase |
| YM20 (FY12096) | h90ade6≪gfp-psy1 | 14 |
| YN104 (FY12332) | h90spo15::ura4+leu1-32 ade6-M210 ura4-D18 | 23 |
| YN372 (FY12550) | h90alp4-gfp::Knrleu1-32 | Y. Nakase |
Accession numbers are from the Yeast Genetic Resource Center of Japan supported by the National BioResource Project (YGRC/NBRP; http:/yeast.lab.nig.ac.jp/nig/). The S. pombe strains constructed in this study have been deposited with the YGRC/NBRP under the accession numbers shown here.
x ≪ y means that gene y is integrated at gene x.
Plasmid construction.
The plasmids used in this study are listed in Table 2. The cam1+ gene was cloned into the pAL-KS vector as described below. The cam1+ gene was amplified by PCR using 5′-GCAGCCTCGAG(XhoI)AGAATAAACTATAAAATCTTTAAA-3′ and 5′-GAGCTAAATAATGGGCATGGGAATTGCAAG-3′ as forward and reverse primers, respectively. The PCR product was digested with XhoI and SpeI and then ligated into the corresponding sites of pAL-KS to yield pAL(cam1). The 2.4-kb ApaI-SacI fragment of the pAL(cam1) was inserted into pBR322 to yield pBR(cam1).
Table 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pAL-KS | ars1 LEU2-based vector | 31 |
| pBR(leu1) | leu1+ in pBR322 | 22 |
| pREP1 | ars1 LEU+-based vector carrying a thiamine-repressible nmt1 promoter | 16 |
| pIL2 | LEU2-based integration vector | 20 |
| pIA | ade6+-based integration vector | K. Tamai |
| pAL(cam1) | cam1+ in pAL-KS | This study |
| pAL(gfp-cam1) | gfp-cam1+ in pAL-KS | This study |
| pIA(gfp-cam1) | gfp-cam1+ in pIA1 | This study |
| pREP1(ade6) | ade6+-based expression vector | T. Nakamura |
| pREP1(ade6)gfp-cam1+ | gfp-cam1+ in pREP1(ade6) | This study |
| pREP1(ade6)gfp-cam1-22 | gfp-cam1-22 in pREP1(ade6) | This study |
| pREP1(ade6)gfp-cam1-22,117 | gfp-cam1-22,117 in pREP1(ade6) | This study |
| pREP1(ade6)gfp-cam1-EF2 | gfp-cam1-EF2 in pREP1(ade6) | This study |
| pREP1(ade6)gfp-cam1-EF3 | gfp-cam1-EF3 in pREP1(ade6) | This study |
| pBR322(spo13-gfp) | spo13+-gfp in pBR(leu1) | Y. Nakase |
| pBR322(spo2-gfp) | spo2+-gfp in pBR(leu1) | Y. Nakase |
| pREP1(spo2) | spo2+ in pREP1 | Y. Nakase |
| pREP1(spo13) | spo13+ in pREP1 | Y. Nakase |
| pREP1(spo15) | spo15+ in pREP1 | T. Nakamura |
| pIA(gfp-psy1) | gfp-psy1+ in pIA | 14 |
| pBR(gfp-cam1+) | gfp-cam1+ in pBR(leu1) | This study |
| pBR(cam1+) | cam1+ in pBR(leu1) | This study |
| pBR(cam1-22) | cam1-22 in pBR(leu1) | This study |
| pBR(cam1-22,117) | cam1-22,117 in pBR(leu1) | This study |
| pBR(cam1-EF1) | cam1-EF1 in pBR(leu1) | This study |
| pBR(cam1-EF2) | cam1-EF2 in pBR(leu1) | This study |
| pBR(cam1-EF3) | cam1-EF3 in pBR(leu1) | This study |
| pBR(cam1-EF4) | cam1-EF4 in pBR(leu1) | This study |
The gfp-cam1+ fusion gene was constructed and integrated at the cam1 locus (19). The gfp-cam1+ fusion gene was amplified by PCR using genomic DNA from the MPF7 strain and the PCR primers 5′-GCAGCCTCGAG(XhoI)AGAATAAACTATAAAATCTTTAAA-3′ (forward) and 5′-GAGCTAAATAATGGGCATGGGAATTGCAAG-3′ (reverse). A 3.0-kb ApaI-SacI fragment was inserted into pAL-KS to yield pAL(GFP-cam1). Plasmid pIA(GFP-cam1) was constructed by inserting the same fragment from pAL(GFP-cam1) into the pIA integration vector (K. Tamai, personal communication).
Western blotting.
The abundance of Spo15 in vegetative and meiotic cells was evaluated by Western blotting with anti-Spo15 antibody. The abundance of GFP-Cam1 in cells was analyzed by Western blotting using anti-GFP antibody. The pIA(GFP-cam1) plasmid carrying the cam1 promoter and a coding region for GFP-Cam1 was linearized by restricting it with BamHI near the center of the ade6+ sequence and then introduced into an ade6 strain (YN24 and YN104). The resulting gfp-cam1 integrant strains, AI248 and AI210, were cultured in MM-N sporulation medium. At intervals, portions of the culture were sampled, and crude cell extracts were prepared as described by Masai et al. (15). Polypeptides were resolved by SDS-polyacrylamide gel electrophoresis on 7.5% gels (for Spo15) or 10% gels (for GFP-Cam1) and then transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA). Membranes were probed with anti-Spo15 antibody (23) at a 1:100 dilution or with mouse anti-GFP antibody (Roche, Basel, Switzerland) at a 1:200 dilution. Blots were also probed with anti-α-tubulin antibody, TAT1 (32), to normalize protein loading. Immunoreactive bands were visualized by chemiluminescence (NEN Life Sciences, Boston, MA) using horseradish peroxidase-conjugated goat anti-mouse IgG (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Fluorescence microscopy.
The SPB was visualized by immunofluorescence microscopy. Cells were fixed with glutaraldehyde and paraformaldehyde (8). The SPB was visualized by using rabbit anti-Sad1 antibody (a gift from O. Niwa, Kazusa DNA Research Institute) and Alexa Fluor 546- or 488-conjugated secondary antibody (Molecular Probes, Eugene, OR). Sad1 is a major SPB component required for bipolar spindle formation (7). The nuclear chromatin region was stained with 4′,6′-diamidino-2-phenylindole (DAPI) at 1 μg/ml or Hoechst 33342 at 1 μg/ml (Nakarai Tesque, Kyoto, Japan). Stained cells were observed under a fluorescence microscope (model BX50; Olympus, Tokyo, Japan). To assess SPB modification quantitatively, stained cells were observed under a fluorescence microscope (model IX-71; Olympus), and SPB width was measured by AQUACOSMOS software (Hamamatsu Photonics, Shizuoka, Japan).
The FSM was observed by GFP-Psy1 fluorescence imaging (20). Psy1 is a plasma membrane-resident t-SNARE protein that is homologous to budding yeast Sso1 and Sso2 (1) and human syntaxin-1 (2). The gfp-psy1 fusion gene was integrated at the ade6 locus on chromosome III (14).
Electron microscopy.
Samples for electron microscopy were prepared as described previously (33), and sections were viewed on an electron microscope (H-7600; Hitachi, Tokyo, Japan) at 100 kV.
Mutants of Cam1 Ca2+-binding sites.
Cam1 contains four EF-hand Ca2+-binding sites (18). Each of the four EF-hand motifs was mutated by replacing a conserved glutamic acid residue with valine. The wild-type cam1+ gene cloned in an integration vector, pBR(leu1), was subjected to site-directed mutagenesis. A commercial in vitro mutagenesis kit was used (QuikChange; Stratagene, La Jolla, CA). The cam1 mutant allele in EF-hand binding site 1 was designated cam1-EF1. The other alleles were designated similarly. Each of the mutant genes (cam1-EF1, cam1-EF2, cam1-EF3, and cam1-EF4) was integrated at the leu1+ locus in a heterozygous cam1+/cam1::ura4+ diploid strain (AI509). The integrant strains were sporulated to isolate meiotic segregants carrying the cam1 point mutation in the cam1::ura4+ background. The resulting strains were AI510 (cam1+), AI520 (cam1-EF1), AI522 (cam1-EF2), AI524 (cam1-EF3), and AI526 (cam1-EF4) (Table 1). The cam1-22 and cam1-22,117 alleles were created by the same method (AI512 and AI518).
GFP-tagged cam1 mutant genes were constructed in an integration vector carrying leu1+ as a selectable marker. The gfp-cam1 fusion gene was expressed under the authentic cam1 promoter. Plasmid pBR(gfp-cam1+) was then subjected to site-directed mutagenesis as mentioned above (QuikChange; Stratagene, La Jolla, CA). In this way, modified pBR(leu1) plasmids containing either gfp-cam1-22,117, gfp-cam1-22, gfp-cam1-EF1, gfp-cam1-EF2, gfp-cam1-EF3, or gfp-cam1-EF4 were created. These plasmids were linearized by digestion with Eco81I and then used to transform the AI16 strain (h90 leu1-32 cam1+). The plasmids were integrated at the leu1 locus by homologous recombination to create strains AI2030 to AI2036 (Table 1).
In order to overproduce the Cam1 proteins, the gfp-tagged cam1 genes in plasmid pBR(gfp-cam1+) were recloned into expression vector pREP1(ade6) (Table 2). The constructed plasmids carrying the gfp-tagged cam1 missense mutant genes were subsequently introduced into the respective cam1 mutant strains (Table 1, AI3001 to AI3012).
Deposition and accession numbers of strains.
The S. pombe strains constructed in this study have been deposited with the YGRC/NBRP under accession numbers FY19908 to FY19951.
RESULTS
GFP-Cam1 localizes to the SPB during meiosis.
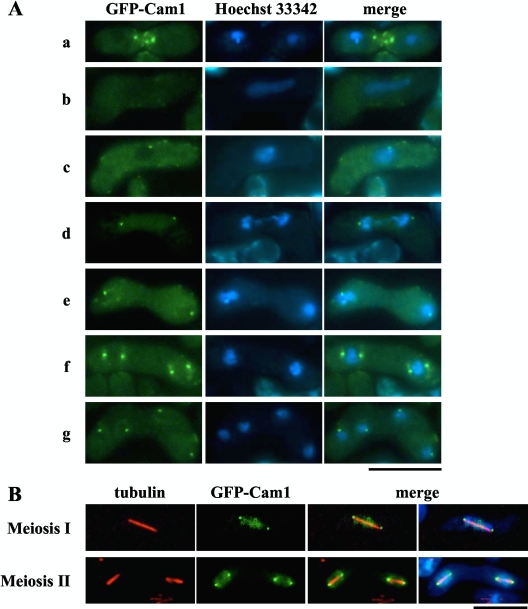
In vegetative cells, GFP-Cam1, which has previously been shown to be functional (19), localizes to both the polarized cell surface at growing tips and the SPB (19). Here, we analyzed localization of GFP-Cam1 during the sexual cycle (Fig. 1), induced by shifting haploid cells to a nitrogen-limiting sporulation medium. After induction, the GFP-Cam1 signal in the SPB and cell poles disappeared, while the fluorescent signal was subsequently detected as several bright patches at the site of cell fusion between cells of opposite mating types (Fig. 1A, row a). After nuclear fusion, meiosis ensues in a diploid zygote. In meiotic prophase I, the nucleus with an elongated shape moves actively back and forth (a so-called horsetail nucleus) (3). Here, the punctate GFP-Cam1 signals disappeared as cells entered the horsetail stage (Fig. 1A, row b). Intense signals appeared again as dots near the nuclear periphery soon after prophase I (Fig. 1A, row c). These dots duplicated in parallel with meiotic divisions (Fig. 1A, rows d to g), suggesting that Cam1 accumulates at the SPB as in vegetative cells. To test this idea, cells expressing GFP-Cam1 were fixed and immunostained with anti-α-tubulin antibody, TAT1 (32). As shown in Fig. 1B, GFP-Cam1 dots were positioned at both ends of the meiotic spindles. Localization of GFP-Cam1 to the SPB was also confirmed by double staining with anti-Sad1 antibody (data not shown). Sad1 is an SPB protein necessary for bipolar spindle formation (7).
Fig. 1.
Cellular localization of Cam1 during mating, meiosis, and sporulation. The homothallic haploid strain AI248 harboring a gfp-cam1 fusion gene was cultured on SSA medium to induce the sexual cycle. (A) Cells were stained with Hoechst 33342 to visualize nuclei. Developmental stages shown are as follows: row a, before karyogamy; row b, horsetail stage; row c, before meiosis I; row d, meiosis I; row e, interphase; row f, early meiosis II; row g, after meiosis II. Bar, 10 μm. (B) Cells were fixed during meiosis. Microtubules and nuclei were visualized by anti α-tubulin antibody, TAT1 (red) (32), and DAPI (blue), respectively. Bar, 10 μm.
cam1 missense mutants fail to form the FSM.
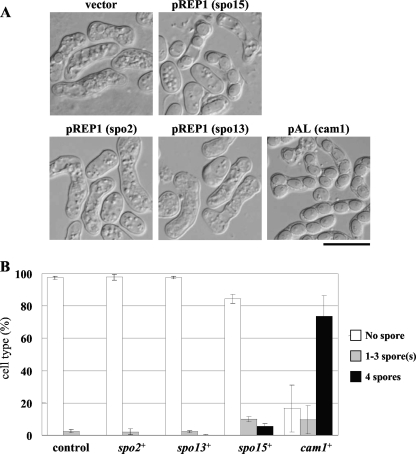
Because cam1+ is essential for growth, the cam1 disruptant is inviable. Thus, the role of Cam1 in sporulation was assessed by using viable missense mutants. The missense mutant allele cam1-22,117 harbors two mutations, D22Y (in which Asp22 is replaced by Tyr) and R117F (in which Arg117 is replaced by Phe). This haploid mutant exhibited no growth defects, but homozygous diploids failed to sporulate. The cam1-22,117 mutant was isolated spontaneously from descendants of the cam1-117 mutant generated by Takeda et al. (29) based on exhibiting a strict Spo− phenotype. The sporulation defect of the cam1-22,117 strain was suppressed by introducing plasmid pAL-KS harboring cam1+ (see Figure 7).
Fig. 7.
Overexpression of Spo15, Spo2, and Spo13 in the cam1-22,117 mutant. (A) The AI22 (cam1-22,117) strain was transformed with a pREP1 expression vector carrying spo15+ spo2+ or spo13+ and a multicopy plasmid carrying cam1+, pAL(cam1). Transformants were cultured on sporulation medium (SSA) for 2 days. Bar, 10 μm. (B) Sporulation was monitored by determining the percentage of asci, which were further classified according to the number of spores per ascus.
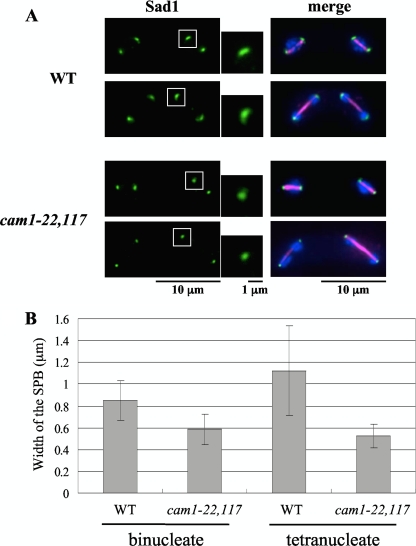
Because the processes of meiosis and spore formation in S. pombe are closely related, we first tested the kinetics of meiosis in the cam1-22,117 mutant. There was no significant difference between the wild-type and cam1-22,117 mutant cultures (data not shown). Sporulation itself proceeds through three steps: (i) structural change in the SPB (SPB modification), (ii) FSM assembly, and (iii) spore wall formation (27). Prior to spore formation, the SPB undergoes a characteristic alteration, as revealed by electron and fluorescence microscopy (7, 10). Our previous studies indicated that SPB modification was essential for initiation of FSM assembly (11, 23). To study defects in sporulation in detail, we first explored structural changes in the SPB during meiosis II, as revealed by immunofluorescence microscopy. The SPB in wild-type cells changes morphology from a compact dot to a crescent form, but such a structural change was not marked in the cam1-22,117 mutant (Fig. 2A). Poor development of the SPB in the cam1 mutant was confirmed quantitatively by measuring the width of the SPB (Fig. 2B).
Fig. 2.
Structural modification of the SPB during sporulation. The TN8 (h90 cam1+) and AI22 (h90 cam1-22,117) strains were cultured on MEA sporulation medium. (A) Immunostaining of the SPB with anti-Sad1 antibody (green). Magnified images of the boxed regions are also presented. Meiotic stages of the cells were examined by staining nuclei with DAPI (blue) and spindle microtubules with anti-α-tubulin antibody, TAT1 (magenta). (B) Comparison of the width of the SPB between wild-type and cam1-22,117 strains. Binucleate and tetranucleate cells were determined separately. The means ± standard deviations of approximately 100 SPBs are shown.
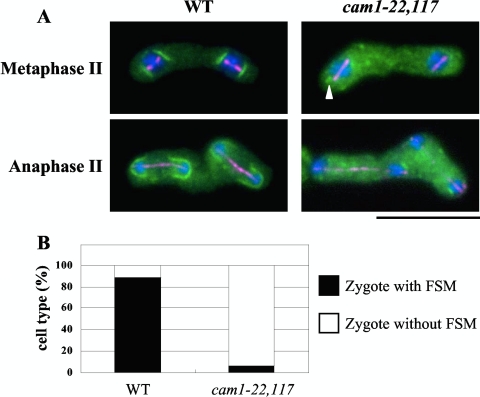
Next, we monitored FSM formation by using GFP-Psy1. Psy1 localizes to the plasma membrane of mother cells in meiosis I and then accumulates in the nascent FSM in meiosis II (14, 20). The FSM was observed as bright lines or arcs near both ends of the spindles of wild-type zygotes at early meiosis II and then extended as the spindles elongated (Fig. 3A). In the cam1-22,117 mutant, cup-shaped GFP-Psy1 signals were barely seen at the termini of short and long spindles. Instead, dots or very short lines of GFP-Psy1 were discernible near the nuclei in some cells. The percentage of zygotes forming any FSM-like structures in the cam1 mutant was only 5%, in contrast to 90% observed in the wild-type strain (Fig. 3B).
Fig. 3.
Initiation of FSM assembly in the cam1-22,117 mutant. (A) The nascent FSM was visualized by GFP-psy1 (green). A cam1+ strain (YM20) and a cam1-22,117 strain (AI52) were cultured on MEA sporulation medium and doubly stained with anti-α-tubulin antibody, TAT1 (magenta), and DAPI (blue). The arrowhead indicates a very short FSM-like structure. Bar, 10 μm. (B) Quantitative assay for zygote formation with an FSM. Only zygotes that assembled meiosis II spindles were counted.
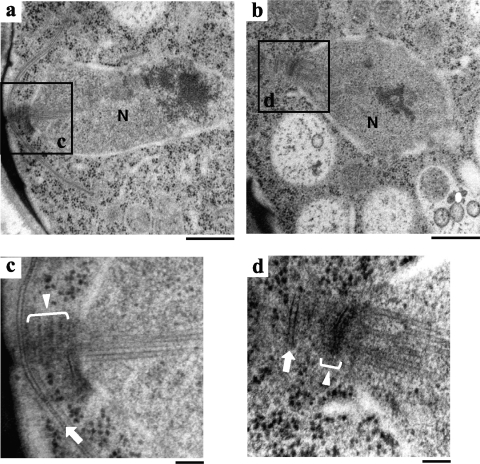
To examine SPB morphology and the FSM-like structures in cam1-22,117 zygotes, the fine structure was observed by electron microscopy. As shown in Fig. 4, the outer plaques of the SPB in the cam1-22,117 mutant were poorly developed. Furthermore, the FSM initiated in some cells, but even in such cells its expansion was severely inhibited. We conclude that FSM formation is blocked at an early stage in the cam1-22,117 mutant.
Fig. 4.
Fine structures of wild-type and cam1-22,117 strains during meiosis II. Cells were fixed after incubation on MEA sporulation medium for 1 day. Electron microscopic images of the wild-type (a and c) and cam1-22,117 (b and d) strains. Panels c and d are magnified images of the boxed regions in panels a and b, respectively. N, nucleus; white lines with triangles, the SPB; white arrows, the expanding FSM (c) and the abortive FSM (d). Bars, 500 nm (a and b) and 100 nm (c and d).
Spo2, Spo13, and Spo15 cannot localize to the SPB in the cam1-22,117 mutant.
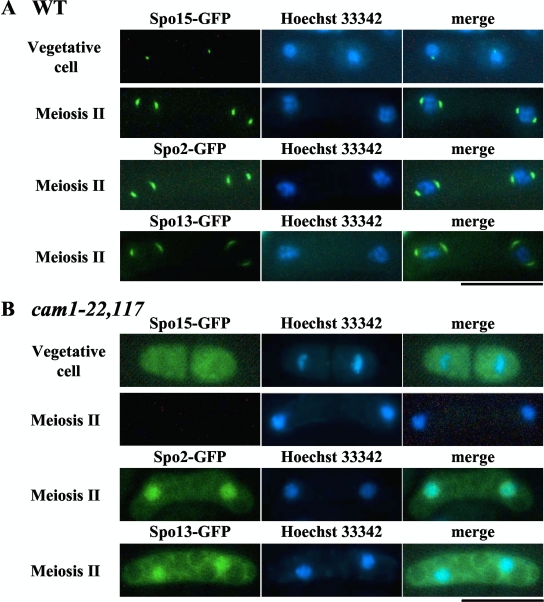
We reported earlier that the coiled-coil protein Spo15 is a constitutive SPB component required for sporulation but dispensable for vegetative growth (11). Two other sporulation-specific SPB components, Spo2 and Spo13, are recruited to the SPB in a Spo15-dependent manner during meiosis (23). Localization of Spo13 to the meiotic SPB relies on Spo2, which physically interacts with both Spo13 and Spo15. Proper recruitment of Spo13 is necessary for construction of the outer plaques on which the FSM is initially assembled. The above results show that Cam1 also localizes to the SPB during meiosis (Fig. 1) and is essential for SPB modification (Fig. 2). We therefore tested whether the sporulation-related SPB components, Spo2, Spo13, and Spo15, localize to the SPB in the cam1-22,117 mutant (Fig. 5).
Fig. 5.
Localization of Spo15, Spo2, and Spo13 in wild-type and cam1-22,117 strains. (A) Homothallic wild-type (cam1+) strains expressing Spo15-GFP (AI154), Spo2-GFP (AI259), or Spo13-GFP (AI258) were cultured on complete (YEA) and sporulation (SSA) media. Cells were stained with Hoechst 33342 (blue). Bar, 10 μm. (B) Similarly, homothallic cam1-22,117 strains, expressing Spo15-GFP (AI156), Spo2-GFP (AI262), or Spo13-GFP (AI263), were cultured on YEA and SSA media. Bar, 10 μm.
In vegetative cells expressing Spo15-GFP, the fluorescent signal localized to the SPB in a wild-type strain (Fig. 5A) as previously reported (11). Notably, the signal was not restricted to the SPB but was diffuse throughout the cytoplasm in vegetative cam1-22,117 cells (Fig. 5B), indicating that the SPB localization of Spo15 requires Cam1 function. Furthermore, the intensity of the Spo15-GFP fluorescence markedly decreased in meiotic cells (Fig. 5B). Spo2-GFP and Spo13-GFP signals did not localize to the SPB but were dispersed in the nucleus in the cam1-22,117 mutant. These observations are consistent with our previous finding that Spo2 and Spo13 in the spo15 deletion strain exhibited nuclear localization (23).
Because the cam1-22,117 mutation impaired localization of SPB components required only for spore formation, we next examined localization of other SPB components essential for growth, namely, Sad1, Cut12, Alp4, Sid4, and Pcp1, a conserved calmodulin target protein closely related to S. cerevisiae Spc110 (6). These proteins localized to the SPB both in the wild-type and the cam1-22,117 strains (see Fig. S1 in the supplemental material). As far as we tested, only sporulation-related SPB components were affected by the cam1 mutation.
We asked whether GFP-Cam1 could localize to the SPB in a spo15Δ mutant in sporulation medium. GFP-Cam1 was observed at the SPB as in wild-type cells (see Fig. S2 in the supplemental material). Nearly the same results were obtained with spo2Δ and spo13Δ (data not shown). These observations suggest that Cam1 localization to the SPB does not depend on Spo15, Spo2, or Spo13 but, rather, that Cam1 governs normal recruitment of these proteins to the SPB.
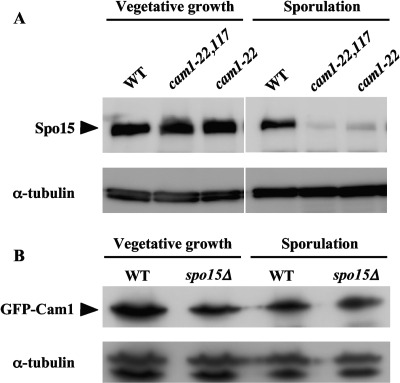
Spo15 is unstable in the cam1-22,117 mutant.
As shown in Fig. 5B, the Spo15-GFP signal was less intense in the cam1-22,117 mutant than in the wild-type strain during meiosis. To verify this observation, the level of Spo15 protein during vegetative growth and sporulation was assayed by Western blotting using an anti-Spo15 antibody. We also examined the effect of the single cam1-22 mutation. The AI510 (cam1+), AI 512 (cam1-22), and AI518 (cam1-22,117) strains were incubated in growth medium for 20 h and then transferred to and incubated in sporulation medium for 8 h. Spo15 protein levels in vegetative cells were roughly the same in these three strains (Fig. 6A). In sporulating cells, however, the cam1-22,117 and cam1-22 mutations severely reduced Spo15 protein levels (Fig. 6A). These results imply that Cam1 is needed to stabilize the Spo15 protein under sporulation conditions. Abundance of the Cam1 protein was then examined in the spo15Δ strain. GFP-Cam1 abundance was not affected by the spo15 deletion in sporulating cells (Fig. 6B).
Fig. 6.
Detection of Spo15 and Cam1 by Western blotting. (A) Western analysis of Spo15. Strains AI510 (cam1+), AI512 (cam1-22), and AI518 (cam1-22,117) were cultured in liquid synthetic medium, MM+N, for 20 h (vegetative growth) and liquid sporulation medium, MM-N, for 8 h (sporulation). Protein extracts were subjected to immunoblotting with anti-Spo15 antibody and with anti-α-tubulin antibody as a loading control. (B) Western analysis of Cam1. AI248 (cam1+) and AI210 (spo15Δ) strains expressing GFP-Cam1 were cultured in MM+N for 8 h and in MM-N for 6 h. Protein extracts were subjected to immunoblotting with anti-GFP antibody and with anti-α-tubulin antibody as a loading control. WT, wild type.
Overexpression of Spo15 partially alleviates the sporulation defect in the cam1-22,117 mutant.
The aforementioned results suggested that the sporulation defect in cam1-22,117 is due to a reduction in Spo15 abundance. We therefore tested whether overexpression of Spo15 might alleviate the sporulation defect in the cam1-22,117 mutant. As shown in Fig. 7, overexpression of Spo15 slightly but significantly enhanced ascus formation in the mutant, whereas overexpression of Spo13 or Spo2 did not. These results suggest that Cam1 is involved in sporulation, at least in part through maintenance of sufficient levels of Spo15.
Other mutations at Cam1 Ca2+-binding sites also affect sporulation.
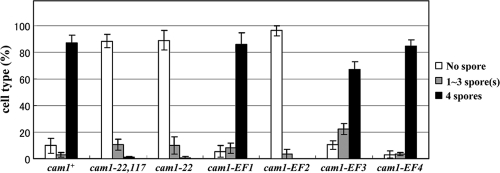
The cam1-22,117 mutant harbors two mutations at Asp22 and Arg117. Asp22 is one of the acidic residues in the EF-hand Ca2+-binding site 1. Because Cam1 has four EF-hand motifs, we tested whether Ca2+-binding site mutants exhibited a sporulation defect. The conserved Glu residue in each of the four EF-hand motifs was replaced with Val. The cam1-EF1, cam1-EF2, cam1-EF3, and cam1-EF4 mutants harbor E33V, E69V, E106V, and E142V substitutions, respectively (18).
We found that the cam1-22 mutant was unable to sporulate although another cam1-EF1 mutant (cam1-E33V) sporulated normally (Fig. 8) as did the cam1-EF4 mutant. In contrast, the cam1-EF2 mutant harboring a missense mutation at the second binding site exhibited a severe sporulation defect, while the cam1-EF3 mutant exhibited a moderate defect. These results suggest that the first, second, and third Ca2+-binding sites of calmodulin play important roles in spore formation.
Fig. 8.
Sporulation ability of the cam1 mutant harboring mutations in EF-hand motifs. The wild-type cam1+ and respective cam1 mutant genes were integrated at the leu1 locus on chromosome II of AI509. Strains used were AI510 (cam1+), AI512 (cam1-22), AI518 (cam1-22, 117), AI520 (cam1-EF1), AI522 (cam1-EF2), AI524 (cam1-EF3), and AI526 (cam1-EF4). Strains were cultured on MEA medium for 2 days, and their sporulation efficiencies were measured. The percentage of asci, classified according to the number of spores per ascus, is presented.
Stability of the mutant Cam1 protein under sporulation conditions.
Takeda et al. (29) reported that the Cam1-117 protein was unstable under nitrogen starvation conditions. It is probable that the defects in spore formation in different cam1 mutants observed here were due to decreased stability of Cam1 in sporulation cultures. To test this possibility, levels of the mutant Cam1 proteins that were tagged with GFP were analyzed by Western blotting. As shown in Fig. S3 in the supplemental material, the quantity of all the mutant proteins was roughly the same as wild-type Cam1 during incubation in sporulation medium. These results suggest that inability of cam1 mutants to complete spore formation is not due to reduced abundance of Cam1. To confirm this conclusion, we tested whether overexpression of the mutant cam1 gene would alleviate sporulation deficiency. As shown in Fig. S4 in the supplemental material, overexpression of mutant Cam1 did not affect sporulation efficiency. The lack of spore formation in these cam1 missense mutants was not due to lability of the mutated Cam1 proteins, except for Cam1-117 (29), but could be due to defective structural features.
DISCUSSION
S. pombe has a typical calmodulin, Cam1, and a calmodulin-like protein, Cam2. Cam1 is essential for proliferation although Cam2 is dispensable for growth (12, 13, 28). Cam2 is a regulatory light chain of type I myosin (13). We previously reported that Cam2 localizes to the leading edge of the FSM and plays a role in sporulation at supraoptimum temperatures (12). In the present study, we report the molecular details of an indispensable role for S. pombe calmodulin Cam1 in spore formation. To determine the biological roles of calmodulin in fission yeast, we undertook a phenotypic analysis of cam1 missense mutants focusing on sporulation processes. We made the following observations: (i) Cam1 preferentially localized to the SPB during meiosis and spore formation; (ii) SPB modification and proper FSM assembly required functional Cam1; (iii) a key SPB component, Spo15, which is necessary for sporulation, was mislocalized and unstable in cam1 mutants; (iv) in addition to Spo15, two other SPB proteins necessary for spore formation, Spo2 and Spo13, failed to localize to the SPB in the cam1 mutant. Taken together, these observations indicate that Cam1 modulates construction of the meiotic SPB that is a prerequisite for FSM assembly.
Cam1 is a constitutive component of the SPB and is necessary for bipolar spindle formation during mitosis (19). In S. pombe, another EF-hand-containing SPB protein is known, referred to as Cdc31, which is an ortholog of a conserved protein centrin (25, 26). Cdc31/centrin in yeasts is essential for SPB duplication (25). As shown in the present study, Cam1 is implicated in meiotic SPB modification required for FSM assembly. To our knowledge, there are no experimental data suggesting that S. pombe Cdc31 is also responsible for spore formation. The question of whether two EF-hand proteins, Cam1 and Cdc31, share a role in structural modification of the meiotic SPB to facilitate spore formation in S. pombe remains to be answered.
In vegetative cells, Cam1 interacts with Pcp1, a conserved calmodulin-binding SPB protein (6). Pcp1 overexpression failed to suppress the sporulation defect in the cam1 mutant (data not shown). It is possible that Cam1 might interact with an additional effector protein in both mitotic and meiotic cells. One candidate protein is Spo15. This protein has features similar to Pcp1 in that it is a high-molecular-weight, coiled-coil SPB protein. Spo15 does not localize to the SPB (Fig. 5) and is unstable in the cam1-22,117 mutant (Fig. 6). Overexpression of Spo15 in the cam1 mutant partially alleviated the sporulation defect. These observations suggest that Cam1 interacts directly or indirectly with Spo15. However, we could not demonstrate a physical interaction between Cam1 and Spo15 by immunoprecipitation or the yeast two-hybrid assay (data not shown). A direct sporulation-related target for Cam1 remains to be elucidated.
S. pombe calmodulin Cam1 has four conserved EF-hand Ca2+-binding sites (18). Conserved acidic residues in the EF-hand motif are important for Ca2+-binding activities. Replacing these acidic residues with Val disrupts Ca2+-binding capacity. A systematic mutagenesis experiment previously demonstrated that at least one of the four binding sites is needed for proliferation (18). Single disruptions of any four EF-hand motifs did not result in growth defects. In contrast, disruption of a specific EF-hand motif, e.g., EF2, caused a sporulation defect, suggesting that dependency of sporulation on Ca2+-binding activity of EF-hand sites might be more specific. Quantitative Ca2+-binding assay and phenotypic analysis indicated that binding site 2 is the most important for the essential mitotic function of Cam1 (18). Consistent with this result, our sporulation assay indicated that a single mutation in Ca2+-binding site 2 caused a sporulation defect.
The single mutation in Ca2+-binding site 1, cam1-22 (D22Y), also conferred a sporulation-deficient phenotype. Surprisingly, replacement of the conserved Glu33 residue in binding site 1 with Val (cam1-E1) did not result in a loss of sporulation. Thus, replacing the two acidic residues, Asp22 and Glu33, with Val has different phenotypic consequences which could reflect the Ca2+-free configurations of these mutant Cam1 proteins.
In summary, fission yeast calmodulin Cam1 plays an indispensable role in the SPB modification essential for FSM assembly. Cam1 stably maintains Spo15 at the SPB, which in turn recruits the other SPB component, Spo2, during meiosis. Finally, Spo13 accumulates at the outer layers of the meiotic SPB by physical interaction with Spo2. Spo13 plays a major role in the initial stage of FSM formation. Fission yeast calmodulin is implicated in this ordered recruitment of three sporulation-specific SPB proteins. To our knowledge, this study provides the first insight into calmodulin function in sexual development in yeast.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Yamamoto of the University of Tokyo for helpful suggestions and S. pombe strains, T. Davis of the University of Washington for strains and plasmids, O. Niwa of the Kazusa DNA Research Institute for affinity-purified antibodies against Sad1, and K. Okazaki of the Kazusa DNA Research Institute for strains. Some of the S. pombe strains and plasmids were provided by the Yeast Genetic Resource Center of Japan (YGRC/NBRP; http://yeast.lab.nig.ac.jp/nig/).
This study was partly supported by a grant from the Asahi Glass Foundation to T.N.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Aalto M. K., Ronne H., Keränen S. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12:4095–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett M. K., Calakos N., Scheller R. H. 1992. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 257:255–259 [DOI] [PubMed] [Google Scholar]
- 3.Chikashige Y., Ding D. Q., Funabiki H., Mashiko S., Yanagida M., Hiraoka Y. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264:270–273 [DOI] [PubMed] [Google Scholar]
- 4.Cyert M. S. 2001. Genetic analysis of calmodulin and its targets in Saccharomyces cerevisiae. Annu. Rev. Genet. 35:647–672 [DOI] [PubMed] [Google Scholar]
- 5.Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. 1986. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell 47:423–431 [DOI] [PubMed] [Google Scholar]
- 6.Flory M. R., Morphew M., Joseph J. D., Means A. R., Davis T. N. 2002. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13:47–58 [PubMed] [Google Scholar]
- 7.Hagan I., Yanagida M. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagan I. M., Hyams J. S. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343–357 [DOI] [PubMed] [Google Scholar]
- 9.Hirata A., Shimoda C. 1994. Structural modification of spindle pole bodies during meiosis II is essential for the normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast 10:173–183 [DOI] [PubMed] [Google Scholar]
- 10.Hirata A., Tanaka K. 1982. Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 28:263–274 [Google Scholar]
- 11.Ikemoto S., Nakamura T., Kubo M., Shimoda C. 2000. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J. Cell Sci. 113:545–554 [DOI] [PubMed] [Google Scholar]
- 12.Itadani A., Nakamura T., Shimoda C. 2006. Localization of type I myosin and F-actin to the leading edge region of the forespore membrane in Schizosaccharomyces pombe. Cell Struct. Funct. 31:181–195 [DOI] [PubMed] [Google Scholar]
- 13.Lord M., Pollard T. D. 2004. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J. Cell Biol. 167:315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda Y., Kashiwazaki J., Shimoda C., Nakamura T. 2009. The Schizosaccharomyces pombe syntaxin 1 homolog, Psy1, is essential in the development of the forespore membrane. Biosci. Biotechnol. Biochem. 73:339–345 [DOI] [PubMed] [Google Scholar]
- 15.Masai H., Miyake T., Arai K. 1995. hsk+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 14:3094–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maundrell K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127–130 [DOI] [PubMed] [Google Scholar]
- 17.Moreno S., Klar A., Nurse P. 1990. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 18.Moser M. J., Lee S. M., Klevit R. E., Davis T. N. 1995. Ca2+ binding to calmodulin and its role in Schizosaccharomyces pombe as revealed by mutagenesis and NMR spectroscopy. J. Biol. Chem. 270:20643–20652 [DOI] [PubMed] [Google Scholar]
- 19.Moser M. J., Flory M. R., Davis T. N. 1997. Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function in chromosome segregation. J. Cell Sci. 110:1805–1812 [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T., Nakamura-Kubo M., Hirata A., Shimoda C. 2001. The Schizosaccharomyces pombe spo3− gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell 12:3955–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura T., Asakawa H., Nakase Y., Kashiwazaki J., Hiraoka Y., Shimoda C. 2008. Live observation of forespore membrane formation in fission yeast. Mol. Biol. Cell 19:3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura-Kubo M., Nakamura T., Hirata A., Shimoda C. 2003. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell 14:1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakase Y., Nakamura-Kubo M., Ye Y., Hirata A., Shimoda C., Nakamura T. 2008. Meiotic spindle pole body bodies acquire the ability to assemble the spore plasma membrane by sequential recruitment of sporulation-specific components in fission yeast. Mol. Biol. Cell 19:2476–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohya Y., Botstein D. 1994. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science 263:963–966 [DOI] [PubMed] [Google Scholar]
- 25.Paoletti B., Bordes N., Haddad R., Schwartz C. L., Chang F., Bornens M. 2003. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell 14:2793–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice L. M., Agard D. A. 2002. Centriole duplication: Centrin in on answers? Curr. Biol. 12:R618–R619 [DOI] [PubMed] [Google Scholar]
- 27.Shimoda C. 2004. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 117:389–396 [DOI] [PubMed] [Google Scholar]
- 28.Shimoda C., Nakamura T. 2003. Control of late meiosis and ascospore formation, 311–327 InEgel R. (ed.), Molecular biology of Schizosaccharomyces pombe. Springer, Berlin, Germany [Google Scholar]
- 29.Takeda T., Imai Y., Yamamoto M. 1989. Substitution at position 116 of Schizosaccharomyces pombe calmodulin decreases its stability under nitrogen starvation and results in a sporulation-deficient phenotype. Proc. Natl. Acad. U. S. A. 86:9737–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda T., Yamamoto M. 1987. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. U. S. A. 84:3580–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka K., Yonekawa T., Kawasaki Y., Kai M., Furuya K., Iwasaki M., Murakami H., Yanagida M., Okayama H. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20:3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods A., Sherwin T., Sasse R., MacRae T., Baines A. J., Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- 33.Ye Y., Fujii M., Hirata A., Kawamukai M., Shimoda C., Nakamura T. 2007. Geranylgeranyl diphosphate synthase in fission yeast is a heteromer of farnesyl diphosphate synthase (FPS), Fps1 and an FPS-like protein, Spo9, essential for sporulation. Mol. Biol. Cell 18:3568–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo B. Y., Calleja G. B., Johnson B. F. 1973. Ultrastructural changes of the fission yeast (Schizosaccharomyces pombe) during ascospore formation. Arch. Mikrobiol. 91:1–10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.