Abstract
Walleye dermal sarcoma virus encodes a retroviral cyclin (rv-cyclin) with a cyclin box fold and transcription activation domain (AD). Co-immune precipitation (co-IP) identified an association of rv-cyclin with cyclin dependent kinase 8 (cdk8). Cdk8 is dependent upon cyclin C and regulates transcription with the Mediator complex, a co-activator of transcription. Mutation of cyclin residues, required for cdk binding, disrupt rv-cyclin-cdk8 co-IP. Mutation or removal of the AD has no effect on cdk8 interaction. Direct rv-cyclin-cdk8 binding is demonstrated by pulldown of active cdk8 and by GST-rv-cyclin binding to recombinant cdk8. Cdk3 is also activated by cyclin C and phosphorylates retinoblastoma protein to initiate entry into the cell division cycle. Co-IP and pulldowns demonstrate direct rv-cyclin binding to cdk3 as well. The rv-cyclin functions as a structural ortholog of cyclin C in spite of its limited amino acid sequence identity with C cyclins or with any known cyclins.
Keywords: WDSV, retroviral cyclin, rv-cyclin, cyclin C, cdk8, cdk3, Mediator, Retinoblastoma protein, Rb, cell cycle
Introduction
Walleye dermal sarcoma virus (WDSV) is a complex retrovirus that encodes three known accessory proteins in addition to the structural and enzymatic proteins encoded by the gag, pro, pol and env genes (Holzschu et al., 1995; Martineau et al., 1992; Martineau et al., 1991). Transcripts from two of these accessory genes, orf a and orf b, are the only WDSV transcripts present during the development of dermal sarcoma (Bowser et al., 1996; Quackenbush et al., 1997), implicating their protein products in tumor induction and progression. The protein product of the orf a transcript contains a predicted cyclin box fold and is referred to as the retroviral cyclin or rv-cyclin protein (LaPierre et al., 1998). The cyclin box fold is a protein-binding domain common to cyclins, transcription factor 2B (TFIIB), and retinoblastoma protein (Rb) (Noble et al., 1997). Each of these contains two copies of the domain, which is characterized by a similar alpha-helical structure, but with remote linear sequence identity. Alignments of the rv-cyclin with cyclins A, C, and D have been made based on combinations of sequence identity and proposed function (LaPierre et al., 1998; Rovnak and Quackenbush, 2002; Zhang and Martineau, 1999).
In addition to its cyclin box fold the rv-cyclin has a functionally separable, transcription activation domain (AD) (Rovnak et al., 2005). The AD directly contacts TATA binding protein (TBP) associated factor 9 (TAF9) in mammalian and piscine cells (Rovnak and Quackenbush, 2006). Mutation of valine to serine at position 260 (V260S) within the TAF9 binding motif interferes physically and functionally with TAF9 binding (Quackenbush et al., 2009; Rovnak and Quackenbush, 2006).
The rv-cyclin localizes in the nucleus and is concentrated in interchromatin granule clusters (IGCs or nuclear speckles) and perichromatin fibrils (Rovnak et al., 2001; Rovnak and Quackenbush, 2002). The rv-cyclin co-localizes and co-purifies with hyperphosphorylated forms of the large subunit of eukaryotic RNA Polymerase II (RNAPII) and is co-precipitated with antibodies against RNAPII (Rovnak and Quackenbush, 2002). RNAPII is phosphorylated predominantly at serines 2 and 5 of the heptad repeat (YSPTSPS)52 in its C-terminal domain (CTD) (reviewed in (Buratowski, 2009)). Progressive phosphorylation and dephosphorylation of the CTD at these sites is associated with transcription initiation and elongation. Serine 5 is highly phosphorylated during transcription initiation and gradually declines during elongation when serine 2 phosphorylation increases (Komarnitsky et al., 2000). Cdks 7 and 8 phosphorylate serine 5 of the heptad repeat (Ramanathan et al., 2001; Rickert et al., 1999). Cdk9 phosphorylates serines 2 and 5, but primarily serine 2 (Price, 2000; Ramanathan et al., 2001).
In co-immune precipitations (co-IP) with antibodies reactive to seven different cdks, rv-cyclin was co-precipitated only with cdk8 (Rovnak and Quackenbush, 2002). Cdk8, its partner, cyclin C, and proteins, Med12 and Med13, are components of the cdk8 submodule of the Mediator complex. Mediator is targeted by activators and inhibitors of transcription and functions via its close association with RNAPII (reviewed in (Taatjes, 2010)). After transcription initiation, cdk8 phosphorylation of the CTD enhances the processivity of elongation (Donner et al., 2010; Donner et al., 2007). Cdk8-Mediator interacts with positive transcription elongation factor b (pTEFb) and affects the recruitment of pTEFb and bromodomain protein, Brd4 (Donner et al., 2010). Cdk8 also phosphorylates transcription factor, E2F-1, repressing its inhibition of β-catenin/T-cell factor-dependent transcription (Morris et al., 2008), and serine 10 of histone H3, which leads to GCN5L acetylation of lysine 14, a mark of active transcription (Meyer et al., 2008).
The dysregulation of either cyclin C or cdk8 has been implicated in cancer. In cases of osteosarcoma and in osteosarcoma cell lines, there is frequent allelic loss of the CCNC gene encoding cyclin C, and over expression of exogenous cyclin C inhibits the continued growth of these cells (Ohata et al., 2006). In contrast, the gene encoding cdk8 resides in a region of the human genome that is often amplified in colon cancers, and over expression of exogenous cdk8 leads to cell transformation of NIH3T3 cells (Firestein et al., 2008).
Although not included in the original screen of rv-cyclin-cdk interaction, cdk3 has since been identified as an alternative partner of cyclin C (Ren and Rollins, 2004). Cyclin C/cdk3 promotes the transition of quiescent cells into the cell division cycle. This transition from G0 to G1 and subsequently to S phase is dependent upon phosphorylation of Rb by cyclin C/cdk3, cyclin D/cdk4/6 and cyclin E/cdk2 complexes as well as by cdk3 complexes with cyclins A and E (Connell-Crowley et al., 1997; Harper et al., 1995). These cyclin/cdk pairs phosphorylate Rb at residues S807 and S811 causing its disassociation from E2F transcription factors to allow E2F-dependent transcription.
The common interaction of the cyclin box of the rv-cyclin with the two cyclin dependent kinases that pair with cyclin C suggests its role as a functional ortholog of this host cyclin, even though their sequence homology is low (Rovnak and Quackenbush, 2002). In spite of their potent gene regulatory and cell division functions, to our knowledge, viral proteins that target cyclin C or its cdk partners have not been identified previously. The results presented here confirm the direct interaction of rv-cyclin with cdk8 and with cdk3 and define it as the first viral ortholog of cyclin C. The outcome of this function includes enhanced cell proliferation and induction of host gene expression.
Results
The rv-cyclin-cdk8 interaction is dependent upon the cyclin box fold
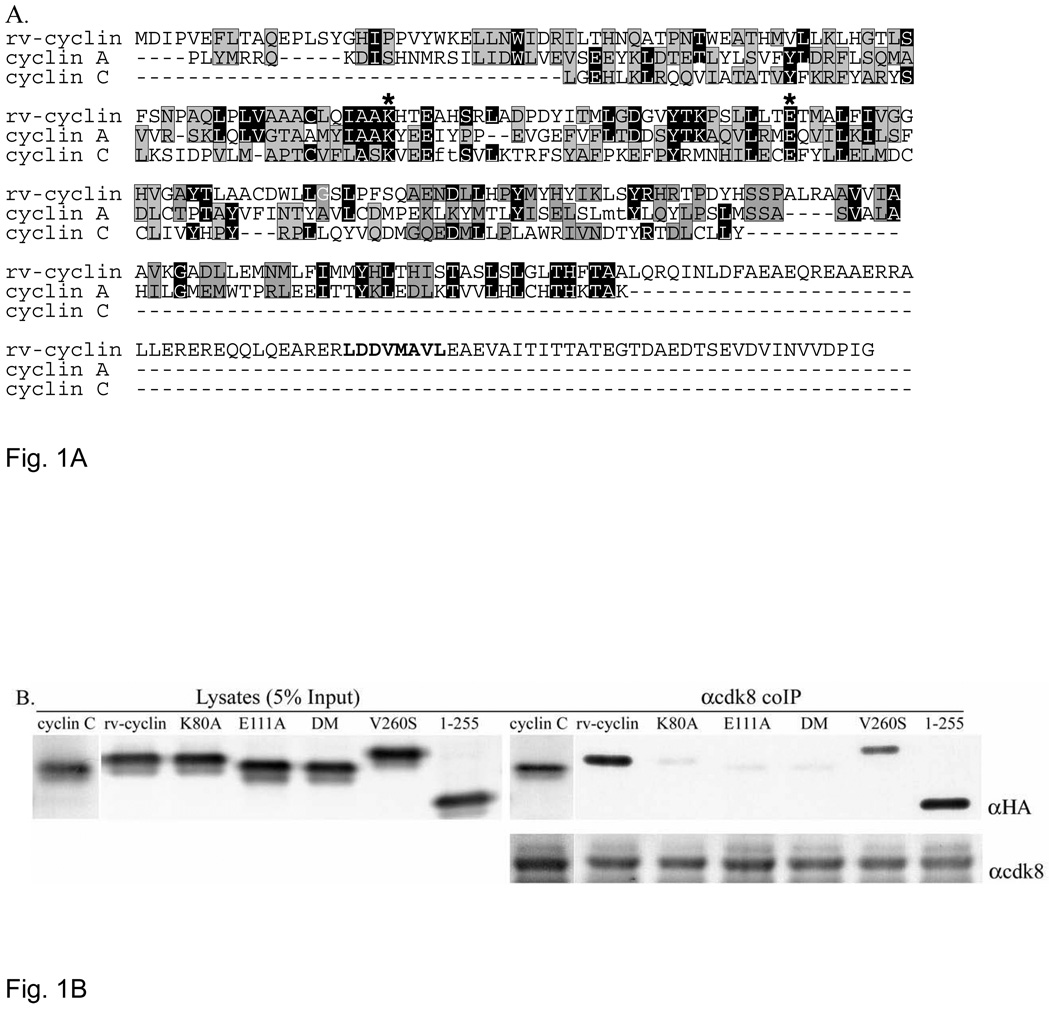
Using the protein homology/analogy recognition engine (Phyre) (Kelley and Sternberg, 2009), rv-cyclin was found to align with 1,000 different cyclins. Cyclin A from Drosophila (D. melanogaster) aligned with rv-cyclin with the highest probability (E-value=4e−43). Figure 1A shows an alignment of the amino acid sequence of rv-cyclin with sequences of Drosophila cyclin A and human cyclin C, which have 19.2% and 14.9% sequence identity with the rv-cyclin, respectively. Cyclin C is included because previous studies showed a physical association of the rv-cyclin with cdk8 and cyclin C is the partner of cdk8 (Rovnak and Quackenbush, 2002).
Fig. 1.
A. Phyre alignment of WDSV rv-cyclin with cyclin A (D. melanogaster)(E-value=4e−43, identity 19.2%) and human cyclin C (E-value=5e−06, identity 14.9%). The alignment was generated with 998 additional cyclins. Black positions are identical; gray are conserved residues. Asterisks indicate conserved cdk contacts and correspond to residues K80 and E111 in the rv-cyclin. The unaligned carboxy terminal region containing the TAF9 binding motif, LDDV260MAVL (bold), extends beyond the cyclin box of all of the aligned cyclins. Small letters, ft in cyclin C and mt in cyclin A, indicate deletions introduced by the alignment software. B. HA-tagged cyclin C, rv-cyclin, rv-cyclin point mutants, K80A, E111A, double mutant (DM; K80A/E111A), and V260S, and a.a.1–255 were transiently co-expressed with FLAG-tagged cdk8 in HeLa cells and whole cell lysates were immune precipitated with anti-cdk8 antibody. Lysates represent 5% of the co-IP input protein (5 µg) (left panel). Expressed and precipitated HA-tagged cyclins were detected on western blots with anti-HA antibody (αHA). The bottom panel shows precipitated cdk8 in each reaction after reprobe with anti-cdk8 antibody (αcdk8).
In order to assess the significance of a cyclin from a walleye virus associating with cdk8 from different species, walleye cdk8 mRNA was cloned and its amino acid sequence was found to be 98% identical to that of human cdk8 (9 amino acid variations out of 464; GenBank accession no.1383538). The walleye cdk8 sequence is 99% identical to zebrafish cdk8 (Danio rerio) with only 4 amino acid variations. Although we have not yet established the sequence of walleye cyclin C, comparisons of known cyclin Cs show that they are the most highly conserved of all cyclins among metazoan species (Hoeppner et al., 2005). The sequence of zebrafish cyclin C is 94% identical to that of human cyclin C. The conservation of cyclin C and cdk8 sequences explains the ability of the rv-cyclin to function across vertebrate species, including HeLa cells, and indicates the importance of this function in gene regulation.
Figure 1A serves to illustrate two important aspects of rv-cyclin structure: one is the remote sequence identity of the rv-cyclin to all known cyclins, none of which has greater than 20% identity with rv-cyclin. Yet structure prediction tools consistently identify its cyclin box fold with high confidence levels. A second important aspect, apparent from the alignment, is the exclusion of the rv-cyclin carboxy-terminal AD from alignment with any cyclin sequence. The AD is a separate functional domain distinct from the cyclin box fold.
Mutations at conserved residues, marked by asterisks in figure 1A, are known to disrupt the binding of cyclins A and C to their cdk partners (Hoeppner et al., 2005). To determine whether the aligned residues, K80 and E111 of rv-cyclin, effect its binding to cdk8, they were each substituted with alanine and the mutated constructs were tested for co-IP with cdk8 (Fig. 1B). In addition, a double mutation, K80A/E111A (DM), a loss-of-function mutation within the rv-cyclin AD, V260S, and a carboxy-truncated form of the rv-cyclin, amino acids 1–255, were also tested. V260S blocks rv-cyclin pulldown of TAF9 and CBP/p300 but not of Mediator component, Med23 (Sur2) (Rovnak and Quackenbush, 2006), and the 1–255 construct contains the entire predicted cyclin box fold and excludes the AD. HA-tagged human cyclin C and wild-type and mutated forms of rv-cyclin were co-expressed with FLAG-tagged human cdk8 in HeLa cells, and whole cell lysates were subjected to immune precipitation with anti-cdk8 antibody. The resultant precipitates were analyzed by western blot for co-IP of individual HA-tagged constructs and for IP of FLAG-tagged cdk8. Anti-cdk8 co-IP of mutants K80A, E111A, and DM were greatly reduced. The V260S mutation and deletion of the carboxy end had little or no effect on anti-cdk8 co-precipitation (Fig. 1B).
The rv-cyclin binds cdk8 directly
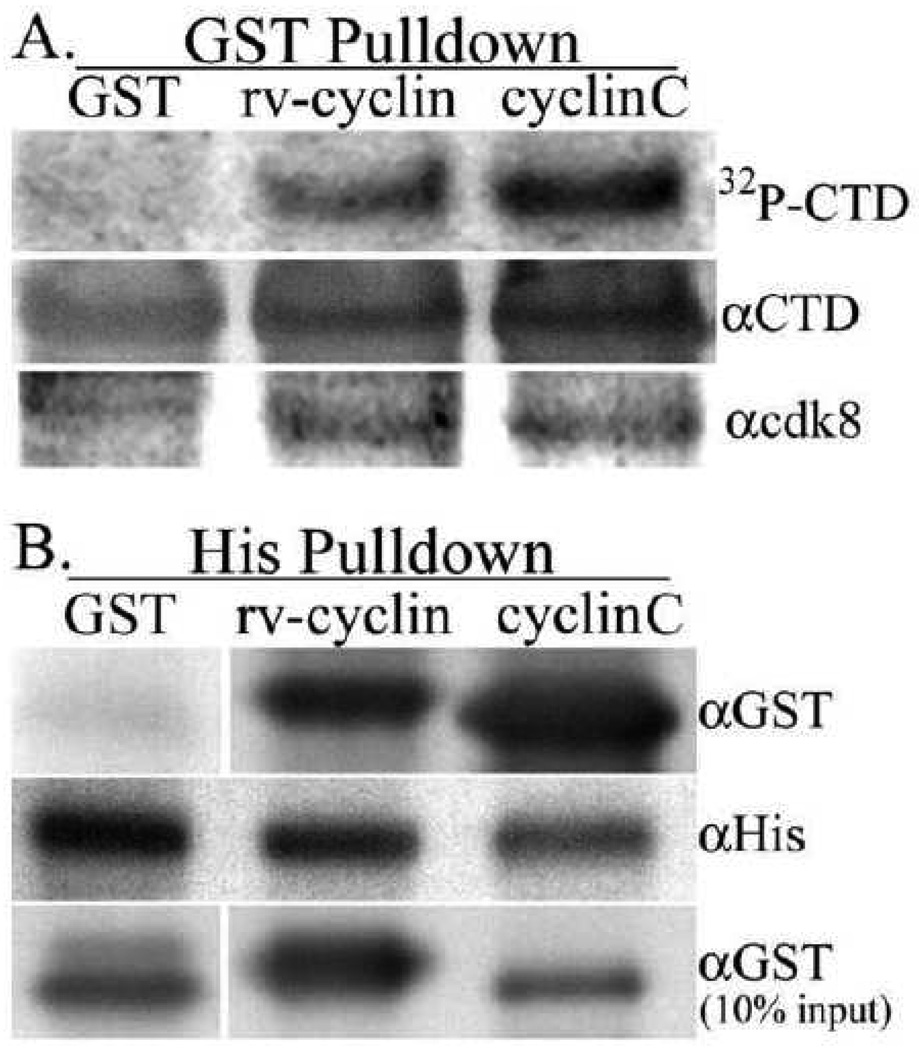
GST fusions with wild-type rv-cyclin and with human cyclin C were prepared and tested for their ability to pull down expressed, FLAG-tagged cdk8 from HeLa cell lysates. After incubation and washing, proteins bound to glutathione Sepharose were tested for kinase activity in vitro with γ32P-ATP and GST-RNAPII CTD fusion protein (GST-CTD) as substrate. After incubation and additional washes, the proteins that were pulled down and the bound GST substrate were eluted in sample buffer, separated on polyacrylamide gels, and blotted on nylon membranes. After autoradiography to determine the phosphorylation status of the CTD, blots were blocked and probed for cdk8 and for input CTD substrate (Fig. 2A). Both GST-rv-cyclin and GST-cyclin C were able to pull down active cdk8 as determined by radiolabeling of the CTD and detection of cdk8 with specific antibody. GST protein alone did not pull down cdk8 or kinase activity. Pulldowns of alternative CTD kinases, cdk7 and cdk9, were not detected by western blot (data not shown). To further confirm direct rv-cyclin-cdk8 interaction, pulldown assays were performed with purified, His-tagged cdk8 protein. In this case the 6-His tagged cdk8, bound to nickel-charged (Ni) Sepharose, was used to pull down soluble GST fusion proteins. Both GST-rv-cyclin and GST-cyclin C, but not GST alone, bound to the cdk8 demonstrating direct interaction of rv-cyclin with cdk8 (Fig. 2B). Soluble GST fusion proteins were pre-cleared with Ni Sepharose to exclude nonspecific binding.
Fig. 2.
A. GST-pulldowns of FLAG-cdk8 from cell lysates with GST, GST-rv-cyclin (rv-cyclin), and GST-cyclin C (cyclinC). GST-CTD served as in vitro substrate in kinase assays of bound cdk8 (top panel: autoradiograph of western blot of kinase reactions (32P-CTD)) followed by sequential antibody detection of input GST-CTD substrate (αCTD; anti-RNAPII CTD) and pulled down cdk8 (αcdk8). B. Ni Sepharose-His-cdk8 pulldown of soluble GST, GST-rv-cyclin (rv-cyclin), or GST-cyclin C (cyclinC) probed with anti-GST antibody (αGST). The blot was reprobed with anti-polyHis (αHis) to detect input His-cdk8. Aliquots of input GSTs were run separately (bottom panel). GST-rv-cyclin and GST-cyclin C are of similar molecular weight (64 KDa vs. 60 KDa); the GST panels show signal for proteins at 27 KDa (GST).
The rv-cyclin displaces cyclin C from cdk8 in transcription complexes
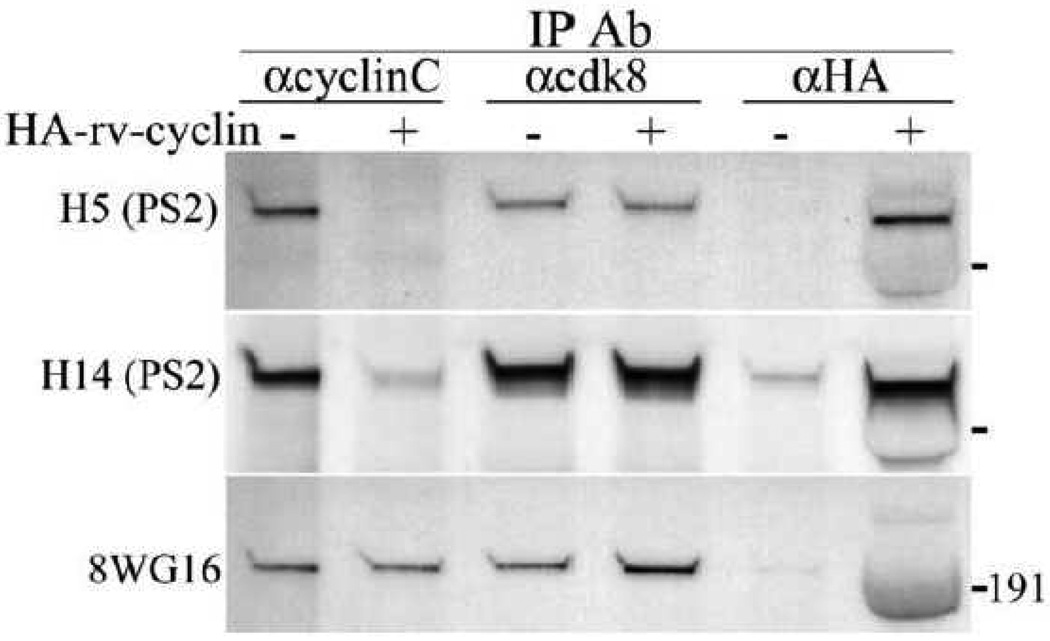
We next examined whether the rv-cyclin affects the association of cyclin C or cdk8 with RNAPII. In the presence of over expressed, HA-tagged rv-cyclin, hyperphosphorylated forms of RNAPII were not effectively co-precipitated with anti-cyclin C antibody, although un-phosphorylated RNAPII was co-precipitated (Fig. 3, lane 2). Hyperphosphorylated forms of RNAPII were precipitated with anti-cdk8 antibody whether or not the rv-cyclin was expressed (Fig. 3 lanes 3 and 4). The co-IP of hyperphosphorylated RNAPII with rv-cyclin confirms previous studies (Fig. 3, lane 6) (Rovnak and Quackenbush, 2002). These results indicate an exclusion of cyclin C specifically from complexes containing hyperphosphorylated RNAPII when the rv-cyclin is present. The continued association of cdk8 with this transcriptionally active form of RNAPII suggests that rv-cyclin/cdk8 pairs supplant cyclin C/cdk8 pairs in transcription complexes.
Fig. 3.
Co-IP of RNAPII with anti-cyclin C (αcyclinC), anti-cdk8 (αcdk8), and anti-HA (αHA) from nuclear extracts of cells without or with expressed, HA-tagged rv-cyclin. Co-precipitates were detected successively on western blots with antibodies specific for phosphorylated forms of the RNAPII CTD: H5 (phospho-Ser2; YSPTSPS), H14 (phospho-Ser5; YSPTSPS), and 8WG16 (un-phosphorylated RNAPII). The position of a 191 KDa marker is indicated to illustrate the size difference between un-phosphorylated RNAPII (predicted molecular weight 220 KDa) and the hyperphosphorylated forms (apparent molecular weight of 240 to 250 KDa).
The rv-cyclin enhances host gene expression
In order to assess the influence of rv-cyclin on host gene expression, radiolabeled cDNAs of transcripts from HeLa cells, with or without transient rv-cyclin expression for 32 hrs posttransfection, were hybridized with a gene array that measures 96 cell-cycle-related genes. The results demonstrated several large increases (>5 fold) and a few modest declines in specific mRNA levels (Fig. 4A). Transcripts encoding cyclins C, D, F, G2, and H, cdks 7 and 8, p19Ink4d, CHK1, and nibrin were present at increased levels versus the levels in control vector-transfected cells. Cyclin A and RAD9 transcript levels were reduced. Cdk3 was not included in the array. The promoter regions of the activated genes were identified in the Database of Transcriptional Start Sites (http://dbtss.hgc.jp) and analyzed for transcription factor binding sites. The activated promoters are TATA-less, have no clear INR or downstream elements, and are rich in SP1 sites between −20 and −40 bp upstream of initiation. Such promoters are common in cell cycle and growth regulation (Yang et al., 2007), and may be regulated by TAF binding sites at initiator and downstream elements (Hilton et al., 2005). Examination of the transcriptional start sites of three non-activated genes identified TATA elements at −20 to −30 bp upstream of initiation. These results identify a subset of host genes and promoters that are subject to activation by rv-cyclin and suggest a specific role for rv-cyclin transcription regulation in host cell proliferation.
Fig. 4.
A. Analysis of the levels of cellular transcripts from HeLa cells at 32 hours post-transfection with control vs. rv-cyclin expression vectors. 96 genes involved with cell cycle regulation were tested by hybridization with radiolabeled cDNA on membrane bound gene arrays (GEArray Q Human Cell Cycle Gene Array). The names of genes with greater than four-fold increases or greater than two-fold decreases are indicated. B. Quantitation of cyclin D and p19Ink4d transcripts at 10 hrs post-transfection with control vs. rv-cyclin expression vector.
Subsequently, assays of expression of cyclin D and p19Ink4d by quantitative RTPCR were performed at 10 hours post-transfection with an rv-cyclin expression vector (Fig. 4B). At this early time point mRNA levels were 7 and 14 times greater, respectively, than levels in control cells. These experiments demonstrate that the rv-cyclin has an immediate affect on the expression of these genes, as the transiently expressed rv-cyclin only first appears in nuclei of transfected cells at 6–8 hours post-transfection (not shown).
The rv-cyclin binds cdk3
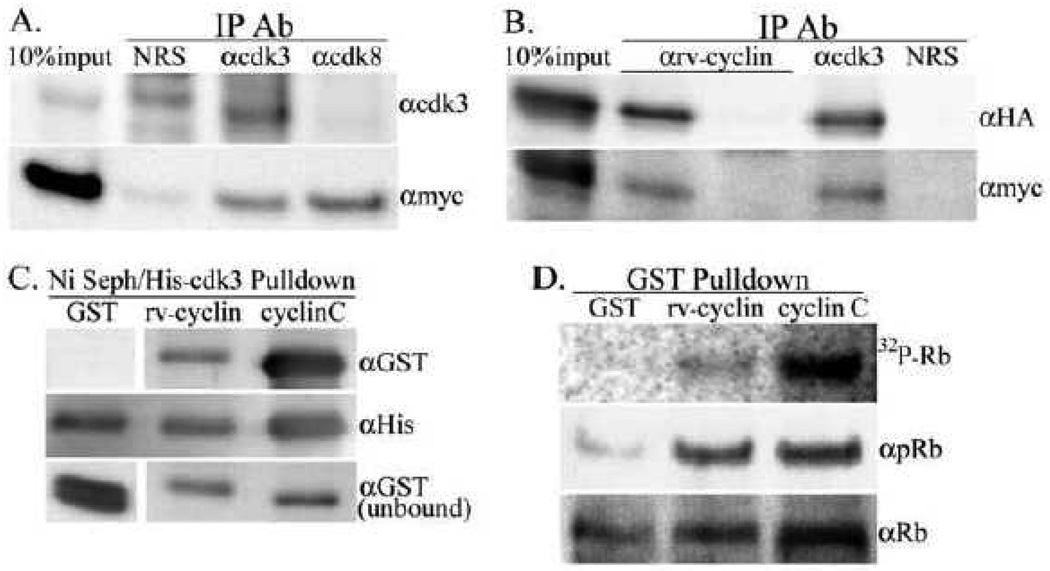
Cyclin C/cdk3 functions in the direct control of cell cycle progression by phosphorylating Rb on residues S807 and S811 to initiate cell cycle and the proliferation of quiescent cells (Ren and Rollins, 2004). Cdk3 expression is transient and is associated with the emergence of cells from quiescence and is poorly expressed in cycling cells (Ren and Rollins, 2004). In order to induce endogenous cdk3 expression, HeLa cells were grown without serum for 72 hours and harvested three hours after serum restoration. The cells used contained a tetracycline regulated (Tet-Off) expression system to produce myc-tagged rv-cyclin. Nuclear extracts from these cells, with or without rv-cyclin expression, were prepared and subjected to immune precipitation with anti-cdk3 and anti-cdk8 antibodies. Co-IP of myc-tagged rv-cyclin was detected with both IP antibodies and anti-cdk8 antibody precipitated the myc-rv-cyclin independently of its interaction with cdk3 (Fig. 5A). To increase cdk3 levels further, HA-tagged cdk3 was over expressed in HeLa cells with or without myc-rv-cyclin induction. Anti-rv-cyclin antisera was able to co-IP HA-cdk3 only from nuclear extracts of cells with expressed myc-rv-cyclin (Fig. 5B). In experiments with HA-tagged rv-cyclin containing cyclin box mutations and FLAG-tagged cdk3, none of the mutations interfered with its co-IP with cdk3 (not shown) indicating alternative contacts between the rv-cyclin and cdk3.
Fig. 5.
Interaction of rv-cyclin and cdk3. A. Nuclear extracts from induced HeLa Tet-Off myc-rv-cyclin cells after 72 hours serum starvation followed by 3 hrs of serum restoration were subject to co-IP with the indicated IP antibodies, normal rabbit serum (NRS), anti-cdk3 (αcdk3) and anti-cdk8 (αcdk8). The western blot was probed first with anti-myc Ab (αmyc) to detect precipitation of myc-tagged rv-cyclin followed by anti-cdk3 Ab (αcdk3) to detect precipitation of endogenous cdk3. B. Over-expressed, HA-tagged cdk3 was subject to co-IP with anti-rv-cyclin (αrv-cyclin) from nuclear extracts of HeLa cells with or without induced expression of myc-tagged rv-cyclin. Blots were probed first with anti-HA (αHA) to detect HA-cdk3 followed by anti-myc (αmyc) to confirm myc-rv-cyclin expression. Co-IPs with anti-cdk3 (αcdk3) and normal rabbit sera (NRS) were from extracts of rv-cyclin-induced cells. C. Ni Sepharose-His-cdk3 pulldown of soluble GST, GST-rv-cyclin (rv-cyclin) or GST-cyclin C (cyclinC) probed with anti-GST antibody (αGST). The blot was reprobed with anti-polyHis (αHis) to detect input His-cdk3. Total unbound GSTs from each reaction were run separately (bottom panels). GST-rv-cyclin and GST-cyclin C are of similar molecular weight (64 KDa vs. 60 KDa); the GST panels show signal for proteins at 27 KDa (GST). D. GST-pulldowns of over-expressed cdk3 were tested in kinase assays for phosphorylation of an Rb fusion protein (MBP-Rb-C, a.a. 379–928). The top panel shows the autoradiograph (32P-Rb) followed by antibodies for phosphorylated residues S807 and S811 of Rb (αpRb, middle panel) and for input MBP-Rb-C substrate (αRb, bottom panel).
The interaction of rv-cyclin with cdk3 was further demonstrated by binding of soluble GST-rv-cyclin and GST-cyclin C fusion proteins to recombinant, His-tagged cdk3 on Ni Sepharose in vitro (Fig. 5C). The direct interaction of rv-cyclin with active cdk3 was detected by pulldown of cdk3 kinase activity from nuclear extracts of HeLa cells with GST-rv-cyclin and GST-cyclin C fusion proteins (Fig 5D). Proteins bound to glutathione sepharose were tested for kinase activity in vitro with maltose binding protein-Rb C-terminal domain fusion protein as substrate (MBP-Rb-C, a.a. 379–928) in the presence of γ32P-ATP. After incubation of the kinase reaction the soluble substrate fraction was analyzed on blots by autoradiography to determine phosphorylation status and with specific antibodies for phosphorylation of residues S807 and S811 and for input Rb substrate. GST-rv-cyclin was able to pulldown kinase activity specific for the phosphorylation of residues S807 and S811 of Rb.
rv-cyclin enhances cell proliferation
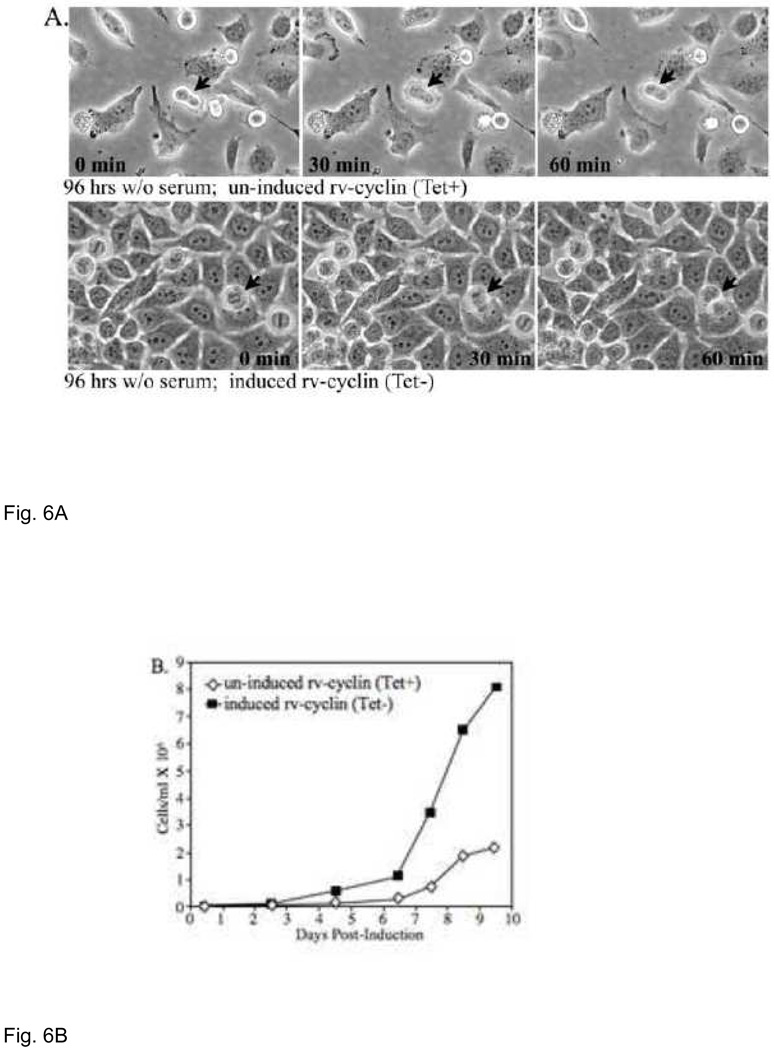
During serum starvation for cdk3 induction, a difference was noted in the number and appearance of HeLa cells with or without induced myc-rv-cyclin expression: cells with rv-cyclin expanded to greater numbers and had a higher percentage of viable cells by trypan blue exclusion. Equal numbers of cells were then plated without serum and monitored for longer periods of time. After 96 hrs, cells expressing rv-cyclin continued to divide without serum stimulation (Fig. 6A). Control, un-induced cells that entered mitosis after 96 hours without serum failed to complete cell division over a one hour period of observation, while cells with rv-cyclin were able to complete cell division in this period as indicated by arrows in figure 6A. Cells expressing rv-cyclin continue to divide without serum stimulation.
Fig. 6.
A. Un-induced and induced HeLa Tet-Off myc-rv-cyclin cells after 96 hrs of serum starvation. Arrows indicate cells undergoing cell division monitored over a 1 hr period and photographed at 0, 30 and 60 minutes. B. Un-induced HeLa Tet-Off myc-rv-cyclin cells were seeded in spinner flasks at 5×104 cells/ml under non-inducing (Tet+) or inducing conditions (Tet−) and viable cells monitored for cell concentration. rv-cyclin protein is detectable between 12 and 24 hrs post-induction (not shown).
HeLa cells with inducible rv-cyclin expression were then adapted to growth in suspension under un-induced conditions and cells were then propagated under un-induced and induced conditions at 5 × 104 cells/ml in 5% serum. Figure 6B charts the viable cell concentration over time with or without rv-cyclin expression. Full expression of induced rv-cyclin was detectable between 12 and 24 hours after removal of tetracycline, and the expression of rv-cyclin remained stable under induced conditions (not shown). Approximately 4 times as many cells were produced with rv-cyclin expression over 9 days. These results demonstrate an accelerated doubling time for these cells, which may reflect shortening of a specific phase, acceleration of all phases, or less time spent out of cycle. Under conditions of serum starvation the latter condition would prevail and suggest a cdk3-dependent mechanism. Alteration of host gene expression via cdk8 function could result in the acceleration of all phases of the cell cycle.
Discussion
Three studies now indicate that the rv-cyclin affects cell growth: 1) rescue of cyclin-deficient yeast from growth arrest (LaPierre et al., 1998), 2) abnormal cell proliferation in transgenic mice after wounding (Lairmore et al., 2000), and 3) enhanced host gene expression and proliferation of cells that express the rv-cyclin (Figures 4 and 6). The specific exclusion of cyclin C from hyperphosphorylated forms of RNAPII when the rv-cyclin is over expressed (Fig. 3) is evidence for the separation of a portion of the cdk8 pool from endogenous cyclin C in favor of the rv-cyclin. We hypothesize that the rv-cyclin, which is concentrated in nuclear speckles and perichromatin fibrils (Rovnak et al., 2001), separates cdk8 from the control of cyclin C and concentrates it at transcription complexes. This mechanism would result in an effective increase of cdk8 activity in regions of active transcription, a process that emulates the over expression of cdk8 observed in human colon cancers (Firestein et al., 2008; Morris et al., 2008) and is supported by the strong activation of transcription resulting from direct binding of a GAL4-cdk8 fusion protein to DNA (Gold et al., 1996). The specific exclusion of cyclin C/cdk8 complexes from hyperphosphorylated RNAPII may mimic the effects of the loss of cyclin C observed in osteosarcoma (Ohata et al., 2006).
The array of gene expression induced in association with rv-cyclin expression identified a putative rv-cyclin-responsive promoter type, specifically a subtype of TATA-less promoter. TATA-less promoters constitute approximately 76% of the promoters identified in the human genome (Yang et al., 2007). The association of rv-cyclin with cdk8 might serve to identify subsets of genes that are particularly responsive to cdk8 function. The gene array results at 32 hrs post-transfection could also represent secondary effects of rv-cyclin action, but quantification of cyclin D and p19Ink4d expression within 2 hrs of rv-cyclin appearance in the nucleus indicates its immediate action on at least two of these gene loci. Interestingly, the seemingly incongruous induction of an inhibitor of cdk4, p19Ink4d, may serve a specific purpose in the survival of a dermal sarcoma; it protects cells from autophagic cell death induced by 1,25-dihydroxyvitamin D3 (Tavera-Mendoza et al., 2006). p19Ink4d also blocks UV-induced apoptosis and enhances DNA repair (Ceruti et al., 2005). The results of the gene array indicate action of the rv-cyclin primarily on the expression of proteins that either regulate gene expression, cyclins C and H and cdks 7 and cdk8, or regulate response to DNA damage, p19Ink4d, CHK1, nibrin, and RAD9. Cyclin F, a Skp1-Cul1-F-box protein (SCF), doesn’t bind to a cdk but instead binds a centrosome protein, CP110, during the G2 phase of the cell cycle causing its degradation and the successful separation of sister chromatids (D'Angiolella et al., 2010). A specific cdk partner for cyclin G2 is also unknown. It binds protein phosphatase 2A, localizes at centrosomes and microtubules and alters microtubule stability (Arachchige Don et al., 2006). Expression of cyclin G2 is up-regulated by DNA damage response and by growth inhibitors and is normally associated with p53-dependent growth arrest. Its expression in a proliferative disease is counter indicated. A broader assessment of immediate rv-cyclin gene regulation is under way.
The ability of so distant a sequence as that of the rv-cyclin to function as a cyclin C ortholog defies prediction. Its low linear sequence identity with any cyclin includes all known cyclins of fish, yet structural predictions show a cyclin box fold for the rv-cyclin with high confidence levels. Clearly the rv-cyclin is not a recent transduction of a host cyclin. The modular nature of this protein, a combination of cyclin box fold and carboxy-terminal transcription activation domain, suggests that recombination events have played a role in its evolution. Similar rv-cyclins in the related walleye epidermal hyperplasia viruses contain only the cyclin box fold domain without the extended C-terminus. A precursor of these viruses may have captured a primitive cyclin or may have acquired an unrelated sequence that converged with host cyclin structure. The high degree of cyclin C conservation across metazoan organisms suggests that it was a process of convergence that yielded a functional cyclin C ortholog. Judging from the low mutation rate of the WDSV genome (Rovnak et al., 2007; Zhang et al., 1996), it would appear that the original acquisition is quite ancient. Analysis of the full extent of cyclin C mimicry will require structural determination of the rv-cyclin paired with either of its cdk partners, cdk3 or cdk8.
Conclusion
The rv-cyclin is the only known viral ortholog of cyclin C. Its association with the cdk8 module of Mediator is provocative in view of the association of cyclin C and cdk8 with established cancer cells, and its interaction with cdk3 offers a model for the initiation of proliferation in otherwise differentiated or quiescent somatic cells. The direct manipulation of these cdks by a known cancer virus offers valuable opportunities for the further delineation of their specific regulatory mechanisms.
Materials and Methods
Cell culture
HeLa cells were maintained in Dulbecco’s modified Eagle medium at 37°C in 5% CO 2. Media was supplemented with 10% fetal bovine sera (HyClone) and 4 mM glutamine. Inducible rv-cyclin cell lines were generated using the Tet-Off Gene Expression System (BD Biosciences). WDSV rv-cyclin was sub-cloned from the pKH3-OrfA vector (Rovnak et al., 2001) into the pTRE-Myc vector at HindIII and XbaI sites after modification of termini of the open reading frame by PCR. The HeLa Tet-Off cell line (BD Biosciences) was co-transfected with pTRE-Myc-rv-cyclin and pTK-Hyg plasmids and selected with G418 and hygromycin. Individual colonies were expanded and screened for inducible expression of myc-tagged rv-cyclin when cultured in the absence of doxycycline (400 µg/ml). Select clones were adapted to suspension culture. Viable cells were quantified with trypan blue exclusion during counting.
Protein expression
Human cyclin C, cdk3, and cdk8 were amplified from HeLa cell mRNA with specific 5’ and 3’ primers containing restriction site sequences appropriate for cloning into p3XFLAG-CMV-10 (Sigma), pKH3 (Rovnak et al., 2001), pGEX-2T and pGEX4T-1 (Pharmacia), and pET-19b (Novagen). All constructs were subject to full sequence analysis. pGEX4T-1-RNAPII-CTD was a generous gift of Philip A. Sharp, Whitehead Institute for Biomedical Research, Cambridge, MA. GST-rv-cyclin (Rovnak et al., 2005), GST-cyclin C, GST-CTD, 6-His-cdk3 and 6-His-cdk8 proteins were expressed in Escherichia coli cells, strain BL21 (DE3)-RIL (Stratagene) by induction with 1 mM IPTG for 2 hours. Bacteria were lysed by sonication and soluble proteins were purified twice by affinity chromatography on glutathione sepharose (Amersham Pharmacia) or immobilized metal ion affinity chromatography on Ni Sepharose (Amersham Biosciences) according to manufacturers’ instructions. For transient protein expression in tissue culture, HeLa cells were co-transfected with indicated eukaryotic expression vectors using FuGENE6 (Roche) according to manufacturers’ instructions.
Preparation of cellular extracts
Whole cell extracts were prepared by cell lysis in IP buffer (1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA pH, 1 mM EGTA, protease inhibitors (PrI; 2 µg/ml leupeptin and aprotinin, 1 µg/ml pepstatin and 0.2 mM PMSF) and phosphatase inhibitors (PhI; 0.2 mM sodium ortho-vanadate, 2 mM sodium pyrophosphate, and 1 mM glycerophosphate). Nuclear extracts from transiently transfected HeLa cells were prepared by hypotonic lysis and KCl extraction of nuclei as described by Mayeda and Krainer (Mayeda and Krainer, 1999). Nuclei from Tet-Off inducible cells were prepared by cell lysis in 0.5% NP-40 in phosphate buffered saline (PBS) (Braun et al., 1983) supplemented with 0.5 mM DTT, PrI and PhI without homogenization. Nuclei were centrifuged at 1500×g for 4 min and isolation was confirmed microscopically after three washes in ice cold PBS. They were then washed once in hypotonic buffer A (10 mM HEPES (pH 8.0), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, PrI, and PhI) prior to salt extraction by suspension in 2.5X packed nuclear volume of buffer C (10 mM HEPES (pH 8.0), 420 mM KCl, 20% glycerol, 0.1 mM EDTA, 0.5 mM DTT, PrI and PhI) for 1 hour rotating at 4°C prior to centrifugation at 21, 000×g for 20 min to yield soluble nuclear extract.
Immune Precipitation, GST Pulldown and Kinase Assays
Whole cell extracts and nuclear extracts at a protein concentration of 2 µg/µl were precleared with 10 µg of goat IgG and 100 µl of protein G Sepharose (50:50) (Pharmacia Biotech) per mg total protein overnight at 4°C. Nu clear extracts were diluted in buffer C without glycerol and with 0.05% NP-40 and whole cell extracts in IP buffer. Precleared extracts (25 µg nuclear extract or 100 µg whole cell extract) were incubated for 1 hr with 0.25–1 µg IP antibody, goat anti-cdk8 (C-19, Santa Cruz), rabbit anti-cyclin C (Ab-1, Oncogene Research Products), goat anti-cdk3 (Y-20, Santa Cruz), rabbit anti-rv-cyclin (Rovnak et al., 2001), or mouse anti-HA (12CA5, Roche). Antibodies were then captured with 20 µl protein G Sepharose (50:50) overnight at 4°C, and the preci pitated complexes were washed four times in dilution buffer prior to separation of precipitated proteins on polyacrylamide gels or use in kinase assays.
For GST pulldowns, extracts were precleared overnight with 10 µg of GST protein and 50 µl of glutathione Sepharose (50:50) (Pharmacia Biotech). 75 µg aliquots were then rotated overnight with 2.5 µg equivalents of glutathione sepharose-bound GST or GST fusion proteins (10–20 µl of a 10% suspension) in siliconized microfuge tubes. Relative quantities of bound input fusion proteins were confirmed by western analysis with anti-GST antibody (Pierce). Glutathione sepharose-bound proteins were washed 5 times prior to gel electrophoresis or kinase assay.
For kinase assays immune precipitates or GST pulldowns were washed once with kinase buffer (50 mM Tris-HCL pH 7.5, 10 mM MgCl2, 0.1 mM EDTA, 0.05 mM DTT, PrI and PhI) supplemented with 0.05% NP40 and suspended in 20 µl kinase reactions with 1 µg of substrate and 0.25–0.5 µCi [γ-32P]ATP (6,000 Ci/mmol; Perkin Elmer) in kinase buffer. Substrates were either GST-RNAPII-CTD fusion protein or maltose binding protein-Rb C-terminal domain fusion protein (MBP-Rb-C, a.a. 379–928, Cell Signaling). Cold kinase reactions used 2.5 mM ATP instead of radiolabeled ATP. Master mixes were dispensed to washed glutathione or protein G sepharose pellets and mixed at setting 1 on a vortexer for 60 min at 37°C. Reactions with GST-CTD substrate were then supplemented with 5 µl of glutathione sepharose and mixed for 15 min at room temperature to capture substrate. All reactants, bound to either the protein G or glutathione sepharose, were washed 2 times with kinase buffer with 0.05% NP40 prior to suspension of beads in loading dye and separation on polyacrylamide gels and western blotting. Only the soluble, unbound fractions of kinase reactions with Mb-Rb-C substrate were subject to gel electrophoresis. After separation proteins were blotted on nylon membranes, which were dried and exposed to phosphor screens for image capture on a BioRad Molecular Imager FX. Phosphorylated GST-CTD from cold kinase assays was detected with monoclonal antibodies reactive to RNAPII CTD, clones 8WG16, H14, and H5 (Covance). Immune precipitates were confirmed with specific antibodies, appropriate secondary antibody-peroxidase conjugates and TMB colorimetric substrate (KPL). Antibody complexes were removed from blots by incubation for 1 hr at 50°C in western strip buffer (62 mM Tris-HCl (pH 6.8), 2% SDS, and 100 mM β-mercaptoethanol), prior to probing with successive antibodies. Western blots were digitized on a Visioneer 9420 reflective scanner with Photoshop 7.0 (Adobe). Composite images were assembled and all parts of each panel adjusted equally for contrast with Photoshop 7.0.
For the in vitro assay of protein-protein interactions, soluble GST fusion proteins were brought to 1µg/100 µl binding buffer (10 mM Tris-HCl (pH 8.0), 100 mM KCl, 10% glycerol, 1 µg/ml bovine serum albumin (BSA), 0.1 mM EDTA, 2 mM DTT, 0.5 mM PMSF, 0.1% NP-40) and precleared with 10 µl of Ni Sepharose (50:50 suspension)(Amersham Biosciences) per ml overnight. 100-µl aliquots were then rotated overnight with 1µg equivalents of Ni Sepharose-bound 6-His-tagged cdk8 or cdk3 (1–5 µl of a 10% suspension) in siliconized microfuge tubes. Sepharose pellets were washed 5 times with binding buffer without glycerol or BSA and analyzed by western blot. Input His-tagged protein was confirmed by western analysis with mouse anti-polyHis (Sigma). Bound GST fusion proteins were detected with anti-GST antibody (Pierce). 10 µl aliquots (0.1 µg) of input GST and GST fusion proteins were run separately and detected with anti-GST antibody. Where proteins of different molecular weights are presented in separated panels, the contrast of the entire image was adjusted prior to assembly of the composite image.
Cloning of walleye cdk8
Total RNA was isolated from a walleye dermal sarcoma, with RNAzol (Tel-Test, Inc.), and reverse transcribed with a Verso cDNA kit (Thermo Scientific) using an anchored oligo-dT primer following the manufacturers’ protocols. Walleye cdk8 was amplified from this cDNA with degenerate primers based on conserved sequences at the start and stop sites of known cdk8 sequences (5’ primer: 5’-ATGGACTAYGAYTTYAARRTG-3’ and 3’ primer: 5’-TCAGTAMCGRTGKGTYTG-3’). The resulting product was cloned into pGEM-T Easy (Promega) and sequenced. Primer site sequences were confirmed by RNA ligase-mediated and oligo-capping rapid amplification of cDNA ends (RACE) (GeneRacer Kit, Invitrogen) according to manufacturer’s instructions.
Gene array
Total RNA was isolated from HeLa cells 32 hrs post-transfection with control vector, pKH3, or rv-cyclin expression vector, pKH3OrfA, and used for cDNA probe synthesis according to manufacturer’s instructions (GEArray Q series, Superarray). Labeled cDNA was hybridized to a GEArray Q Series Human Cell Cycle Gene Array consisting of 112 cDNAs spotted in quadruplicate on a nylon membrane. After hybridization and washing membranes were dried and exposed to phosphor screens and image captured on a Cyclone Phosphorimager (Packard) using OptiQuant software. Images were analyzed with GEArrayAnalyzer software version 1.0. Genes in numerical position on blot and graphically in figure 4 included: c-abl, Apaf-1, ATM, bax, bcl-2, BRCA1, cyclin A1, cyclin A, cyclin B, cyclin B2, cyclin C, cyclinD1, cyclinD2, cyclin D3, cyclin E1, cyclin E2, cyclin F, cyclin G, cyclin G2, cyclin H, Cdc16, cdk1, p55cdc, CDC25a, Cdc27, CDC34, CDC37, CDC45-like1, CDC6, CDC7, cdk2, cdk4, cdk6, cdk7, cd8, p21Waf1, p27Kip1, p57Kip2, p16Ink4, p15Ink2b, p18Ink4, p19Ink4d, chk1, Cks1p9, CKS2, Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5, E2F, E2F-2, E2F, E2F-3, E2F-4, E2F-5, E2F-6, MPP2, GADD45, Hus1, MAD2L1, MAD2L2, MCM2, MCM3, MCM4, MC5, MCM6, MCM7, mdm2, Ki67, MRE11A, MRE11B, nibrin, Nedd8, PCNA, PRC1, RAD17, RAD50, RAD51, RAD53, RAD9, Rb, p107, p130, Rbx1, rpa, skp1, skp2, DP1, DP2, TIMP3, p53, Ubiquitin C, UBE1, E6-AP, SUMO-1, pUC18, pUC18, pUC18, blank, blank, blank, GAPDH, GAPDH, cyclophilin A, cyclophilin A, cyclophilin A, cyclophilin A, RPL13A, RPL13A, β-actin, β-actin.
Quantitative reverse transcriptase PCR
Total RNA was prepared using Trizol reagent (Invitrogen) according to manufacturer's instructions. One microgram of RNA was reverse transcribed using random hexamers and the Verso cDNA kit (Thermo Scientific). cDNAs were analyzed by quantitative PCR using iQ SYBR green Supermix (Bio-Rad) with a CFX96 Real-Time PCR System and C1000 thermal cycler (Bio-Rad). Threshold cycles for triplicate PCRs were determined using Bio-Rad CFX Manager Software, and relative transcript abundance was determined by normalizing against Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. All primer sets generated unique products, as determined by melt curve analysis. Primer sets for quantitative PCR analysis included those for Cyclin D1 mRNA (Ccnd1) (5’ CGGAGGAGAACAAACAGA 3’ and 5’ TGAGGCGGTAGTAGGACA 3’), p19INK4d mRNA (Cdkn2d) (5’ GCTGCAGGTCATGATGTTTG 3’ and 5’CTGCCAGATGGATTGGAAGT 3’), Glyceraldehyde-3-phosphate dehydrogenase mRNA (GAPDH) (5’ GCCATCAATGACCCCCAT 3’ and 5’ CGCTCCTGGAAGATGGTG 3’).
Analysis of protein sequences
Sequences for comparative alignments were retrieved from the NCBI nr database (National Center for Biotechnology Information (GenBank), http://www.ncbi.nlm.nih.gov.) and included the WDSV rv-cyclin (ORF-A protein, accession no. AAA99528), human cyclin C and cdk8 (accession nos. AAC50825 and CAA59754) and zebrafish cyclin C and cdk8 (Danio rerio, accession nos. AAR20478 and AAI53308). Protein sequences were aligned with MacVector multiple sequence alignment software (Accelrys) using the Clustal W algorithm (Thompson et al., 1994) with BLOSUM 30 matrix and an open gap penalty of 10.0 and extend gap penalty of 0.1.
Discovery of proteins that align with the rv-cyclin protein sequence were made by submission to the protein homology/analogy recognition engine (Phyre, version 0.2; http://www.sbg.bio.ic.ac.uk/~phyre/ (Kelley and Sternberg, 2009)). Additional analyses of the rv-cyclin sequence were made by submission to web-based servers, I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/)(Roy et al., 2010), PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/)(Lobley et al., 2009), and Robetta (http://robetta.bakerlab.org/) (Kim et al., 2004).
Acknowledgements
This work was supported by Public Health Service grant CA095056 from the National Cancer Institute to S.L.Q. and College Research Council grants from the College of Veterinary Medicine and Biomedical Sciences, Colorado State University to J.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Connie D. Brewster, Email: connie.brewster@colostate.edu.
Claire H. Birkenheuer, Email: claire.birkenheuer@colostate.edu.
Megan B. Vogt, Email: megan.vogt@colostate.edu.
Sandra L. Quackenbush, Email: sandra.quackenbush@colostate.edu.
References
- Arachchige Don AS, Dallapiazza RF, Bennin DA, Brake T, Cowan CE, Horne MC. Cyclin G2 is a centrosome-associated nucleocytoplasmic shuttling protein that influences microtubule stability and induces a p53-dependent cell cycle arrest. Exp. Cell Res. 2006;312:4181–4204. doi: 10.1016/j.yexcr.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser PR, Wooster GA, Quackenbush SL, Casey RN, Casey JW. Comparison of fall and spring tumors as inocula for experimental transmission of walleye dermal sarcoma. J. Aquat. Anim. Health. 1996;8:78–81. [Google Scholar]
- Braun DK, Pereira L, Norrild B, Roizman B. Application of denatured, electrophoretically separated, and immobilized lysates of herpes simplex virus-infected cells for detection of monoclonal antibodies and for studies of the properties of viral proteins. J. Virol. 1983;46:103–112. doi: 10.1128/jvi.46.1.103-112.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceruti JM, Scassa ME, Flo JM, Varone CL, Canepa ET. Induction of p19INK4d in response to ultraviolet light improves DNA repair and confers resistance to apoptosis in neuroblastoma cells. Oncogene. 2005;24:4065–4080. doi: 10.1038/sj.onc.1208570. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L, Harper JW, Goodrich DW. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS, Loda M, Shivdasani RA, Meyerson M, Hahn WC. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MO, Tassan J-P, Nigg EA, Rice AP, Herrmann CH. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucl. Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, et al. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton TL, Li Y, Dunphy EL, Wang EH. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol. Cell Biol. 2005;25:4321–4332. doi: 10.1128/MCB.25.10.4321-4332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner S, Baumli S, Cramer P. Structure of the mediator subunit cyclin C and its implications for CDK8 function. J. Mol. Biol. 2005;350:833–842. doi: 10.1016/j.jmb.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Holzschu DL, Martineau D, Fodor SK, Vogt VM, Bowser PR, Casey JW. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J. Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structur prediction and analysis using the Robetta server. Nucl. Acids Res. 2004;32 Suppl 2:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho E-J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes and Develop. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairmore MD, Stanley JR, Weber SA, Holzschu DL. Squamous epithelial proliferation induced by walleye dermal sarcoma retrovirus cyclin in transgenic mice. Proc. Natl. Acad. Sci. USA. 2000;97:6114–6119. doi: 10.1073/pnas.110024497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre LA, Casey JW, Holzschu DL. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J. Virol. 1998;72:8765–8771. doi: 10.1128/jvi.72.11.8765-8771.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobley A, Sadowski MI, Jones DT. pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition an dsuperfamily discrimination. Bioinformatics. 2009;25:1761–1767. doi: 10.1093/bioinformatics/btp302. [DOI] [PubMed] [Google Scholar]
- Martineau D, Bowser PR, Renshaw RR, Casey JW. Molecular characterization of a unique retrovirus associated with a fish tumor. J. Virol. 1992;66:596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau D, Renshaw R, Williams JR, Casey JW, Bowser PR. A large unintegrated retrovirus DNA species present in a dermal tumor of walleye Stizostedion vitreum. Dis. Aquat. Org. 1991;10:153–158. [Google Scholar]
- Mayeda A, Krainer AR. Preparation of Hela cell nuclear and cytosolic S100 extracts for in vitro splicing. Meth. Molec. Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, Taatjes DJ. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. Nature. Vol. 455. 2008. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8; pp. 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble MEM, Endicott JA, Brown NR, Johnson LN. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends in Biol. Sci. 1997;22:482–487. doi: 10.1016/s0968-0004(97)01144-4. [DOI] [PubMed] [Google Scholar]
- Ohata N, Ito S, Yoshida A, Kunisada T, Numoto K, Jitsumori Y, Kanzaki H, Ozaki T, Shimizu K, Ouchida M. Highly frequent allelic loss of chromosome 6q16-23 in osteosarcoma: involvement of cyclin C in osteosarcoma. Int. J. Mol. Med. 2006;18:1153–1158. [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin -dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush SL, Holzschu DL, Bowser PR, Casey JW. Transcriptional analysis of walleye dermal sarcoma virus (WDSV) Virology. 1997;237:107–112. doi: 10.1006/viro.1997.8755. [DOI] [PubMed] [Google Scholar]
- Quackenbush SL, Linton A, Brewster CD, Rovnak J. Walleye dermal sarcoma virus rv-cyclin inhibits NF-κB-dependent transcription. Virology. 2009;386:55–60. doi: 10.1016/j.virol.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe'ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- Ren S, Rollins BJ. CyclinC/Cdk3 Promotes Rb-Dependent G0 Exit. Cell. 2004;117:239–251. doi: 10.1016/s0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- Rovnak J, Casey JW, Quackenbush SL. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin) Virology. 2001;280:31–40. doi: 10.1006/viro.2000.0731. [DOI] [PubMed] [Google Scholar]
- Rovnak J, Casey RN, Brewster CD, Casey JW, Quackenbush SL. Establishment of productively infected walleye dermal sarcoma explant cells. J. Gen. Virol. 2007;88:2583–2589. doi: 10.1099/vir.0.82967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Hronek BW, Ryan SO, Cai S, Quackenbush SL. An activation domain within the walleye dermal sarcoma virus retroviral cyclin protein is essential for inhibition of the viral promoter. Virology. 2005;342:240–251. doi: 10.1016/j.virol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus cyclin interacts with components of the Mediator complex and the RNA polymerase II holoenzyme. J. Virol. 2002;76:8031–8039. doi: 10.1128/JVI.76.16.8031-8039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovnak J, Quackenbush SL. Walleye dermal sarcoma virus retroviral cyclin directly contacts TAF9. J. Virol. 2006;80:12041–12048. doi: 10.1128/JVI.01425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavera-Mendoza L, Wang TT, Lallemant B, Zhang R, Nagai Y, Bourdeau V, Ramirez-Calderon M, Desbarats J, Mader S, White JH. Convergence of vitamin D and retinoic acid signalling at a common hormone response element. EMBO Rep. 2006;7:180–185. doi: 10.1038/sj.embor.7400594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Du Tremblay D, Lang BF, Martineau D. Phylogenetic and epidemiologic analysis of the walleye dermal sarcoma virus. Virology. 1996;225:406–412. doi: 10.1006/viro.1996.0616. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Martineau D. Walleye dermal sarcoma virus: OrfA N-terminal end inhibits the activity of a reporter gene directed by eukaryotic promoters and has a negative effect on the growth of fish and mammalian cells. J. Virol. 1999;73:8884–8889. doi: 10.1128/jvi.73.10.8884-8889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]