Abstract
Meiotic genes in budding yeast are repressed during vegetative growth but are transiently induced during specific stages of meiosis. Sin3p represses the early meiotic gene (EMG) by bridging the DNA binding protein Ume6p to the histone deacetylase Rpd3p. Sin3p contains four paired amphipathic helix (PAH) domains, one of which (PAH3) is required for repressing several genes expressed during mitotic cell division. This report examines the roles of the PAH domains in mediating EMG repression during mitotic cell division and following meiotic induction. PAH2 and PAH3 are required for mitotic EMG repression, while electrophoretic mobility shift assays indicate that only PAH2 is required for stable Ume6p-promoter interaction. Unlike mitotic repression, reestablishing EMG repression following transient meiotic induction requires PAH3 and PAH4. In addition, the role of Sin3p in reestablishing repression is expanded to include additional loci that it does not control during vegetative growth. These findings indicate that mitotic and postinduction EMG repressions are mediated by two separate systems that utilize different Sin3p domains.
Meiosis is the process that produces haploid gametes from diploid parental cells. Similar to other developmental pathways, many genes required for meiosis and spore formation in the budding yeast Saccharomyces cerevisiae display a transient transcription profile (7, 25). During vegetative growth, their mRNA levels are low but increase dramatically at precise stages in meiosis. This expression is usually followed by an equally rapid repression that returns the mRNA to mitotic levels.
The vegetative repression of a group of genes designated “early meiotic genes” (EMG) requires the recruitment of the histone deacetylase (HDAC) Rpd3p (13) and the chromatin-remodeling factor Isw2p (10) by the Ume6p DNA binding protein (31). Ume6p binds an element termed upstream repressor sequence 1 (URS1) that is responsible for the full repression and activation of several early meiotic genes (3, 5, 37). The Ume6p-Rpd3p association occurs through the global corepressor Sin3p (13). Similarly, the last known member of this repression complex, Ume1p (30), associates with Rpd3p in an Sin3p-dependent manner (17). The function of Ume1p in this complex is currently unknown, but it is suggested to be a tightly associated cofactor (41).
The interactions between the URS1 regulatory element and its associating factors are complex. For example, URS1 is also required for the repression of several vegetative genes (22, 27, 32, 38). Of these loci, only CAR1 is repressed by Ume6p (23), while Sin3p alone regulates HO (6). Given the diverse loci regulated by URS1, it is likely that specificity is introduced through the interaction of additional factors targeted to the various promoters. Indeed, Abf1p helps stimulate transcription of the URS1-regulated meiotic gene HOP1 (37). Similarly, an element termed the auxiliary repression element (ARE) has been identified genetically that contributes to vegetative repression of the meiosis-specific heat shock gene HSP82 (34). Therefore, the context in which URS1 is found may allow this single element to respond to different stimuli and function in a positive or negative manner.
Sin3p belongs to a conserved gene family that contains four paired amphipathic helix (PAH) protein-protein interaction domains (see reference 29 for a review). Mutational analysis in yeast revealed that of the four PAH domains, PAH3 is required for the repression of several genes, including HO, PHO5, and IME2 (43). Functionally, PAH3 helps recruit the HDAC complex and other corepressors, while PAH2 mediates the interaction with Ume6p in yeast (44). The roles of the PAH domains in transcriptional repression appear conserved in the human Sin3 (hSin3). For example, hSin3p also associates with the histone deacetylase HDAC1 (15), while PAH2 binds transcription factors (2, 28). Less is known about the other two PAH domains. PAH1 recruits a variety of corepressors, depending on the gene context (29), while PAH4 is reported to bind the enzyme O-acetylglucosamine transferase (OGT) to help repress transcription in higher eukaryotes (45). These results indicate that the PAH domains perform separate, but complementary, roles in mediating transcriptional repression.
Although it regulates diverse gene sets, mutants lacking SIN3 do not display significant growth defects (39, 42). However, Sin3p is required for the execution of the first meiotic nuclear division (30), with mutants arresting in meiotic prophase I (11). This present study explores the role of Sin3p in controlling meiotic gene expression. We find evidence for two separate Sin3p-dependent regulatory systems, one repressing EMG transcription during mitotic cell division, and the other functioning to reestablish repression as the cells complete meiosis.
MATERIALS AND METHODS
Strains, media, and plasmids.
The strains used in this study are listed in Table 1. The PAH deletion strains were constructed by first subcloning the different SIN3 PAH mutant alleles (a gift from D. Stillman, University of Utah) into the integrating vector YIplac22 (9). These constructs were used to replace the wild-type SIN3 gene using the pop-in/pop-out strategy (26). The successful introduction of all mutant alleles was verified by sequencing genomic PCR fragments. All growth and sporulation procedures have been described previously (8). Mutant derivatives of URS1 were introduced into the spo13-lacZ reporter plasmid p(spo13)40 (5) by site-directed mutagenesis, and lacZ activity was assayed as described previously (5). Single-stranded oligonucleotides (see Fig. 3E) were used to mutate a 215-bp EcoRI-BstEII fragment (−170 to +45) of SPO13 in vector pVZ1 (Bio-Rad), and this fragment was cloned into the EcoRI-BstEII fragment in p(spo13)40.
Table 1.
Strains and genotypes
| Straina | Genotype |
|---|---|
| RSY10 | MATα ade2 ade6 can1-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 |
| RSY301 | MATα ade2 ade6 can1-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 ume6::LEU2 |
| RSY305 | MATα ade2 ade6 can1-100 leu2-3,112 his3-11,15 trp1-1 ura3-1 sin3::HIS3 |
| RSY272 | MATα can1-100 his4 leu2-3,112 trp1-1 ura3-1 ume6::LEU2 |
| RSY273 | MATα can1-100 his4 leu2-3,112 lys2 trp1-1 ura3-1 |
| RSY274 | MATα can1-100 his3-11,15 leu2-3,112 lys2 trp1-1 ura3-1 sin3::TRP1 ume6::LEU2 |
| RSY275 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sin3::TRP1 |
| RSY276 | MATacyh2rleu1-c met-13B tyr1-2 |
| RSY403 | MATacyh2rleu1-c met-13B tyr1-2 sin3::HIS3 |
| RSY404 | MATα can1-100 leu1-12 lys2 tyr1-1 sin3::HIS3 |
| RSY427 | MATacyh2rleu1-C met13-B tyr1-2 sin3-pahΔ1 |
| RSY428 | MATacyh2rleu1-C met13-B tyr1-2 sin3-pahΔ2 |
| RSY429 | MATacyh2rleu1-C met13-B tyr1-2 sin3-pahΔ3 |
| RSY430 | MATacyh2rleu1-C met13-B tyr1-2 sin3-pahΔ4 |
| RSY877* | MATa/MATα lys2 trp1::hisG ura3 LYS2::hoΔ |
| RSY1028* | MATa/MATα lys2 trp1::hisG ura3 LYS2::hoΔ sin3::URA3 |
| RSY1029* | MATa/MATα lys2 trp1::hisG ura3 LYS2::hoΔ sin3-pahΔ1 |
| RSY1030* | MATa/MATα lys2 trp1::hisG ura3 LYS2::hoΔ sin3-pahΔ2 |
| RSY1031* | MATa/MATα lys2 trp1::hisG ura3 LYS2::hoΔ sin3-pahΔ3 |
| RSY1032* | MATa/MATα lys2 trp1::hisG ura3 LYS2::hoΔ sin3-pahΔ4 |
All strains were generated in this study except RSY10 (27). An asterisk indicates diploid strains that are homozygous for all alleles shown. Only one is shown for clarity.
Fig. 3.
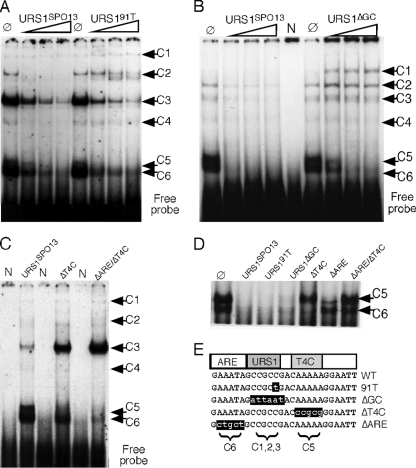
Multiple protein binding elements exist in URS1SPO13. (A) Unlabeled wild-type (URS1SPO13) or mutant (URS191T) oligonucleotides were added (50-fold, 150-fold, and 300-fold excess) to standard EMSA with wild-type extracts prepared from mid-log-phase cells. The six complexes are indicated by black arrows. Ø, no competitor added. (B) Competition EMSA as described for panel A with the unlabeled URS1ΔGC oligonucleotide added. N, no extract added; Ø, no competitor added. (C) 32P-labeled wild-type (URS1SPO13) and two mutant URS1 oligonucleotide probes (URS1ΔT4C, URS1ΔT4C/ΔARE) were incubated with wild-type mid-log-phase extracts in standard EMSA reactions. N, no extract added. The six specific complexes observed under wild-type conditions are indicated by black arrows. (D) EMSA conducted with radiolabeled URS1SPO13 and the indicated unlabeled competitors. Complexes 5 and 6 are indicated. Ø, no competitor added. (E) The DNA sequences of wild-type URS1SPO13 and mutant derivatives are indicated. Proposed locations of the repressor ARE and activator T4C domains are indicated. The complexes corresponding to each domain are indicated below the sequences. The ΔT4C and ΔARE double-mutant oligonucleotide represented in panel C is not shown for clarity.
Meiotic progression/recombination assays.
Synchronous meiotic cultures were generated and analyzed as described previously (8). The recombination assays were performed as follows. Haploid strains containing one of the PAH deletion derivatives (RSY427 to RSY430; see Table 1) were mated to RSY404 (sin3Δ). Eight individual pahΔ/sin3Δ isolates, along with SIN3/sin3Δ (RSY276 × RSY404) and sin3Δ/sin3Δ (RSY404 × RSY404) controls, were induced to enter meiosis by using standard protocols. Samples were taken prior to the shift and 12 h after. Initial experiments identified 12 h following the transfer to sporulation medium (SPM) as the time point that recombination was complete (data not shown). The cells were lightly sonicated, serially diluted (1:10), and then plated on solid rich medium to determine total viable cell numbers and minimal medium lacking leucine to monitor intergenic exchange at the leu1 locus. The samples taken prior to transfer to SPM were used to exclude any isolates that had a high level of Leu+ prototrophs prior to meiotic induction due to mitotic recombination. The plates were incubated for 3 days at 30°C, and the number of colonies was determined. The unpaired Student t test was used to determine the statistical significance of these results.
Northern/S1 nuclease protection/qPCR analyses.
Northern blot analyses were performed as described previously (18) with 25 μg of total RNA. Probes were obtained by PCR amplification of the gene in question and labeled with [32P]dCTP using the PrimeIt random priming kit (Stratagene Inc.). S1 nuclease protection studies were conducted as described earlier (30) using 20 μg of total RNA and [32P]UTP continuously labeled strand-specific riboprobes. Northern blot signals (see Fig. 5) were quantitated by phosphorimaging (Kodak Inc.), and the values presented were first corrected for loading differences using ENO1 levels as internal controls. The corrected signal from each time point was plotted as a percentage of the peak accumulation value within each time course. The values plotted represent the averages of results from two separate experiments. The standard deviation within the different trials was 18% or less. Quantitative PCR (qPCR) analyses were conducted using total RNA isolated from 10 ml of sporulation culture as described previously (30). Precipitated RNA samples were treated with DNase I (New England Biolabs [NEB]), and reverse transcription was performed using oligo(dT) priming and avian myeloblastosis virus (AMV) reverse transcriptase (NEB). TaqMan reactions were conducted using an Applied Biosystems StepOne thermal cycler with 2× TaqMan gene expression master mix (Applied Biosystems) using primers described in Table 2. Relative mRNA levels were calculated by comparative threshold cycle (CT) methodology using ENO1 as an endogenous control. Values shown are means with standard deviations from technical triplicates.
Fig. 5.
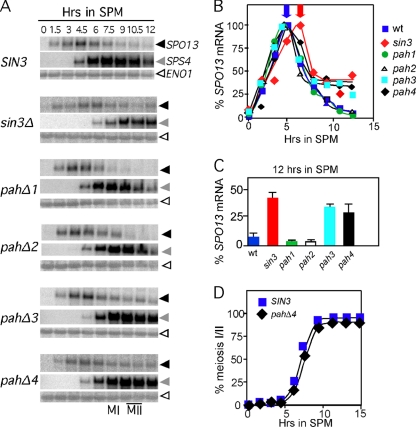
PAH3 and PAH4 are required to reestablish SPO13 repression in meiotic cells. Isogenic wild-type (RSY877), sin3Δ (RSY1028), pahΔ1 (RSY1029), pahΔ2 (RSY1030), pahΔ3 (RSY1031), or pahΔ4 (RSY1032) strains were induced to enter meiosis, and time points were taken as indicated. Northern blots of total RNA were probed with SPO13 (black arrowheads) and then stripped and probed sequentially with probes directed toward SPS4 (gray arrowheads) and ENO1 (white arrowheads). (B) Quantitation of SPO13 signals in panel A following standardization with ENO1. The time point exhibiting the maximum signal from each time course was set at 100%. (C) The percentage of maximum induction of the 12-h signal from panel B is shown for the SIN3 alleles indicated. (D) Meiotic progression in the wild type (RSY877) and pahΔ4 mutant (RSY1032) was determined by monitoring nuclear divisions using DAPI analysis. The percentage of each population executing at least one nuclear division is shown.
Table 2.
Quantitative PCR primers
| Gene | Forward primer | Reverse primer | Probea |
|---|---|---|---|
| ENO1 | GCCGCTGCTGAAAAGAATGT | TGGAGAGGTCTTAGACAA | (FAM)CATTATACAAGCACTTGGC |
| SPO13 | AAGCCCACATCCAGGATTAAATT | CGAACATCTCCAGCCTTTGAG | (FAM)TGTAAGTGATGCTCAACAGT |
| SPS4 | GCACAAACAAAGCCTAGAATCGA | CACTGGTGCTACGGCTTGAA | (FAM)AAGTGGTAGTGTCACGTCC |
| IME1 | TCCCCTAGAAGTTGGCATTTTG | CCAAGTTCTGCAGCTGAGATGA | (VIC)AAGTCTGACAAAATTG |
FAM, 6-carboxyfluorescein.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays (EMSAs) were conducted and extracts prepared as described previously (1) from cells harvested in mid-logarithmic growth. A 26-bp oligonucleotide containing the SPO13 URS1 sequence (GAAATAGCCGCCGACAAAAAGGAATT; designated URS1SPO13) or derivatives as indicated in the text was end-labeled with [γ32P]ATP using polynucleotide kinase and then hybridized to a 3-fold molar excess of an unlabeled complement to drive the labeled probe into a duplex state. The probe was separated from unincorporated nucleotides by either column chromatography or polyacrylamide gel purification (18). Reaction mixtures containing 10 to 20 μg of crude extract, 20,000 dpm of probe (approximately 0.1 ng), and nonspecific competitors [2.5 μg poly(dI-dC) and 1.0 μg poly(dA-dT)] were incubated at 16°C for 20 min and then loaded directly onto a 6% nondenaturing polyacrylamide gel and electrophoresed at 10 V/cm for 2.5 h at 25°C. The gels were dried and exposed to X-ray film with intensifying screens. Competition assays were conducted as described above except that 33-, 100-, and 300-fold excess duplex oligonucleotides were added prior to the introduction of the extract. The reaction mixtures were vortexed and equilibrated at 16°C for 10 min, and then the extract was added. Complex quantitation was conducted using phosphorimaging (Kodak).
Chromatin immunoprecipitation (ChIP) analysis.
Chromatin solutions were prepared essentially as previously described (19) with the following modifications. A 50-ml dextrose culture was grown to a final density of ∼2 × 107 cells/ml and cross-linked for 15 min using 1% formaldehyde with occasional swirling. Cross-linking reactions were quenched with 50 mM glycine for 5 min, harvested, and washed twice with ice-cold 1× phosphate-buffered saline (PBS). Immunoprecipitations of spheroplasted, sonicated material were conducted using 250 μg of total chromatin (25 μg for inputs), diluted in immunoprecipitation (IP) dilution buffer. The chromatin was first precleared with a protein A/G slurry for 1 h at 4o. Following preclearing, 5 μl of polyclonal α-Ume6p (Open Biosystems) and the no-antibody (Ab) control were immunoprecipitated overnight. After harvesting the immunoprecipitations with protein A/G slurry for 1 h, beads were washed (4) and eluted (19) as described. Cross-linking was reversed, and precipitated DNA was then treated with proteinase K, organically extracted, and used in qPCRs. qPCR primers (forward, GCT AGT TAG TAC CTT TGC ACG GAA A; reverse, TCT TAT TGC GCT AAT TGT CTG TTA GAC) were directed at the SPO13 promoter using SYBR green reactions (Applied Biosystems Power SYBR master mix). Data collected were first normalized to input signal. These values (percent input) were then compared to enrichment over a no-Ab control. Wild-type enrichment was set to 100%. Error bars in the figures represent the means from biological triplicates, and error bars are the standard errors of the means.
RESULTS
PAH2 and PAH3 are required for mitotic SPO13 repression.
Previous studies indicated that Sin3p-dependent repression of the IME2 EMG predominantly utilizes PAH3 but also PAH2 to a lesser extent (43). To determine if a similar pattern of activity was observed for another EMG, SPO13 mRNA levels were examined in vegetative cultures expressing the different PAH deletion alleles. A strain deleted for SIN3 was transformed with plasmids harboring the wild-type gene, internal deletions of each individual PAH domain, or the vector alone. The protein levels of these deletion derivatives are similar to the wild-type Sin3p (43). As observed previously (30), the loss of Sin3p activity derepressed SPO13 transcripts about 10-fold during mitotic cell division (Fig. 1A, compare lanes 2 and 3). The derepression observed with sin3 mutants is still far below that observed when UME6 is deleted (lane 8), indicating that Sin3p mediates only part of the vegetative repression of early meiotic genes. The analysis of the individual PAH deletion mutants revealed that PAH2 and to a lesser extent PAH3 are required for SPO13 repression (lanes 5 and 6). No aberrant SPO13 derepression was observed in strains expressing the pahΔ1 or pahΔ4 mutants. These results indicate that PAH2 and to a lesser extent PAH3 mediate SPO13 repression in vegetative cells.
Fig. 1.
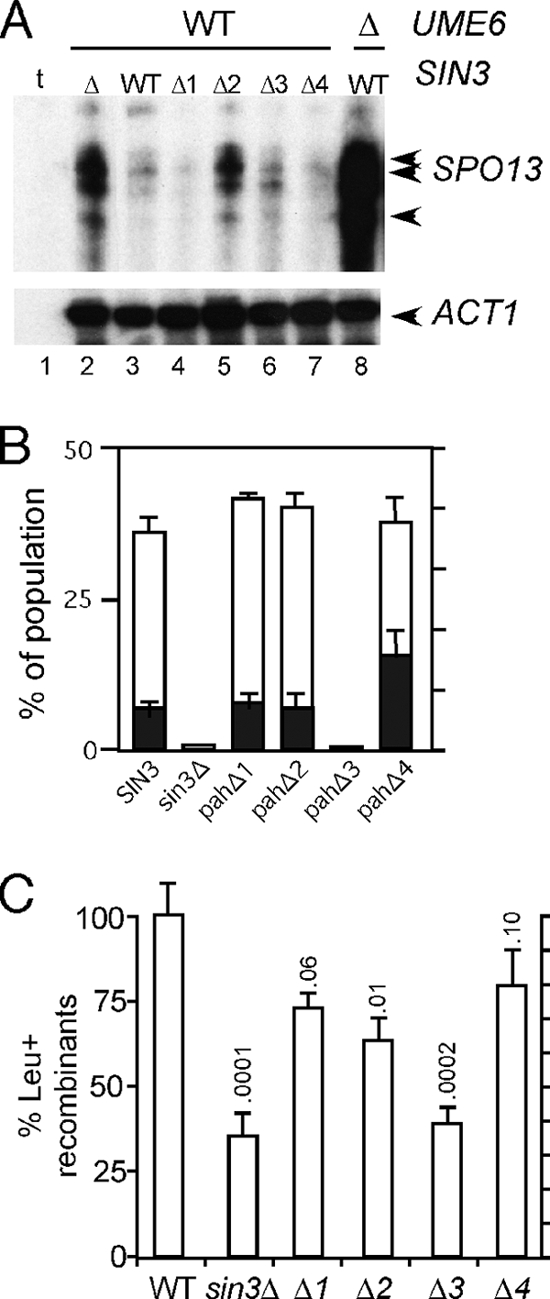
PAH regulation during meiotic and mitotic cell division. (A) S1 nuclease protection assays were performed on total RNA prepared from RSY278 transformed with plasmids expressing the indicated SIN3 alleles. The SPO13-specific bands corresponding to the 3′ end of the mRNA are indicated by the arrows. ACT1 mRNA levels serve as a control for the amount of poly(A)+ RNA in each sample. t, tRNA control for nonspecific self-annealing of the single-stranded riboprobe. (B) PAH deletion derivatives (RS427 to RS430) mated to RSY404 (sin3Δ) were induced to enter meiosis in liquid culture for 24 h. The cells were fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI), and the percentages of binucleated (black columns) and tetranucleated (white columns) cells were determined. The values presented represent the averages of results from two independent trials for each strain. (C) The percentage of Leu+ prototrophs were established for independent isolates for each strain (n = 8 isolates, except pahΔ3 n = 7 isolates) with wild-type levels set at 100%. The standard deviations from the means are shown with corresponding P values.
PAH3 is required for meiotic nuclear divisions.
Previous studies found that mutants lacking Sin3p failed to progress past the first meiotic division (11, 30). Another study found that the pahΔ3 mutant exhibited a 33-fold reduction in sporulation efficiency when assaying the haploidization of recessive drug resistance markers (43). To further explore the role of the PAH domains in meiotic progression, we first monitored meiotic nuclear divisions in diploid strains heterozygous for the various deletion mutations and the null allele (pahΔ/sin3Δ). These diploids were sporulated in liquid culture, and the percentages of cells undergoing one (binucleated) or both (tetranucleated) meiotic divisions were determined by DAPI [4′,6-diamidino-2-phenylindole] analysis. Similar to the sin3Δ/sin3Δ-null strain, the pahΔ3/sin3Δ mutant failed to produce bi- or tetranucleated cells (Fig. 1B; n = 200). Strains lacking either of the other PAH domains executed the meiotic divisions at frequencies similar to that of the wild-type control. The only exception was an increase in the number of binucleated cells in the pahΔ4 mutant (33%) compared to that in the wild type (11%). In addition, the kinetics of bi- and tetranucleate production in the pahΔ1/sin3Δ, pahΔ2/sin3Δ, and pahΔ4/sin3Δ mutants were similar to the kinetics of the SIN3/sin3Δ control diploid (data not shown). These results indicate that PAH3 is required for the execution of meiotic nuclear divisions. The remaining PAH domains are largely dispensable for meiosis and spore formation.
PAH2 and PAH3 are required for normal meiotic recombination.
The PAH mutant diploids just described also contained heteroalleles at the leu1 locus (see Table 1). To test the requirements of the PAH domains for recombination, each diploid was examined for the production of Leu+ prototrophs (see Materials and Methods for details). With wild-type levels set at 100%, these studies found that pahΔ1 and pahΔ4 mutants produced Leu+ prototrophs near wild-type levels (Fig. 1C). However, compared to the wild type, the sin3-null strain and pahΔ2 and pahΔ3 mutants displayed a reduction in the number of recombinants (P < 0.01). This defect does not appear to be the result of delayed recombination kinetics, as the production of prototrophs reached a maximum between 12 and 24 h in all strains tested (data not shown). These experiments revealed that PAH2 and PAH3 are required for the normal execution of meiotic recombination. As meiotic DNA synthesis appears normal in an sin3Δ mutant (reference 11 and our unpublished results), these findings point to a requirement of PAH2 and PAH3 during prophase I of the meiotic program.
PAH2 is required for Ume6p-URS1 promoter complex formation in vitro.
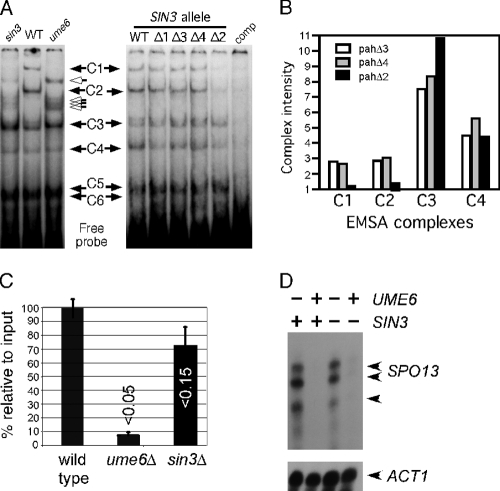
We previously demonstrated that six complexes (C1 to C6) are observed in electrophoretic mobility shift assays (EMSAs) when crude vegetative extracts are incubated with an oligonucleotide probe containing the SPO13 URS1 element (URS1SPO13) (31). This study also demonstrated that two of these complexes, C1 and C2, were absent in extracts prepared from ume6 mutant cultures (see Fig. 2A, left). In addition, new species were detected in the ume6 extract that were not present in the wild type (Fig. 2A, open arrows). To determine if Sin3p is required for normal complex formation in vitro, EMSAs were performed with an extract derived from an sin3-null strain. Interestingly, sin3 mutant extracts produced an EMSA pattern similar to that observed with ume6 strains. In addition to the loss of C1 and C2 in the sin3 mutant, the new complexes observed in the ume6 mutant are also present in extracts lacking sin3 (open arrows).
Fig. 2.
Sin3p is required for Ume6p-dependent complex formation. (A) Left, extracts prepared from RSY10 (WT), RSY305 (sin3Δ), and RSY301 (ume6Δ) were incubated with the 32P-labeled SPO13 URS1 promoter element (URS1SPO13; see Materials and Methods for details). Six protein-DNA complexes (C1 to C6) are indicated by the black arrows. The open arrows indicate new complexes that are observed in the sin3 and ume6 extracts. The C5 and C6 complexes do not resolve under these gel conditions (see Fig. 3). Right, the experiments were repeated with extracts prepared from mid-log-phase RSY278 (sin3Δ) harboring plasmids containing the wild-type SIN3 gene or the different PAH mutant derivatives as indicated. comp, 150-fold excess of unlabeled competitor URS1SPO13 double-stranded oligonucleotide added to the reaction. (B) Complex intensities of C1, C2, C3, and C4 were quantitated from pahΔ2, pahΔ3, and pahΔ4 extracts as indicated. Complex intensities are given in arbitrary units. (C) Chromatin immunoprecipitations were conducted with Ume6p antibodies in extracts prepared from three independent wild-type (RSY10), ume6Δ (RSY291), or sin3Δ (RSY579) cultures. Enrichment over no antibody control was established for each sample with the wild type set to 100%. The data are presented as the means ± standard deviations (SD) of results from three independent experiments (P values are indicated; t test). (D) S1 nuclease protection assays were conducted with total RNA prepared from wild-type (RSY273), ume6Δ (RSY272), sin3Δ (RSY275), and sin3Δ ume6Δ (RSY274) vegetative cultures. SPO13-specific bands are indicated by the arrows. ACT1 mRNA levels were used to control for the poly(A)+ percentages in the total RNA preparations.
To determine if any of the PAH domains are required for C1 and C2 formation, these experiments were repeated in an sin3-null strain harboring plasmids expressing wild-type SIN3 or the different PAH deletion derivatives. Of the PAH mutants, only pahΔ2 demonstrated an alteration in the EMSA pattern (Fig. 2A, right). In this extract, levels of C1 and C2 are reduced compared to those of the other pah mutant extracts, while C3 to C6 are unaffected (quantitated in Fig. 2B). Of note is that the new complexes observed in either ume6- or sin3-null extracts (open arrows) are not observed in the pahΔ2 mutant strain. The requirement of PAH2 for C1 and C2 formation is consistent with our finding that the pahΔ2 mutant allows SPO13 derepression in vegetative cultures (Fig. 1A). To determine whether a similar loss in Ume6p-URS1SPO13 association is observed in vivo, chromatin immunoprecipitation (ChIP) studies were performed. Wild-type, ume6Δ, and sin3Δ log-phase cultures were harvested and cross-linked samples immunoprecipitated with Ume6p-specific antibodies (see Materials and Methods for details). Quantitative PCR was utilized to calculate the relative occupancy of Ume6p at URS1SPO13. These studies found that Ume6p occupancy was reduced but not strongly (P < 0.15) compared to that of the wild-type control in this assay (Fig. 2C). The differences in these results compared to those of the EMSA studies may reflect the in vivo versus in vitro environment. Ume6p may be more stably associated with URS1 in its natural chromatin context than when binding a 26-bp probe. Conversely, the EMSA conditions may amplify subtle differences in protein-DNA interactions.
Sin3p is not required for SPO13 mRNA derepression in a ume6 mutant.
A previous study found that Sin3p plays a positive and negative role in gene transcription (40). For example, Sin3p represses the acid phosphatase PHO5 in a high-phosphate environment but is also necessary for maximal expression under limiting phosphate conditions. To determine whether Sin3p is required for SPO13 mRNA derepression associated with the ume6 mutation, isogenic strains were constructed containing the sin3Δ or ume6Δ alleles individually or in combination. Mid-log-phase SPO13 mRNA levels were determined in each strain background by S1 nuclease protection analysis as described above. This experiment revealed similar SPO13 mRNA levels in an ume6 mutant regardless of SIN3 status (Fig. 2D). In addition, a similar analysis conducted with the individual pahΔ mutants revealed identical results (data not shown). Taken together, these results indicate that Sin3p is not required for SPO13 derepression in the absence of Ume6p.
Three distinct protein binding domains exist at URS1SPO13.
The results presented in the previous section indicate that C1 and C2 represent Ume6p-URS1SPO13 interactions in a repressed configuration, while their loss correlates with SPO13 derepression. However, the complicated pattern of six complexes observed for the relatively small probe (26 bp) prompted further investigation of this promoter element. A previous study identified a single nucleotide substitution (C91T) that disrupted the URS1 core consensus element and allowed mitotic derepression of SPO13 (5). An oligonucleotide was synthesized (URS191T) (Fig. 3 E) to include this single base change and used as an unlabeled competitor in EMSAs employing the wild-type probe. The URS191T oligonucleotide was able to compete for C3 to C6 binding as effectively as the wild-type control (Fig. 3A). However, complex C2 was not as effectively competed with URS191T. In these experiments, C1 was not effectively competed by either the wild-type or 91T probe. These results suggest that the derepression observed with the C91T mutation is due to reduced Ume6p binding to URS1. Next, the entire URS1 GC core consensus element was mutated (URS1ΔGC) (Fig. 3E) and used as a competitor in EMSAs using URS1SPO13 and wild-type vegetative extracts. These experiments found that the formation of the C1 to C3 complexes was not affected by this competitor, indicating that, along with the Ume6p-dependent C1 and C2 complexes, the C3 complex also recognizes the GC-rich URS core element (Fig. 3B). The C5 and C6 complexes, however, were still effectively competed by URS1ΔGC, indicating that they recognize another element(s) in the probe.
To determine what sequences direct C5 and C6 complex formation, we examined the flanking sequences more carefully. 5′ to URS1 is a motif (GAAATA) with significant homology to the auxiliary repression element (ARE) described for the meiotic gene HSP82 (34). This element helps maintain full repression of HSP82 during vegetative growth and is found in the promoters of several early meiotic genes (data not shown). The other flanking region contains sequences similar to the T4C motif that is required for full transcriptional activation of the meiotic gene IME2 (3). To test if either one, or both, of these motifs was directing the binding of C5 or C6, mutant oligonucleotides were synthesized with changes in either T4C (URS1ΔT4C) or T4C and ARE (URS1ΔARE/ΔT4C) (see Fig. 3E). The duplex oligonucleotides were radiolabeled and incubated with wild-type mitotic extracts in a standard EMSA. The alteration of T4C eliminated C5 formation, while changing both ARE and T4C dramatically reduced C5 and C6 assembly (Fig. 3C). Enhanced binding of C3 was observed for both mutant oligonucleotides. The underlying reason for this observation is unclear but may be the result of the mutations introduced into these derivatives providing a better binding substrate for this unknown protein (or proteins). To strengthen these conclusions, standard unlabeled competitor assays were performed as described in the legend to Fig. 3A. In these assays, gel conditions and exposure times were employed that allowed better separation and visualization of C5 and C6. Oligonucleotides 91T and ΔGC were able to compete with C5 and C6 in a manner similar to that of the wild-type probe (Fig. 3D). However, the ΔT4C mutant probe failed to compete for C5, while ΔARE did not compete for C6. As expected, the double mutant failed to compete for either complex. These results indicate that three separate domains exist within URS1SPO13.
Three functional elements are present at URS1SPO13.
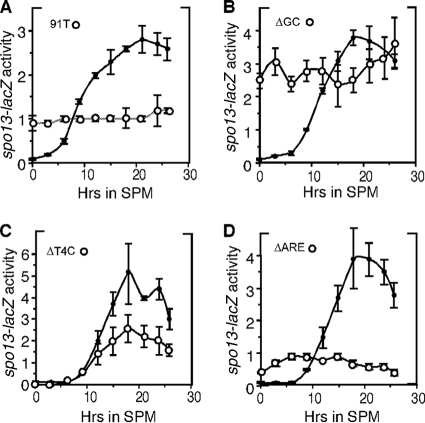
To test the physiological significance of the elements identified in vitro, the different mutations represented in Fig. 3 were introduced into an spo13-lacZ reporter gene. Cultures harboring the different reporter genes were transferred to SPM and samples taken at various time points. Protein extracts were prepared from samples collected prior to the switch to SPM (0 h) and at subsequent times as indicated in Fig. 4. For each experiment, the wild-type spo13-lacZ expression pattern was determined in parallel cultures. Mutating this reporter gene at position −91 (91T) allowed constitutive β-galactosidase expression in vegetative cells but prevented full induction during meiosis (Fig. 4A), suggesting that partial repression of Ume6p was still retained and that induction was reduced. Altering the entire GC core sequence, which failed to compete for Ume6p complex formation, resulted in high vegetative spo13-lacZ expression that was similar to its fully induced levels observed in wild-type meioses (Fig. 4B). These observations indicate that URS1 participates in both repression and activation of SPO13 transcription in meiosis.
Fig. 4.
Three functional domains exist at URS1SPO13. Strain S104 harboring the wild-type spo13-lacZ reporter expression plasmid or derivatives containing URS1 mutations as indicated was induced to enter meiosis. β-Galactosidase activity was determined from samples taken at the times indicated. Each point represents the average of values for that time point from 2 to 3 independent experiments measured in triplicate or quadruplicate. Error bars represent standard deviations of the means (Miller units). The closed and open circles denote the wild-type and mutant spo13-lacZ expression patterns, respectively.
The T4C and ARE DNA elements regulate SPO13 meiotic induction and vegetative repression.
The A-rich sequence resembling the T4C coactivator sequence required for complex 5 formation was mutated (ΔT4C) in the spo13-lacZ reporter gene. Analysis of β-galactosidase expression revealed that, while mitotic repression was unaffected by this mutation, peak meiotic expression was reduced (Fig. 4C), indicating that this sequence is required for full derepression of spo13-lacZ. Next, we tested the functionality of the sequence resembling the ARE repression element. Mutating this sequence (ΔARE) revealed two defects in the spo13-lacZ expression profile. First, a low level of derepression was observed in vegetative cells (Fig. 4D) that was similar to the levels observed in sin3Δ mutants. Next, no meiotic induction was observed, as the activity remained constant throughout the time course. These results indicate that the ARE serves two roles in both maintaining vegetative repression and directing normal induction. Taken together, the in vitro and in vivo results indicate the presence of two additional functional elements in the SPO13 promoter. The T4C element is required for full induction, while the URS1 and ARE control both repression and activation.
PAH3 and PAH4 are required to reestablish transcriptional repression of early meiotic genes.
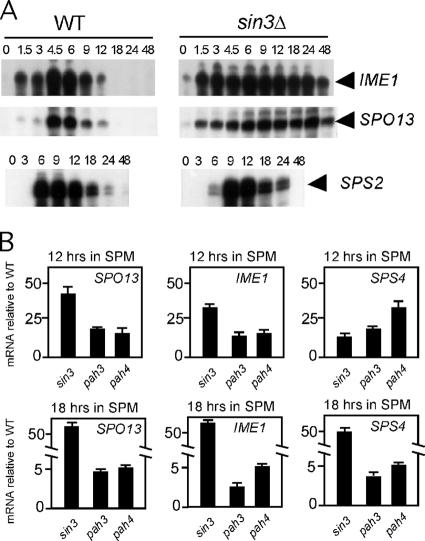
The meiotic transcription program consists of several waves of transient expression that are partitioned into three general classes termed early, middle, and late (reviewed in reference 20). Most analyses of meiotic gene repression have focused on regulation during mitotic cell division. However, the repression of these genes is reestablished (meiotic repression) to their vegetative levels late in meiosis. To determine if Sin3p is required for meiotic repression, diploid strains were constructed that are homozygous for either the wild-type, null, or different PAH mutant alleles in a strain background (SK1) that exhibits synchronous meiotic progression (see Table 1). These strains were induced to undergo meiosis, and SPO13 transcription profiles were followed by Northern blot analysis. Compared to the wild type, the sin3 mutant displayed two distinct differences. First, peak SPO13 mRNA accumulation was delayed (Fig. 5A, black arrowheads; quantitated in Fig. 5B) compared to that in the wild type. Second, rerepression was not fully established in the sin3 mutant strain, as SPO13 mRNA levels at 12 h posttransfer to SPM were reduced to only 40% of the peak expression value compared to <5% for the wild-type control (Fig. 5C). These results indicate that Sin3p is required for both normal induction and efficient establishment of meiotic repression.
To determine which, if any, of the PAH domains is required for either normal induction or meiotic repression, the experiments were repeated with the pah deletion diploids. These experiments revealed that no single PAH domain was required for the timely induction of SPO13 mRNA (Fig. 5A and B). However, both PAH3 and PAH4 mutants exhibited a defect in reestablishing SPO13 repression as the cells completed the meiotic program. The magnitudes of the defect, when normalized to peak induction values, were similar between the null and pahΔ3 or pahΔ4 strains (Fig. 5C). These results suggest that these two PAH domains do not contribute a portion of the rerepression activity independently. Rather, the data suggest that each domain provides an essential function.
The inability to reestablish repression in sin3Δ and pahΔ3 strains may be a consequence of the meiotic arrest associated with these mutations. However, the pahΔ4 mutant, which also displays a failure to reestablish repression, executes meiosis with kinetics identical to those of the wild type (Fig. 5D). These findings suggest that Sin3p-dependent repression of SPO13 is independent of meiotic progression. In addition, the delayed SPO13 mRNA accumulation profile observed in the null strain suggests that Sin3p has an activity that is independent of any individual PAH domain.
The next question we posed is whether the continued expression of SPO13 in an sin3Δ mutant was a terminal phenotype or part of a delayed transient transcription profile. To test these possibilities, an extended meiotic time course was performed. Total RNA was prepared from samples collected from the wild type and the sin3-null mutant for 48 h following the shift to sporulation medium. In addition, more sensitive S1 nuclease protection assays were performed to allow the detection of even a relatively small defect in late meiotic repression. As observed above, SPO13 mRNA levels remain elevated in the sin3Δ mutant even 48 h following meiotic induction (Fig. 6A). These findings indicate that Sin3p is required for reestablishing EMG repression.
Fig. 6.
Sin3p displays expanded meiotic regulatory targets. (A) S1 nuclease protection assay monitoring IME1, SPO13, and SPS2 mRNA levels in wild-type (RSY887) and sin3 (RSY1028) cultures in extended meiotic time courses. Arrows indicate the protected riboprobe for each mRNA. (B) Quantitative PCR was utilized to monitor mRNA levels of the indicated genes at either 12 h (top) or 18 h (bottom) in wild-type (RSY877), sin3Δ (RSY1028), pahΔ3 (RSY1031), or pahΔ4 (RSY1032) strains. The graphs are expressed as fold increase over wild-type mRNA levels at a given time point. Note the split axes for the 18-h time points.
Sin3p is required for the meiotic repression of additional expression classes.
A previous study reported that Sin3p is required for the normal induction of SPS4, a member of the “middle” gene expression group (21). To test whether the PAH domains are required for SPS4 induction, the Northern blots shown in Fig. 5 were stripped and reprobed with SPS4 sequences. These experiments revealed a reduction and delay in SPS4 mRNA accumulation in the null strain compared to that in the wild-type control (Fig. 5A). However, the examination of the PAH mutant strains did not reveal a defect in SPS4 induction. Therefore, similar to SPO13 regulation, the delay in SPS4 transcript accumulation appears to be independent of any one PAH domain. In addition, meiotic repression of SPS4 was also defective in the sin3Δ strain as well as both pahΔ3 and pahΔ4 mutants (Fig. 5A, compare the 12-h time points). These results suggest that Sin3p couples the timings of early and middle gene expression.
We next tested the role of Sin3p in regulating the transcription of two additional meiosis-specific genes. First, the expression profile of another middle gene (SPS2) (24) was examined. In this case, we expanded the time course to more fully evaluate the role of Sin3p in meiotic repression. Interestingly, SPS2 induction was also delayed approximately 3 h (Fig. 6A) as observed for SPS4 mRNA. However, unlike SPS4 transcription, SPS2 mRNA levels returned to their preinduction levels with kinetics similar to those of the wild type (note the different time points for SPO13 and SPS2 experiments). These findings indicate that SIN3 is required for the normal timing and magnitude of middle-gene induction but not the meiotic repression of all loci in this expression group.
We next examined whether Sin3p was required for meiotic repression of IME1, a gene transcribed prior to the EMG (reviewed in reference 12). In the sin3Δ mutant, a low level of IME1 transcript is observed prior to transfer to SPM (Fig. 6A, compare the 0 h time points). However, IME1 mRNA is often observed in wild-type cells growing in acetate-based medium (14), making any conclusions about Sin3p repressing IME1 expression difficult. However, compared to those of the wild type, IME1 mRNA levels remain elevated throughout the duration of the experiment in the sin3Δ mutant. These results indicate that Sin3p is also required for reestablishing IME1 repression as cells progress through meiosis.
To further explore the role of the PAH domains in meiotic repression, the mRNA levels of IME1, SPS4, and SPO13 were monitored in the wild-type, sin3Δ, pahΔ3, and pahΔ4 strains. For these studies, qPCR was utilized to quantitate the mRNA levels during meiosis (see Materials and Methods for details). Time points were taken at 12 and 18 h following transfer to SPM to focus on the meiotic repression activities of these Sin3p derivatives. The expression levels of each mRNA were first normalized to ENO1 transcript levels and then compared to those of the wild-type strain. At 12 h, the sin3Δ, pahΔ3, and pahΔ4 mutants exhibited significant increases in mRNA levels for all loci tested. By 18 h, the sin3Δ mutant still displayed meiotic repression defects, as observed in Fig. 6A. Although still exhibiting elevated mRNA levels, the requirement for PAH3 or PAH4 for meiotic repression was more modest at this later time point. Similar results were observed for a 36-h time point (data not shown). Taken together, our findings provide evidence for an expanded role for Sin3p in reestablishing repression of meiotic genes from different expression classes.
DISCUSSION
This report dissects the contributions of the four Sin3p PAH domains in controlling both in vivo meiotic gene expression and in vitro promoter-protein complex formation. PAH2 and PAH3, but not PAH1 and PAH4, are required for EMG transcription during vegetative growth and efficient recombination during meiosis. In addition, Sin3p is required for both the normal timing of induction and reestablishing repression of meiotic genes from several expression classes. The latter activity requires only PAH3 and PAH4, suggesting that a different repression mechanism is installed in postinduction meiotic cells than the one active during mitotic cell division.
Our data indicated that the PAH domains played separable roles in repressing meiotic gene transcription. Only PAH2 and PAH3 are required for repression in vegetative cells, while PAH3 and PAH4 are solely responsible for meiotic repression as the cells complete the sporulation program. As outlined earlier, the roles of PAH2 and PAH3 have been elucidated and found to be conserved. PAH2 associates with DNA binding proteins, while PAH3 tethers the HDAC to promoters. Much less is known about PAH4 function, although the finding that this domain recruits the O-linked N-acetylglucosamine (O- GlcNac) transferase (OGT) to promoters provides some clues to its role. This study found that PAH4 interacted with the tetratricopeptide (TRP) protein-protein interaction domain on OGT (45). This interaction is required for establishing the repression of SP1-activated genes in HepG2 cell culture. However, this enzyme is not found in yeast, suggesting that another TRP protein may be bound by PAH4. Analysis of the yeast proteome identified several TRP proteins that function in a variety of processes, including protein trafficking and destruction. Interestingly, a prominent corepressor, Cyc8p, also possesses TRP repeats. Determining what role, if any, Cyc8p and other TRP proteins play in meiotic repression may provide insight into the mechanism by which PAH4 exerts its control.
A poorly understood facet of meiotic gene regulation is how transcriptional repression is reestablished following normal induction. Several findings in the present and previous studies argue that the system controlling mitotic repression of the EMG is different than that installed in meiotic cells. First, Ume6p is not required for reestablishing repression. This is demonstrated in several ways. We have previously shown that Ume6p destruction is required for EMG induction (16). However, Ume6p levels do not return until spore wall assembly, which occurs after EMG repression is reestablished. Consistent with this conclusion, we demonstrate that PAH2, the Ume6p interaction domain, is not required for reestablishing repression. In addition, other factors that mediate EMG repression in vegetative cells are not required for reestablishing repression. For example, loss of Ume1p (17), cyclin C (Ssn3p) (8), or Cdk8p (Ssn8p) (33) activity does not effect this process. We have not directly tested the role of Rpd3p in this regulation, but the requirement of PAH3, the HDAC-interacting domain, strongly suggests this possibility. These results indicate that only Sin3p and Rpd3p are required for EMG repression before and after induction. Furthermore, Sin3p utilizes PAH2 and PAH3 for vegetative repression but PAH3 and PAH4 for meiotic repression.
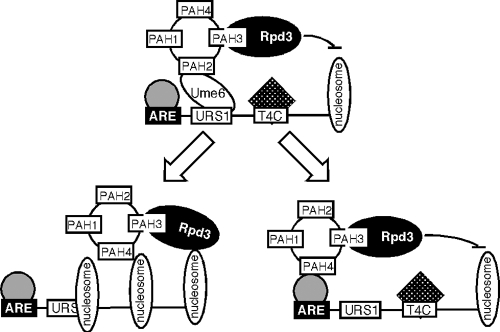
How does Sin3p mediate meiotic repression? Most studies to date in several systems indicate that Sin3p is recruited to the promoter by a DNA binding factor. For EMG vegetative repression, Ume6p serves this role. However, Ume6p is not involved in Sin3p-dependent meiotic repression. One possibility is that Sin3p jumps to one of the other two DNA binding factors identified in this report that recognize the ARE or T4C sequences (see Fig. 7, bottom right). In support of this model, EMSA analysis found that complexes C5 and C6 are maintained throughout meiosis and spore formation (data not shown). An alternative possibility is that Sin3p does not utilize a transcription factor to reestablish repression (Fig. 7, bottom left). For example, chromatin immunoprecipitation studies revealed that the human Sin3 associates with additional loci independent of known DNA binding factors following myogenic differentiation (35). In addition, the Sin3p-Rpd3p complex can stably associate with chromatin in vitro (36). A careful mapping of Sin3p-Rpd3p locations before and after induction, as well as the identification of the ARE and/or T4C binding proteins, will be necessary to answer this question.
Fig. 7.
Model for Sin3p-dependent mitotic (top) and two potential meiotic (bottom) repression systems. During vegetative growth, Sin3p bridges Rpd3p to Ume6p, utilizing PAH3 and PAH2, respectively. Meiotic repression occurs in the absence of Ume6p and requires PAH3 and PAH4 by associating to the promoter in a transcription factor-independent (left) or -dependent (right) fashion.
Why employ two systems to repress EMG expression? During vegetative growth, the meiotic genes are silent but must be ready to be activated upon the correct environmental cues. However, following the induction of these genes during meiosis, the cell completes the program and the haploid nuclei are encapsulated into spores. These spores can remain dormant for extended periods without losing viability. Therefore, a mechanism that maintains repression in spores has to be sturdy but perhaps not as responsive. Then, as the spore germinates and returns to mitotic cell division, the responsive, PAH2-dependent system would be installed at the early meiotic promoters. Understanding the nature of this system may shed light onto gene silencing that occurs in terminally differentiated cells in higher systems.
ACKNOWLEDGMENTS
We thank E. Winter, K. Cooper, and M. Henry for helpful discussions and critical readings of the manuscript. We thank D. Stillman for the PAH deletion series and valuable discussion throughout the course of this work.
This work was supported by public health service grants GM-086788 and CA-099003 from the General Medicine and National Cancer Institute, respectively.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Arcangioli B., Lescure B. 1985. Identification of proteins involved in the regulation of yeast iso-1-cytochrome C expression by oxygen. EMBO 4:2627–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer D. E., Lawrence Q. A., Eisenman R. N. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767–776 [DOI] [PubMed] [Google Scholar]
- 3.Bowdish K. S., Mitchell A. P. 1993. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592–604 [DOI] [PubMed] [Google Scholar]
- 5.Buckingham L. E., Wang H.-T., Elder R. T., McCarroll R. M., Slater M. R., Esposito R. E. 1990. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 87:9406–9410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrozza M., Florens L., Swanson S., Shia W., Anderson S., Yates J., Washburn M., Workman J. 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731:77–87; discussion 75-76 [DOI] [PubMed] [Google Scholar]
- 7.Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P. O., Herskowitz I. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705 [DOI] [PubMed] [Google Scholar]
- 8.Cooper K. F., Strich R. 2002. Saccharomyces cerevisiae C-type cyclin UME3/SRB11 is required for efficient induction and execution of meiotic development. Eukaryot. Cell 1:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz R. D., Sugino A. 1988. Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Gene 74:527–534 [DOI] [PubMed] [Google Scholar]
- 10.Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433 [DOI] [PubMed] [Google Scholar]
- 11.Hepworth S. R., Friesen H., Segall J. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honigberg S. M., McCarroll R. M., Esposito R. E. 1993. Regulatory mechanisms in meiosis. Curr. Opin. Cell Biol. 5:219–225 [DOI] [PubMed] [Google Scholar]
- 13.Kadosh D., Struhl K. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365–371 [DOI] [PubMed] [Google Scholar]
- 14.Kassir Y., Granot D., Simchen G. 1988. IME1, a positive regulator of meiosis in S. cerevisiae. Cell 52:853–862 [DOI] [PubMed] [Google Scholar]
- 15.Laherty C. D., Yang W.-M., Sun J.-M., Davie J. R., Seto E., Eisenman R. N. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349–356 [DOI] [PubMed] [Google Scholar]
- 16.Mallory M. J., Cooper K. F., Strich R. 2007. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell 27:951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallory M. J., Strich R. 2003. Ume1p represses meiotic gene transcription in S. cerevisiae through interaction with the histone deacetylase Rpd3p. J. Biol. Chem. 278:44727–44734 [DOI] [PubMed] [Google Scholar]
- 18.Maniatis T., Fritsch E. F., Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19.Meluh P. B., Koshland D. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pak J., Segall J. 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6417–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H.-O., Craig E. A. 1989. Positive and negative regulation of basal expression of a yeast HSP70 gene. Mol. Cell. Biol. 9:2025–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H. D., Luche R. M., Cooper T. G. 1992. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 20:1909–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percival-Smith A., Segall J. 1986. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 6:2443–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., Hwang S. Y., Davis R. W., Esposito R. E. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415–423 [DOI] [PubMed] [Google Scholar]
- 26.Rothstein R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast, p. 281–301InAbelson J. N., Simon M. I. (ed.), Methods in enzymology, vol. 194 Academic Press Inc., San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 27.Russell D. W., Jensen R., Zoller M. J., Burke J., Errede B., Smith M., Herskowitz I. 1986. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory region. Mol. Cell. Biol. 6:4281–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber-Agus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi A. I., DePiho R. A. 1995. An amino-terminal domain of Mxi1 mediates anti-myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN33. Cell 80:777–786 [DOI] [PubMed] [Google Scholar]
- 29.Silverstein R. A., Ekwall K. 2005. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47:1–17 [DOI] [PubMed] [Google Scholar]
- 30.Strich R., Slater M. R., Esposito R. E. 1989. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc. Natl. Acad. Sci. U. S. A. 86:10018–10022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strich R., Surosky R. T., Steber C., Dubois E., Messenguy F., Esposito R. E. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8:796–810 [DOI] [PubMed] [Google Scholar]
- 32.Sumrada R. A., Cooper T. G. 1985. Point mutation generates constitutive expression of an inducible eukaryotic gene. Proc. Natl. Acad. Sci. U. S. A. 82:643–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surosky R. T., Strich R., Esposito R. E. 1994. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol. Cell. Biol. 14:3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szent-Gyorgyi C. 1995. A bipartite operator interacts with a heat shock element to mediate early meiotic induction of Saccharomyces cerevisiae HSP82. Mol. Cell. Biol. 15:6754–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Oevelen C., Wang J., Asp P., Yan Q., Kaelin W. G., Jr., Kluger Y., Dynlacht B. D. 2008. A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 32:359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeulen M., Walter W., Le Guezennec X., Kim J., Edayathumangalam R. S., Lasonder E., Luger K., Roeder R. G., Logie C., Berger S. L., Stunnenberg H. G. 2006. A feed-forward repression mechanism anchors the Sin3/histone deacetylase and N-CoR/SMRT corepressors on chromatin. Mol. Cell. Biol. 26:5226–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vershon A. K., Hollingsworth N. M., Johnson A. D. 1992. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol. 12:3706–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal M., Buckley A. M., Yohn C., Hoeppner D. J., Gaber R. F. 1995. Identification of essential nucleotides in an upstream repressing sequence of Saccharomyces cerevisiae by selection for increased expression of TRK2. Proc. Natl. Acad. Sci. U. S. A. 92:2370–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal M., Gaber R. F. 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:6317–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vidal M., Strich R., Esposito R. E., Gaber R. F. 1991. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol. Cell. Biol. 11:6306–6316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogelauer M., Wu J., Suka N., Grunstein M. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495–498 [DOI] [PubMed] [Google Scholar]
- 42.Wang H., Clark I., Nicholson P. R., Herskowitz I., Stillman D. J. 1990. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol. Cell. Biol. 10:5927–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Stillman D. J. 1993. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol. Cell. Biol. 13:1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Washburn B. K., Esposito R. E. 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21:2057–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X., Zhang F., Kudlow J. E. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O- GlcNAcylation to transcriptional repression. Cell 110:69–80 [DOI] [PubMed] [Google Scholar]