Abstract
Race, family history and age are the unequivocally accepted risk factors for prostate cancer (PCa). Androgen receptor (AR)-dependent signaling is an important element in prostate carcinogenesis and its progression to metastatic disease. We examined the possibility of genomic changes in the AR in association with familial PCa in African Americans who have a higher incidence and mortality rate and a clinically more aggressive disease presentation than Caucasians. Genomic DNAs of 60 patients from 30 high-risk African American and Caucasian families participating in the Louisiana State University Health Sciences Center genetic linkage study of PCa were studied. Exon-specific polymerase-chain reaction, bi-directional automated sequencing and restriction enzyme genotyping were used to analyze for mutations in the coding region of the AR gene. We identified a germline AR (A1675T) (T559S) substitution mutation in the DNA-binding domain in three PCa-affected members of an African-American family with a history of early-onset disease. The present study describes the first AR germline mutation in an African-American family with a history of familial PCa. The AR (T559S) mutation may contribute to the disease by altering AR DNA-binding affinity and/or its response to androgens, non-androgenic steroids or anti-androgens. Additional studies will be required to define the frequency and contribution of the AR (A1675T) allele to early-onset and/or familial PCa in African Americans.
Keywords: African Americans, androgen receptor, familial prostate cancer, germline mutation
Introduction
Prostate cancer (PCa) aggregation in families has led many investigators to believe that it has a strong hereditary component. Linkage analyses in families with hereditary PCa have identified several possible susceptibility or genetic predisposition loci suspected to harbor gene mutations conferring an increased PCa risk. These include highly penetrant susceptibility genes such as HPC1, HPC2 and CAPB 1, 2, 3, 4. In addition, mutations in the BRCA1 and BRCA2 genes and polymorphic variants of candidate genes such as the 5α-reductase and vitamin D receptor have been suggested to influence the risk for PCa 5, 6, 7, 8, 9.
The androgen receptor gene (AR) has also been suggested as a PCa susceptibility gene. Alterations in the AR gene can have profound effects on AR expression, the activity of its target genes, and its responsiveness to androgen, non-androgenic steroids and anti-androgens during the natural history of PCa. Somatic AR mutations are rarely found in untreated localized PCa (< 2%), but are detected at a higher frequency after androgen ablation therapy and in hormone-refractory or metastatic tumors 10, 11. In contrast, germline AR mutations are rarely found. So far, the R726L germline AR mutation has been reported to be the only mutation contributing to 2% of both sporadic and familial PCa in the Finnish population 12, 13.
We evaluated the possibility of genomic changes in the AR in African Americans and Caucasian families with a history of familial PCa, which is defined as having equal to or more than three patients with PCa. We screened the AR coding region in 60 PCa cases from 15 African American and 15 Caucasian families. In an African American family, we identified a novel germline AR missense mutation in three siblings with early-onset PCa, referring to the X-linked transmission pattern.
Materials and methods
Study subjects
We screened the genomic DNAs of two PCa patients from each of the 15 African American and 15 Caucasian families with at least three PCa-affected members. Recruitment of these families began in March 2000 as an effort to initiate a genetic linkage study of PCa at Louisiana State University Health Sciences Center, New Orleans 14. In each family, a reliable and informative contact person (proband) was communicated to discuss the nature of the study with both affected and non-affected family members. Family history was obtained and a report on any type of cancer was documented. A family pedigree was prepared from information provided by the proband and verified by communicating with other relatives. Written informed consent was obtained from all patients and their family members. All study-related protocols were approved by the Institutional Review Board. Medical records were reviewed to confirm the primary site of cancer.
Analysis of AR sequence in familial PCa cases
Whole blood samples were used to extract genomic DNA using a Puregene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN, USA). Polymerase chain reaction (PCR) amplification was performed by using the previously well-characterized exon-specific primer sequences covering AR gene intron/exon boundaries 15.
For exon 2 (the site of AR mutation), we used 5′-GCCTGCAGGTTAATGCTGAAGACC-3′ (sense; position 99133–99156) and 5′-CCTAAGTTATTTGATAGGGCCTTGCC-3′ (antisense; position 99511–99486, Accession No. NC_000023). In all, 50 ng of genomic DNA was amplified by 35 cycles of PCR in 50 μL containing 0.2 μmol L−1 of each primer, 0.2 mmol L−1 of dNTPs , 1.5 mmol L−1 of MgCl2 and 2.5 units of GoTaq DNA polymerase (Promega, Madison, WI, USA). PCR conditions were 95°C for 5 min, followed by 95°C for 45 s, 55°C for 45 s and 72°C for 1 min, with a 10-min extension at 72 °C after the last cycle. The correct band size was verified by running a 1.2% agarose gel. PCR products were gel-purified by using a Qiagen PCR-cleaning kit (Qiagen, Inc., Valencia, CA, USA). Sequencing was performed in both directions and repeated independently to ensure the accuracy of the data. The reported sequence was examined by Chromas LITE software (version 2.0) and compared with AR gene in the NCBI gene database (Accession No. NM_000044). After detecting the AR (A1675T) mutation in two PCa-affected brothers, we examined available genomic DNAs from five other siblings from this specific African American family.
Establishment of BsbI restriction enzyme assay for AR (A1675T) genotyping
We developed a rapid enzymatic assay to detect AR (A1675T) genotype using an amplified PCR product and a BsbI restriction enzyme to digest the constitutive BsbI sites in the normal A-allele. The PCR condition included a hot start (95°C/10 min) and 35 cycles of 95°C for 45 s, 56°C for 45 s and 72°C for 40 s, followed by a final extension at 72°C for 10 min. The primers sense, 5′-AAGACCTGAGACTTCACTTGC-3′, and AR (A1675T) antisense, 5′-AAGACCTTGCAGCT-TCCACAT-3′, were used to amplify the DNA fragment containing the allele of interest at the AR-1675 position. Gel-purified PCR products were cleaned and, before digestion with the enzyme, the accuracy of the PCR products was confirmed by sequencing. BsbI restriction enzyme digestion was performed in a 15 μL volume including 1.5 μL of 10 × NEB Buffer 2, 1.5 μL of 10 × BSA solution, 1 μL of BsbI (5U, New England Biolabs, Ipswich, MA, USA) and 11 μL of amplified PCR product. Digestion products were incubated at 37°C for 3 h and loaded in a 2.5% agarose gel. BsbI enzyme sites are 5′...GAAGAC(N)2...3′ and 3′...CTTCTG(N)6...5′.
Determination of African ancestry
The African ancestry of the family members with AR (A1675T) mutation was confirmed by using the StyI restriction enzyme and PCR for FY-null genomic marker, a population-specific allele for Africans as described by Tournamille et al.16 and Parra et al. 17.
Analysis of polymorphic AR CAG and GGC repeats
Several studies have suggested an association between the polymorphic (CAG)n/(GGC/N)n repeat length of AR and PCa risk 18. To evaluate the possibility of a linkage disequilibrium between the CAG/GGC repeat length and AR (A1675T) allele, we genotyped the study subjects by PCR and sequencing using the A1/A2 and A7/A8 primers, as described by Lubahn et al. 15. PCR was performed under the following conditions: an initial denaturation step at 95°C for 5 min for CAG repeat and 95°C for 10 min for GGC repeat (as a hot-start), followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 55°C (for CAG) or 60°C (for GGC) for 45 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min.
Determination of PSA-ARE1 (−158 G/A) genotype
PSA-ARE1 (−158 G/A) single-nucleotide polymorphism (SNP; rs266882) alone or in association with the polymorphic CAG repeat of AR has been the subject of numerous studies to identify its association with PCa risk or susceptibility 19, 20, 21, 22. To analyze the possibility of a linkage disequilibrium between PSA-G/A SNP and the AR (A1675T) allele, we genotyped all our study subjects by sequencing the PCR product and using the NheI restriction enzyme digestion, as described by Binnie et al. 23.
Statistical methods
Marker allele frequencies from sibship data were estimated using MENDEL v9.0 (UCLA, Los Angeles, CA, USA) 24. Linkage analyses for PSA SNP were carried out using the reconstruction-combination transmission disequilibrium test (RC-TDT). Linkage analyses for AR-(A1675T) allele were performed using the X-linked reconstruction combination-transmission/disequilibrium test (XLRC-TDT) 25. Reconstruction combination tests were used because parental genotypes were not available. The AR gene is on the X-chromosome and PCa is limited to males; thus, the usage of XLRC-TDT procedure is justified. Comparisons of average CAG and GGC repeats by race and AR status were performed using the MIXED procedure of SAS v9.1 (SAS Institute Inc.,Cary, NC, USA) to account for the sibship membership to nuclear families; P-values were adjusted for multiple testing where necessary to maintain a family-wise significance level of 0.05.
Results
Identification of AR (A1675T) (T559S) mutation in a high-risk African American family
Sequencing studies conducted in 15 African American and 15 Caucasian families with a history of familial PCa identified a single base substitution (ACC to TCC) of the AR gene in two PCa-affected members in an African American family. This single base substitution led to a missense mutation changing the amino acid, threonine, to serine at codon 559 of the DNA-binding domain (exon 2) of AR. A normal A-allele, a mutant T-allele and a normal female with heterozygous AT alleles are shown in of Figure 1. This family had nine PCa cases with five cases confirmed by medical records and four by description by relatives (Figure 2). Genomic DNAs were available from family members 1, 3, 12, 13, 14, 15 and 16. Of these, members 1, 3, 13 and 16 showed a mutant T-allele, member 12 showed a normal A-allele, and female members 14 and 15 were heterozygous at AT-alleles with no history of cancer (Figure 1C). Although family member 16 showed the mutant T-allele and was reported by family members to have severe urological problems, no medical confirmation of urological malignancy exists because of refusal of medical attention. The alignment of amino acid sequences of the human AR before and after the AR (A1675T) (T559S) position showed that the mutant amino acid is located in the N-terminal portion of the DNA-binding domain, which is highly conserved and showed 100% homology with mouse, rat and monkey. Our analysis of genomic DNA from 150 normal unrelated individuals (75 African Americans and 75 Caucasians) excluded the possibility for AR (A1675T) as a polymorphic variant. Overall, the AR (A1675T) (T559S) might present as a PCa-predisposing germline mutation and/or a novel SNP on the X-chromosome in familial PCa.
Figure 1.
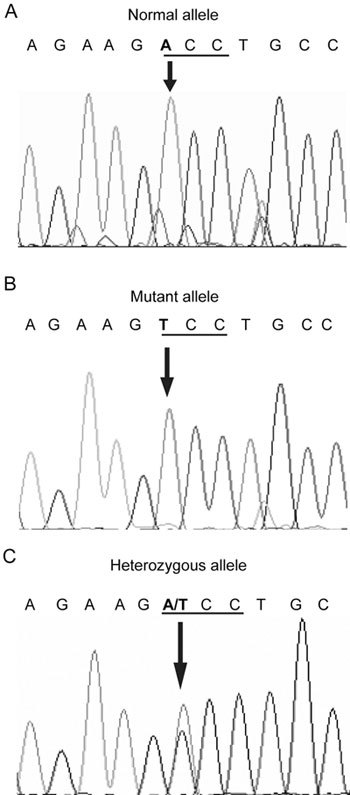
The AR (A1675T) (T559S) mutation. PCR amplification of genomic DNA with exon-specific primers and bi-directional sequence analysis revealed the presence of a single base-substitution mutation, which resulted in threonine to serine amino-acid change in the early region of DNA-binding domain of AR. (A): A normal male with A-allele; (B): An African-American patient with familial PCa and a mutant T-allele; (C): His sister with normal phenotype and heterozygous AT alleles.
Figure 2.
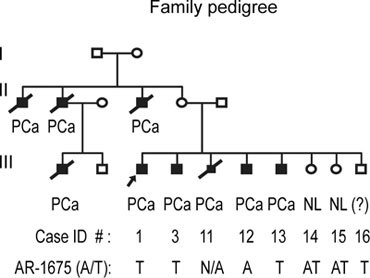
The patients' family tree. The family members with the AR (A1675T) (T559S) are indicated, as are those who presented with heterozygous AT-alleles (female). Affected siblings of the proband (contact person), except family member 12, showed a mutant T-allele and both females had heterozygous AT-alleles. Family member 11 had confirmed medical diagnosis of PCa and died of the disease before being recruited into the study. Family member 16, a mutation carrier, had severe urological problems; no medical confirmation of urological malignancy exists because of refusal of medical attention. Squares indicate male family members; circles, female family members; solid symbols, affected family members; slash, deceased. The proband is indicated by an arrow.
AR (A1675T) genotype identification by BsbI restriction digestion
As shown in Figure 3A, digestion with BsbI identified three possible genotypes: a mutant T-allele with a 206-bp band, a heterozygous AT-allele showing 70-, 136- and 206-bp bands and existing only in normal females (carrier of the mutant T-allele), and a normal A-allele with two bands of 70 and 136 bp.
Figure 3.
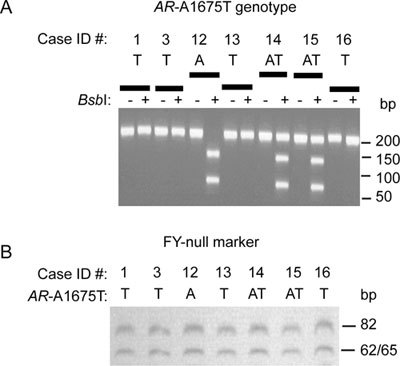
(A) Identification of AR (A1675T) genotype by BsbI restriction enzyme digestion. Genomic DNAs were amplified and digested with 5 U of BsbI to destroy a constitutive BsbI site in AR containing the normal A-allele. Digested PCR products were loaded on a 2.5% agarose gel, visualized and photographed. Using BsbI digestion, the mutant allele in family members 1, 3, 13 and 16 leads to a single 206-bp band, whereas the PCa-affected member 12 with normal A-allele shows two bands (70 and 136 bp). Family members 13 and 14, two normal females with heterozygous AT-alleles (one mutant and one wild type), produce three bands (70, 136 and 206 bp). (B): Determination of African ancestry by restriction analysis of the PCR-amplified FY-null locus in a PCa-affected family with AR (A1675T) mutation. African ancestry was analyzed by StyI-restriction analysis of the PCR-amplified FY-null locus as a population-specific allele in all available members of the PCa-affected family with AR (A1675T) mutation.
Determination of African ancestry
The African ancestry of the PCa-affected family members with AR (A1675T) mutation was confirmed by StyI restriction enzyme digestion and by PCR for FY-null genomic marker, a population-specific allele for Africans 16, 17. As shown in Figure 3B, all members in this specific family showed a similar restriction banding pattern.
Correlation with demographics and clinical data
The clinical features of the AR (A1675T) mutation-positive PCa patients are shown in Table 1. The average age at PCa diagnosis was 57.6 years, which is considered early-onset. Serum-PSA level at PCa diagnosis was also low (3.5–7.4 ng mL−1). The Gleason score for patients with the mutant T-allele was also at the intermediate level.
Table 1. Demographics and allelic distribution of common polymorphic sites in the African American family with germline AR (A1675T) mutation1.
| Case ID #2 | 1 | 3 | 115 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|
| PCa 3 | + | + | + | + | + | Normal Female | Normal Female | ? 4 |
| AR (A1675T) | T | T | N/A | A | T | TA | TA | T |
| CAGn | 23 | 23 | N/A | 20 | 23 | 23 | 23 | 23 |
| GGCn | 16 | 16 | N/A | 20 | 16 | 16 | 16 | 16 |
| ARE1-PSA | AG | AG | N/A | AG | AG | AG | AG | AG |
| Age6 (years) | 58 | 62 | 56 | 51 | 57 | 60 | 57 | 55 |
| PSA (ng mL−1) | 6.2 | 3.5 | N/A | 7.4 | 4.3 | N/A | N/A | N/A |
| Gleason score | 6 | 7 | N/A | 9 | 7 | N/A | N/A | N/A |
Abbreviations: PCa, prostate cancer; N/A, denotes not available; ARE1, Androgen-response element 1; PSA, Prostate-specific antigen
This family was the only one out of 15 high-risk African American families identified with germline AR (A1675T) mutation
Case ID numbers are based on family pedigree
PCa diagnosis was confirmed by medical records
This family member carries mutation and has reported urological problems, but no additional medical information was available
Family member 11 had confirmed medical diagnosis of PCa and died of the disease before being recruited for the study
Age at which PCa was diagnosed.
Distribution of AR (A1675T) genotype and polymorphic AR-CAG/GGC repeat and PSA-ARE1 (G/A) sites
The polymorphic CAG and GGC repeats of AR gene alone or in combination with PSA-ARE1 (G/A) SNP have been suggested as the contributing factors for PCa susceptibility or risk 18, 19, 20, 21, 22. We tested the CAG/GGC repeat length in all our cases and in particular in the members of the family presented with the AR (A1675T) mutation. With the exception of family member 12 with CAG20 and GGC20, all other members of this family showed the CAG23 and GGC16 repeat length (Table 1). This difference might be a reflection of the inheritance of different alleles from the maternal X-chromosome containing the normal A-allele.
The number of CAG repeats varied from 15 to 27 in African Americans, with 23 repeats being the most frequent (n = 9), and from 18 to 28 in Caucasians, with 21, 23 and 26 repeats being the three frequent alleles (n = 5). The number of CAG repeats was 21.3 ± 0.5 (mean ± SE) in African Americans and 23.2 ± 0.6 (mean ± SE) in Caucasians, which was significantly different (P = 0.01). The number of GGC repeats varied from 14 to 25 in African American families, with 23 repeats being the most frequent (n = 9), and from 17 to 26 in Caucasian families, with 23 repeats being the most frequent (n = 15). The mean number of GGC repeats was 20.7 ± 0.5 in African Americans, which was significantly different from that in Caucasians 23.3 ± 0.5 (P < 0.001). All members of the family showed the AG genotype for PSA-ARE1 (−158 G/A) SNP (Table 1). In our data, no association existed between the PSA-ARE1 (G/A) polymorphism and CAG (≤ 22) and GGC (≤ 16) repeats or AR (A1675T) alleles, or between the AR (A1675T) mutation and the GGC repeat length in African Americans (Table 2). However, a linkage may exist (P = 0.06) between the AR (A1675T) mutation and the CAG repeat length (≤ 22 vs. ≥ 23). Considering that the investigation was performed on a random set of African American families with at least three PCa-affected members, it is estimated that the mutant T-allele may exist with a prevalence of 16.2% in the previous generation and in 4.2% of the current generation among nuclear families of African Americans, with at least two sibs being affected with PCa.
Table 2. AR CAG and GGC and PSA (G/A) frequency distribution and association with AR (A1675T) mutation in African Americans and Caucasian Americans with familial prostate cancers.
| Characteristics | African American | Caucasians |
|---|---|---|
| No. of high-risk families | 15 | 15 |
| No. of PCa-affected males1 | 37 | 30 |
| PSA allele frequency2 | ||
| A | 0.484 | 0.548 |
| G | 0.516 | 0.452 |
| AR (A1675T) allele frequency3 | ||
| A | 0.958 | N/A |
| T | 0.042 | N/A |
| PSA genotype linkage4 with: | ||
| AR (A1675T) mutation status | 1.0 | N/A |
| CAG repeats (≤ 22) | 0.857 | 1.0 |
| GGC repeats (≤ 16) | 1.0 | 1.0 |
| AR mutation linkage5 with: | ||
| CAG repeats (≤ 22) | 1.0 | N/A |
| GGC repeats (≤ 16) | 0.20 | N/A |
Abbreviations: PCa, prostate cancer; AR, androgen receptor; PSA, prostate-specific antigen; N/A, not applicable.
PCa-affected males are brothers within a nuclear family
Estimated with Mendel v9.0, autosomal
Estimated with Mendel v9.0, X-linked
Exact P-value using Reconstruction-combination transmission disequilibrium test procedure assuming full sibships
Exact P-values using X-linked reconstruction combination-transmission/disequilibrium Test procedure assuming full sibships.
Discussion
African American men present with a higher incidence and mortality rate and more aggressive PCa at a younger age than other ethnic groups 26. A family history of PCa significantly increases their risk 27. AR genetic aberrations in the form of mutation, amplification or polymorphisms have been considered major contributors in pathophysiology of PCa and its progression to advanced stages. AR protein expression is estimated to be 22% higher in the benign prostate and 81% higher in the malignant prostate of African Americans compared with Caucasians 28.
In this study, we report the first germline AR (A1675T) (T559S) mutation in a high-risk nuclear family of African Americans with nine PCa-affected cases. This mutation may present as an interesting PCa-susceptibility allele in African Americans with familial inheritance. The localization of the T559S mutation in the DNA-binding domain makes it a likely candidate to affect the AR-binding affinity to its target genes. However, substitution of an uncharged polar amino acid (threonine) for another uncharged polar amino acid (serine) might not affect the AR surface charge, expression or its interactions with coactivators.
The existing reports on AR germline mutations in PCa are limited to Caucasian patients. The R726L mutation was reported in Finnish patients with sporadic or familial PCa 12, 13. Additional reports include those on two unrelated PCa patients with G2T and C214A mutations within the 5′-UTR (non-coding) region of the AR 29. One final report showed the AR-Q798E mutation in both the PCa tissue and the genomic DNA of a patient 30.
In vitro biofunctional assays have shown an inverse relationship between polymorphic CAG and GGC repeats and AR activity status. Based on these data, it has been widely hypothesized that the CAG/GGC repeat length inversely correlates with PCa risk or early-onset disease 18, 20, 31. In our study, the difference between mean CAG and GGC repeat in African Americans and Caucasian was significant (P < 0.05). This might be due to racial difference or a characteristic of familial PCa. The absence of linkage disequilibrium between the X-linked AR (A1675T) mutation and GGC repeat might indicate the lack of their significance or contribution at least in familial PCa in African Americans. It is noteworthy that a recent comprehensive analysis on a large nested case-control study in the Physicians' Health Study, together with a meta-analysis of the published studies, did not find any significant association between PSA-ARE1 SNP and PCa risk 32. In contrast to GGC repeats, there may be a linkage between the AR (A1675T) mutation and CAG repeat length. Owing to the relatively small number of families investigated and the detection of AR (A1675T) mutation in only one out of 15 African American families, our results should be interpreted with caution.
Analysis of the European genomic contribution of 10 different populations of African descent in the United States revealed that the level of European admixture, mainly French, is the highest in New Orleans, which is the source of our study population 17. In future large-scale studies, determination of African Ancestry and admixture population might be necessary to map this specific AR germline mutation to a founder PCa allele of a specific African or European descent.
Additional large-scale population studies are currently being conducted to determine the frequency of AR (A1675T) germline mutation in both sporadic and familial PCa and to elucidate its biological significance and potential as a predisposing genetic marker or a modifier variant in African Americans versus Caucasians.
Acknowledgments
This study was supported by the Stanley S Scott Cancer Center and Louisiana Cancer Research Consortium (to S.K.) at the LSU-Health Sciences Center, New Orleans, LA, USA; a pilot study grant for the Clinical and Translational Research, Education, and Commercialization Project (CTRECP) from the LSU-Health Sciences Center and Tulane University Health Sciences Center (to S.K.); NCI/NIH grant R21 CA149137 (to S.K.); and a grant from the NIH/NCRR (1P20 RR021970; to Augusto Ochoa/S.K.). Dr Diptasri Mandal was supported by a grant from NCI (1 R03 CA97778-01). We are indebted to the patients and their families and physicians for contributing to the study. We thank Ms Jonna Ellis for editorial assistance. We specially thank Drs Augusto Ochoa (Stanley S Scott Cancer Center, LSUHSC-NO) and Jay K. Kolls (Department of Genetics, LSUHSC-NO) for their critical review and helpful comments on the manuscript.
References
- Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–4. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, et al. Predisposing gene for early-onset prostate cancer, localized on chromosome 1q42.2–43. Am J Hum Genet. 1998;62:1416–24. doi: 10.1086/301879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M, Stanford JL, Mclndoe RA, Jarvik GP, Kolb S, et al. Evidence for a rare prostate cancer-susceptibility locus at chromosome 1p36. Am J Hum Genet. 1999;64:776–87. doi: 10.1086/302287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel-Tassin G, Cussenot O. Genetic susceptibility to prostate cancer. BJU Int. 2005;96:1380–5. doi: 10.1111/j.1464-410X.2005.05836.x. [DOI] [PubMed] [Google Scholar]
- Uchida T, Wang C, Sato T, Gao J, Takashima R, et al. BRCA1 gene mutation and loss of heterozygosity on chromosome 17q21 in primary prostate cancer. Int J Cancer. 1999;84:19–23. doi: 10.1002/(sici)1097-0215(19990219)84:1<19::aid-ijc4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Johannsson O, Loman N, Moller T, Kristoffersson U, Borg A, et al. Incidence of malignant tumours in relatives of BRCA1 and BRCA2 germline mutation carriers. Eur J Cancer. 1999;35:1248–57. doi: 10.1016/s0959-8049(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Reichardt JKV, Makridakis N, Henderson BE, Yu MC, Pike MC, et al. Genetic variability of the human SRD5A2 gene: implications for prostate cancer risk. Cancer Res. 1995;55:3973–5. [PubMed] [Google Scholar]
- Feldman D. Androgen and vitamin D receptor gene polymorphisms: the long and short of prostate cancer risk. J Natl Cancer Inst. 1997;89:109–11. doi: 10.1093/jnci/89.2.109. [DOI] [PubMed] [Google Scholar]
- Ingles SA, Ross RK, Yu MC, Irvine RA, Pera LG, et al. Association of prostate cancer risk with genetic polymorphisms in vitamin D receptor and androgen receptor. J Natl Cancer Inst. 1997;89:166–70. doi: 10.1093/jnci/89.2.166. [DOI] [PubMed] [Google Scholar]
- Gottlieb B, Beitel LK, Wu JH, Trifiro M. The androgen receptor gene mutations database (ARDB): 2004 update. Hum Mutat. 2004;23:527–33. doi: 10.1002/humu.20044. [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Mononen N, Syrjäkoski K, Matikainen M, Tammela TLJ, Schleutker J, et al. Two percent of Finnish prostate cancer patients have a germ-line mutation in the hormon-binding of the androgen receptor gene. Cancer Res. 2000;60:6479–81. [PubMed] [Google Scholar]
- Koivisto PA, Hyytinen ER, Matikainen M, Tammela TL, Ikonen T, et al. Germline mutation analysis of the androgen receptor gene in Finnish patients with prostate cancer. J Urol. 2004;171:431–3. doi: 10.1097/01.ju.0000089774.99728.ef. [DOI] [PubMed] [Google Scholar]
- Mandal DM, Sartor O, Halton SL, Mercante DE, Bailey-Wilson JE, et al. Recruitment strategies and comparison of prostate cancer specific clinical data on African-American and Caucasian males with and without family history. Prostate Cancer Prostatic Dis. 2008;11:274–9. doi: 10.1038/pcan.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubahn DB, Brown TR, Simental JA, Higgs HN, Migeon CJ, et al. Sequence of the intron/exon junctions of the coding region of the human androgen receptor gene and identification of a point mutation in a family with complete androgen insensitivity. Proc Natl Acad Sci USA. 1989;86:9534–8. doi: 10.1073/pnas.86.23.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeegers MP, Kiemeney LA, Nieder AM, Ostrer H. How strong is the association between CAG and GGN repeat length polymorphisms in the androgen receptor gene and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2004;13:1765–71. [PubMed] [Google Scholar]
- Bratt O, Borg A, Kristoffersson U, Lundgren R, Zhang QX, et al. CAG repeat length in the androgen receptor gene is related to age at diagnosis of prostate cancer and response to endocrine therapy, but not to prostate cancer risk. Br J Cancer. 1999;81:672–6. doi: 10.1038/sj.bjc.6690746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy DO, Scher HI, Bogenreider T, Sabbatini P, Zhang ZF, et al. Androgen receptor CAG repeat lengths in prostate cancer: correlation with age of onset. J Clin Endocrinol Metab. 1996;81:4400–5. doi: 10.1210/jcem.81.12.8954049. [DOI] [PubMed] [Google Scholar]
- Cramer SD, Sun J, Zheng SL, Xu J, Peehl DM. Association of prostate-specific antigen promoter genotype with clinical and histopathologic features of prostate. Cancer Epidemiol Biomarkers Prev. 2008;17:2451–7. doi: 10.1158/1055-9965.EPI-08-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Kedda MA, Hinze K, Smith RLG, Yaxley J, et al. PSA/KLK3 ARE1 promoter polymorphism alters androgen receptor binding and is associated with prostate cancer susceptibility. Carcinogenesis. 2007;28:1032–9. doi: 10.1093/carcin/bgl236. [DOI] [PubMed] [Google Scholar]
- Binnie MC, Alexander FE, Heald C, Habib FK. Polymorphic forms of prostate specific antigen and their interaction with androgen receptor trinucleotide repeats in prostate cancer. Prostate. 2005;63:309–15. doi: 10.1002/pros.20178. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48:22–5. [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Laird NM, Knapp M. The transmission/disequilibrium test and parental-genotype reconstruction for X-chromosomal markers. Am J Hum Genet. 2000;66:1161–7. doi: 10.1086/302823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society. Cancer Facts and Figures 2008. Accessed 13 August 2008 atURL: : http://www.cancer.org/docroot/STT/content/STT_1x_Cancer_Facts_ and_Figures_2008.asp?from=fast ).
- Powell IJ. Epidemiology and pathophysiology of prostate cancer in African-American men. J Urol. 2007;177:444–9. doi: 10.1016/j.juro.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Gaston KE, Kim D, Singh S, Ford OH, Mohler JL. Racial differences in androgen receptor protein expression in men with clinically localized prostate cancer. J Urol. 2003;170:990–3. doi: 10.1097/01.ju.0000079761.56154.e5. [DOI] [PubMed] [Google Scholar]
- Crocitto LE, Henderson BE, Coetzee GA. Identification of two germline point mutations in the 5′ UTR of the androgen receptor gene in men with prostate cancer. Cancer Res. 1997;158:1599–601. [PubMed] [Google Scholar]
- Evans BA, Harper ME, Daniells CE, Watts CE, Matenhelia S, et al. Low incidence of androgen receptor gene mutations in human prostatic tumors using single strand conformation polymorphism analysis. Prostate. 1996;28:162–71. doi: 10.1002/(SICI)1097-0045(199603)28:3<162::AID-PROS3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Rajender S, Singh L, Thangaraj K. Phenotypic heterogeneity of mutations in androgen receptor gene. Asian J Androl. 2007;9:147–79. doi: 10.1111/j.1745-7262.2007.00250.x. [DOI] [PubMed] [Google Scholar]
- Jesser C, Mucci L, Farmer D, Moon C, Li H, et al. Effects of G/A polymorphism, rs266882, in the androgen response element 1 of the PSA gene on prostate cancer risk, survival and circulating PSA levels. Br J Cancer. 2008;99:1743–7. doi: 10.1038/sj.bjc.6604690. [DOI] [PMC free article] [PubMed] [Google Scholar]


