Summary
Background
From yeast to human, TOR (Target Of Rapamycin) kinase plays pivotal roles in coupling extracellular stimuli to cell growth and metabolism. TOR kinase functions in two distinct protein complexes, TOR complex 1 (TORC1) and 2 (TORC2), which phosphorylate and activate different AGC-family protein kinases. TORC1 is controlled by the small GTPase Rheb, but little is known about TORC2 regulators.
Results
We have identified the Ryh1 GTPase, a human Rab6 ortholog, as an activator of TORC2 signaling in the fission yeast Schizosaccharomyces pombe. Mutational inactivation of Ryh1 or its guanine nucleotide exchange factor compromises the TORC2-dependent phosphorylation of the AGC-family Gad8 kinase. In addition, the effector domain of Ryh1 is important for its physical interaction with TORC2 and for stimulation of TORC2 signaling. Thus, GTP-bound Ryh1 is likely to be the active form stimulatory to TORC2–Gad8 signaling. Consistently, expression of the GTP-locked mutant Ryh1 is sufficient to promote interaction between TORC2 and Gad8 and to induce Gad8 hyper-phosphorylation. The loss of functional Ryh1, TORC2 or Gad8 brings about similar vacuolar fragmentation and stress sensitivity, further corroborating their involvement in a common cellular process. Human Rab6 can substitute Ryh1 in S. pombe and therefore, Rab6 may be a potential activator of TORC2 in mammals.
Conclusions
In its GTP-bound form, Ryh1, an evolutionarily conserved Rab GTPase, activates TORC2 signaling to the AGC kinase Gad8. The Ryh1 GTPase and the TORC2–Gad8 pathway are required for vacuolar integrity and cellular stress resistance in S. pombe.
Introduction
The target of rapamycin (TOR) is a serine/threonine-specific protein kinase that is structurally related to phosphatidylinositol kinases. First discovered in the budding yeast Saccharomyces cerevisiae, the TOR protein kinase has been identified widely among eukaryotic species [1]. TOR kinases from yeast to humans form two distinct protein complexes referred to as TOR complex (TORC) 1 and 2 (TORC2), which play discrete roles in cellular signaling partly through activation of different AGC-family protein kinases, such as mammalian S6K1 and Akt/protein kinase B [2]. TORC2 is required for activating phosphorylation of the C-terminal hydrophobic motif in Akt kinase, which is also phosphorylated within its activation loop by the 3-phosphoinositide-dependent protein kinase 1 (PDK1).
Metazoan TORC2–Akt signaling is activated in response to insulin and growth factors that induce activation of the phosphatidylinositol 3-kinase pathway [3]. Despite the pivotal roles of the TORC2–Akt pathway in insulin response and cellular proliferation, very little is known about the regulators that control TORC2 signaling. On the other hand, it has been found that TORC1 is activated by the Ras-family Rheb GTPase, which is negatively regulated by the Tsc1-Tsc2 complex that acts as Rheb GTPase-activating protein (GAP) [4]. Mutations to Tsc1 or Tsc2 bring about hyper-activation of TORC1 and its downstream AGC kinase, S6K1, leading to tumorigenic cell proliferation. Despite the extensive studies, however, the exact mechanism of TORC1 activation by Rheb is controversial and remains to be established [5–7].
In this study, we set out to search for activators of TORC2 signaling, using the fission yeast Schizosaccharomyces pombe that has the TOR pathways highly homologous to those in higher eukaryotes. The subunit composition of TORC2 is well conserved in S. pombe [8], including the TOR kinase Tor1, Sin1, and Ste20, an ortholog of mammalian Rictor and Avo3 in budding yeast. A recently identified mammalian TORC2 component, Protor/PRR5 [9,10], is structurally related to Bit61 in S. pombe TORC2 [8]. S. pombe strains lacking functional TORC2 are defective in phosphorylating the C-terminal hydrophobic motif of an AGC-family kinase, Gad8 [11,12]. Activation of Gad8 also requires phosphorylation of its T-loop by Ksg1, an ortholog of mammalian PDK1 [11]. Thus, the S. pombe TORC2–Gad8 pathway appears to parallel the TORC2–Akt pathway in higher eukaryotes. The TORC2- and Gad8-deficient mutants are viable, but show increased stress sensitivity as well as sterility [11,13–15].
Here we show that a Rab-family GTPase, Ryh1, regulates TORC2–Gad8 signaling in S. pombe. In strains lacking Ryh1 or its putative guanine nucleotide exchange factor (GEF), TORC2-dependent phosphorylation and activation of Gad8 kinase is compromised. On the other hand, expression of the GTP-locked Ryh1 promotes the physical interaction between TORC2 and Gad8, leading to increased Gad8 phosphorylation. Thus, the nucleotide-binding state of Ryh1 controls TORC2–Gad8 signaling. Moreover, cells lacking Ryh1, TORC2 or Gad8 share common phenotypes of vacuole fragmentation associated with hypersensitivity to osmostress and Ca2+. Together, these results suggest that the Ryh1 GTPase positively regulates TORC2–Gad8 signaling which is important for vacuolar integrity and stress resistance. Finally, the Ryh1 function in S. pombe can be substituted by expression of human Rab6, and therefore, evolutionarily conserved Rab GTPase may also be involved in the regulation of the mammalian TORC2–Akt pathway.
Results
S. pombe sat1, sat4 and sat7 mutants are defective in TORC2 signaling
S. pombe strains lacking functional TORC2 show increased stress sensitivity as well as sterility due to their failure to arrest in G1 after nitrogen starvation [11,13–15]. Similar phenotypes were reported with sat (starvation-induced arrest) mutants, sat1-11, sat4-86 and sat7-110 [16]. To test whether these sat mutants are defective in TORC2–Gad8 signaling, we raised antibodies that recognize the TORC2-dependent phosphorylation of Ser-546 within the hydrophobic motif of Gad8 kinase (Figure S1A). Indeed, phosphorylation of Gad8 Ser-546 was significantly reduced in the sat1-11, sat4-86 and sat7-110 strains (Figure 1A), indicating compromised TORC2–Gad8 signaling in these mutants. In parallel, we screened a S. pombe haploid mutant library that includes the null mutants of ~2,800 genes, about 60% of the protein-coding genes in S. pombe. Stress-sensitive mutants identified from this library were subsequently tested for Gad8 Ser-546 phosphorylation by immunoblotting (see Supplemental Experimental Procedure). This unbiased, two-step screen re-isolated sat1 and sat7/ryh1 but no other genes that significantly affected Gad8 Ser-546 phosphorylation.
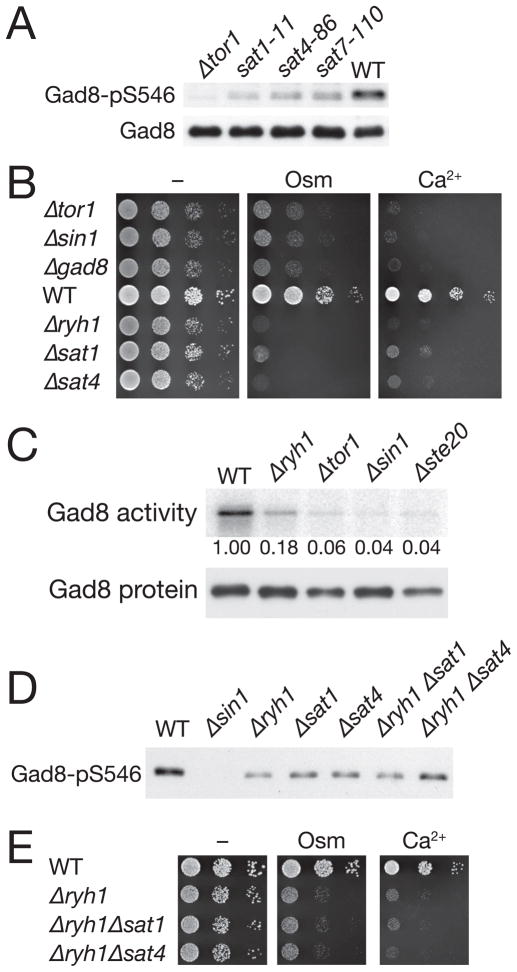
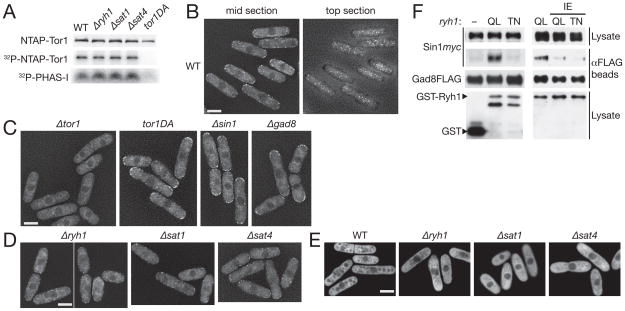
Figure 1. Sat1, Sat4 and Ryh1/Sat7 function in the same pathway that stimulates TORC2–Gad8 signaling.
(A) TORC2-dependent phosphorylation of Gad8 Ser-546 is reduced in sat1-11, sat4-86 and sat7-110 cells. Wild type, Δtor1, sat1-11, sat4-86 and sat7-110 strains carrying the gad8:6HA allele were grown at 30°C and their crude cell lysate was analyzed by immunoblotting with anti-phospho-Ser546 and anti-HA antibodies. (B) Like TORC2 and gad8 mutants, Δryh1/sat7, Δsat1 and Δsat4 mutants are hypersensitive to high osmolarity and calcium. Wild type, Δtor1, Δsin1, Δgad8, Δryh1, Δsat1 and Δsat4 cells were spotted in serial dilution onto YES agar containing 0.4 M KCl (“Osm”) or 50 mM CaCl2 (“Ca2+”) at 28°C. (C) Gad8 kinase activity is decreased in Δryh1 cells. Wild type, Δryh1, Δtor1, Δsin1 and Δste20 strains carrying the gad8:3HA allele were grown in YES medium. Gad8 kinase was immunoprecipitated and assayed for its activity using the GST-Fkh2 protein as substrate. Quantified phosphorylation levels of GST-Fkh2 (relative to that of WT) are shown below. (D) The reduced Gad8 Ser-546 phosphorylation in the Δryh1 mutant is not further decreased by the Δsat1 or Δsat4 mutations. Wild-type, Δtor1, Δsin1, Δryh1, Δsat1, Δsat4, Δryh1 Δsat1 and Δryh1 Δsat4 strains were grown in YES medium. Gad8 Ser-546 phosphorylation was examined by anti-phospho-Ser546 immunoblotting. (E) The high osmolarity- and Ca2+-sensitive phenotypes of the Δryh1 Δsat1 and Δryh1 Δsat4 double mutants are similar to those of the Δryh1 single mutant.
Sat7/Ryh1 is a Rab GTPase regulated by the Sat1-Sat4 GEF complex
Cloning of the sat7+ gene by genetic complementation determined that sat7 is allelic to ryh1+, which encodes a Rab-family small GTPase orthologous to budding yeast Ypt6 and mammalian Rab6 [17]. sat7-110 contained a nucleotide change (Table S1) that resulted in aspartate substitution of Ryh1 Gly-18, the glycine residue conserved among the Ras superfamily GTPases [18]. Like sat7-110 cells, the ryh1 null (Δryh1) mutant was sensitive to stress conditions (Figure 1B; [19]), and Gad8 kinase isolated from Δryh1 cells showed notably decreased activity (Figure 1C) and Ser-546 phosphorylation (Figure 1D). The sat1+ gene encodes an ortholog of the budding yeast Rgp1 protein, which forms a complex with Ric1 to function as GEF for the Ypt6 GTPase [20], whereas sat4-86 turned out to be a mutation of a Ric1 ortholog in S. pombe (SPAC1851.04c; Table S1). Therefore, we hypothesized that, like their orthologs in budding yeast, Sat1 and Sat4 form a protein complex that acts as GEF for the Ryh1 GTPase. The following genetic and biochemical evidence supported this hypothesis. First, like sat1-11 and sat4-86, the Δsat1 and Δsat4 mutants showed phenotypes very similar to those of Δryh1 [17,19,21], including sterility and increased sensitivity to high temperature (data not shown), high osmolarity, Ca2+ and high pH (Figure 1B and Figure S1B). Gad8 Ser-546 phosphorylation was also significantly decreased in the Δsat1 and Δsat4 mutants (Figure 1D). Second, the stress-sensitive Δryh1 phenotypes were not enhanced in the Δryh1 Δsat1 and Δryh1 Δsat4 double mutants, consistent with the idea that Sat1 and Sat4 function in the same pathway as Ryh1 (Figure 1E). Similarly, the Δsat1 and Δsat4 mutations did not further decrease the low level of Gad8 Ser-546 phosphorylation in the Δryh1 mutant (Figure 1D). Third, affinity purification of the Sat1 protein from the S. pombe cell lysate co-purified Sat4 and vice versa, demonstrating the complex formation of the two proteins as predicted (Figures 2A and B; Figure S2A). Analogous to the Rgp1-dependent localization of Ric1 to Golgi/endosomes in budding yeast [20], the characteristic punctate signals of Sat4 fused to GFP were disrupted by the Δsat1 mutation (Figure 2C). Fourth, expression of Ryh1Q70L, a constitutively GTP-bound mutant of Ryh1 (Figure S2; [21]), alleviated the Δsat1 and Δsat4 phenotypes (Figures 2D). Lastly, Sat4 was co-precipitated with the Ryh1G23A mutant protein (Figure 2E), an equivalent of RasG15A that cannot bind GTP/GDP (Figure S2B); the nucleotide-free form of GTPase is an intermediate of the nucleotide exchange reaction and therefore, stably binds to its GEF [22]. Together, these results indicate that the Ryh1 GTPase and its putative GEF, the Sat1-Sat4 complex, positively regulate TORC2–Gad8 signaling.
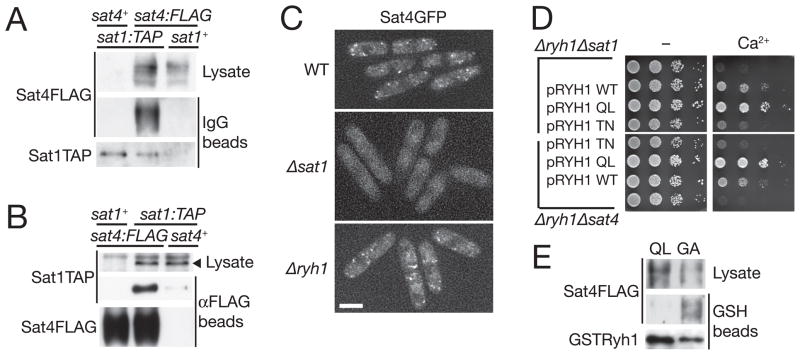
Figure 2. Sat1 and Sat4 form a complex that may function as GEF for the Ryh1 GTPase.
(A, B) Physical association of the Sat1 and Sat4 proteins. TAP-tagged Sat1 (A) and FLAG-tagged Sat4 (B) were affinity-purified from the cell lysate of sat1:TAP sat4:FLAG, sat1:TAP and sat4:FLAG strains, and the Sat1 and Sat4 proteins in the lysate and the purified fractions were detected by immunoblotting. (C) Sat4 is delocalized in the Δsat1 mutant. The chromosomal sat4 gene was tagged with the GFP sequence in wild type, Δsat1 and Δryh1 strains for fluorescence microscopy. Scale bar, 5 μm. (D) The GTP-locked mutant of ryh1 complements the Δsat1 and Δsat4 phenotype. Serial dilutions of the Δryh1 Δsat1 and Δryh1 Δsat4 strains transformed by plasmids carrying the wild-type, Q70L or T25N mutant of ryh1 were spotted onto YES agar with or without 50 mM CaCl2. (E) The nucleotide-free G23A mutant of Ryh1 can stably bind to Sat4. Ryh1Q70L (“QL”) and Ryh1G23A (“GA”) fused to GST were expressed in the sat4:FLAG strain and purified by glutathione-beads, and the proteins were detected by immunoblotting.
GTP-bound form of Ryh1 stimulates TORC2–Gad8 signaling
Reduced TORC2-dependent phosphorylation of Gad8 in strains lacking the Ryh1 GTPase or its GEF suggested that the GTP-bound form of Ryh1 is stimulatory to TORC2 signaling. In accord with such a model, ectopic expression of the GTP-locked Ryh1Q70L protein was sufficient to increase Ser-546 phosphorylation of Gad8 (Figure 3A). On the contrary, expression of Ryh1T25N, a GDP-bound mutant form [21], reduced the Gad8 phosphorylation. In strains that lack the essential TORC2 subunits, Ste20 or Sin1 [12], Ryh1Q70L could not induce the Gad8 phosphorylation (Figure 3B, left panels), corroborating that Ryh1 stimulates the TORC2-dependent phosphorylation of Gad8. As expected, a mutation to the Raptor-like TORC1 subunit, mip1-310 [23,24], did not compromise Gad8 activation by Ryh1Q70L (Figure 3B right panels; Figure S3A). In addition, Δryh1 and ryh1QL mutations did not significantly affect phosphorylation of the ribosomal S6 proteins (Figure S3B), which is under the regulation by TORC1 [25]. These results suggest that the Ryh1 GTPase specifically regulates TORC2, but not TORC1 signaling, at least under the conditions tested.
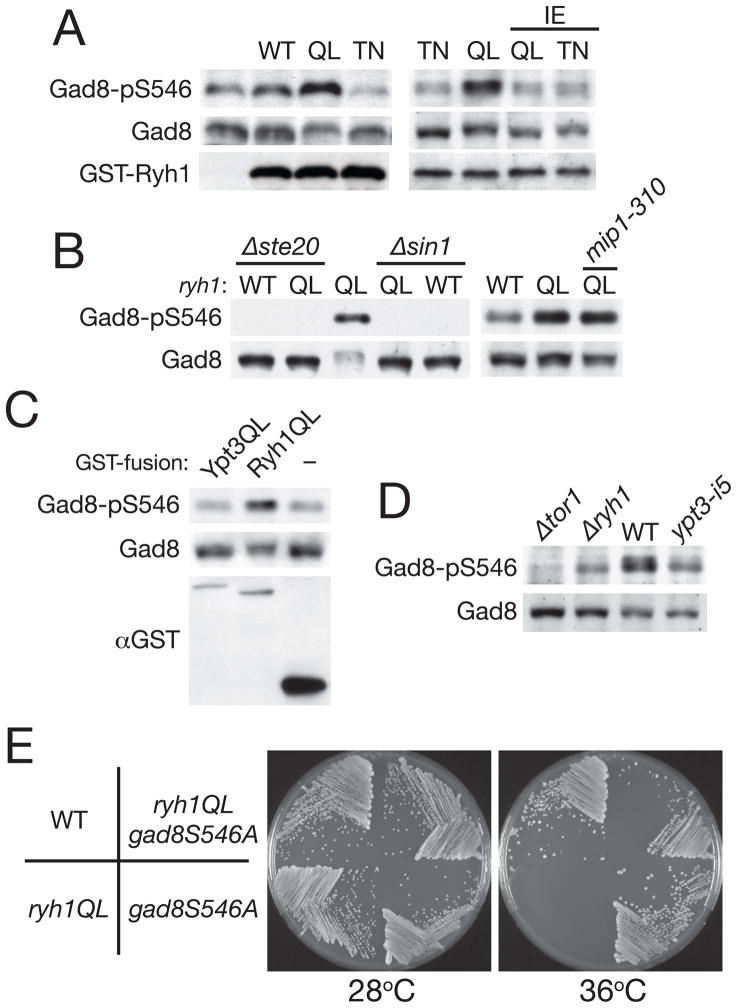
Figure 3. The GTP-bound form of Ryh1 stimulates TORC2–Gad8 signaling.
(A) TORC2-dependent phosphorylation of Gad8 Ser-546 is stimulated by the GTP-bound form of Ryh1 through its effector domain. A wild type strain was transformed with pREP1-derived plasmids to express GST, or GST fused to wild-type and mutant Ryh1 proteins. The transformants were cultured in thiamine-free medium to induce the plasmid genes from the thiamine-repressible nmt1 promoter, and their lysate was analyzed by anti-phospho-Ser546, anti-Gad8 and anti-GST immunoblotting. GST-Ryh1 mutants used were GTP-locked Q70L (“QL”), GDP-locked T25N (“TN”), effector domain defective I44E (“IE”), as well as Q70L I44E and T25N I44E double mutants. (B) Ryh1 regulates TORC2-dependent phosphorylation of Gad8.Δ ste20, Δste20 ryh1Q70L, ryh1Q70L, ryh1Q70L Δsin1, and Δsin1 strains were grown in YES medium at 30°C, and their lysate was analyzed by immunoblotting to detect Gad8 phosphorylated at Ser-546 (left panels). Wild type, ryh1Q70L and ryh1Q70L mip1-310 strains were grown at 28°C and examined for Gad8 phosphorylation at Ser-546 (right panels). (C) Ypt3 GTPase does not stimulate TORC2-dependent phosphorylation of Gad8. Expression of GST, GST-Ryh1Q70L or GST-Ypt3Q69L was induced in wild-type cells using the thiamine-repressive nmt1 promoter, followed by immunoblotting with anti-phospho-Ser546, anti-Gad8 and anti-GST antibodies. (D) TORC2 signaling is compromised in the ypt3-i5 mutant. Δtor1, Δryh1, wild-type, and ypt3-i5 strains grown in YES medium at 25°C were examined for Gad8 phosphorylation at Ser-546. (E) The temperature-sensitive phenotype of the ryh1Q70L mutant is complemented by the gad8S546A mutation. Wild-type, ryh1Q70L, gad8S546A, and ryh1Q70L gad8S546A strains were grown on YES agar medium at 28°C and 36°C.
Although genetic interaction implied an overlapping function between Ryh1 and another Rab-family GTPase, Ypt3 [21], the GTP-locked Ypt3Q69L failed to increase the phosphorylation of Gad8 (Figure 3C), suggesting that GTP-bound Ypt3 cannot directly stimulate TORC2 signaling. On the other hand, Gad8 Ser-546 phosphorylation was reduced in the ypt3-i5 mutant [26], like in the Δryh1 mutant (Figure 3D); the aberrant cellular localization of Ryh1 in the ypt3-i5 mutant [21] might hamper the normal regulation of Ryh1 and its function in TORC2–Gad8 signaling. Consistent with such a possibility, the compromised Gad8 phosphorylation in ypt3-i5 can be restored by expressing GTP-bound Ryh1Q70L (Figure S3C).
Cycling between the GDP- and GTP-bound forms, Rab GTPases mediate multiple steps of intracellular membrane traffic [27]. In contrast, we found that activation of TORC2 signaling was not dependent on the nucleotide cycle of Ryh1, because phosphorylation of Gad8 Ser-546 was augmented in a strain of which chromosomal ryh1+ locus was replaced by ryh1Q70L (“QL” in Figure 3B). This strain, which expresses only GTP-locked Ryh1Q70L, showed a temperature-sensitive growth defect (Figure 3E). However, this phenotype was suppressed by alanine substitution of Gad8 Ser-546, suggesting that hyper-phosphorylation of Gad8 is responsible for the growth phenotype of the ryh1QL mutant. This genetic interaction between ryh1QL and gad8S546A further confirms the role of Ryh1 in TORC2–Gad8 signaling in vivo.
Physical interaction of Ryh1 GTPase with TORC2
As predicted from the Ryh1-mediated TORC2 regulation described above, physical interaction between Ryh1 and TORC2 was detectable; affinity-purification of Ryh1 expressed as GST fusion in S. pombe resulted in co-purification of Tor1 (Figure 4A) as well as the other TORC2 subunits, Sin1 and Ste20 (Figure 4B). Interaction of GST-Ryh1 with Tor1 was not dependent on the other TORC2 subunits (Figure 4A). Co-precipitation of Tor1 with GTP-locked Ryh1Q70L and GDP-locked Ryh1T25N was also detected, with more Tor1 bound to Ryh1Q70L in multiple experiments (Figure 4C). However, the enhanced interaction of Ryh1Q70L with Tor1 was not apparent in some experiments, possibly due to the fluctuation in the expression level of the plasmid-borne GST-Ryh1 genes. Therefore, to further examine whether the Ryh1-Tor1 interaction involves GTP-dependent binding of the Ryh1 GTPase to its “effector”, we next introduced the I44E mutation in the conserved effector domain [28] of Ryh1 (Figure S2B). This mutation has been successfully used to disrupt interaction between Rab-family GTPases and their effectors [29,30]. The I44E mutation in Ryh1 and Ryh1Q70L reproducibly decreased the amount of bound Tor1 (Figure 4C); signal quantification indicated that Tor1 co-precipitated with GST-Ryh1I44E and GST-Ryh1Q70L/I44E was only 30~40% of that bound to wild-type GST-Ryh1. On the other hand, the interaction of GDP-locked Ryh1T25N with Tor1 was not affected by the I44E mutation, consistent with the idea that the effector domain mediates GTP-dependent interaction of small GTPases with their effectors. Importantly, the I44E mutation completely blocked the ability of Ryh1Q70L to induce the Gad8 phosphorylation (Figure 3A, right panel), suggesting that the interaction mediated by the effector domain of Ryh1 is required to stimulate TORC2–Gad8 signaling.
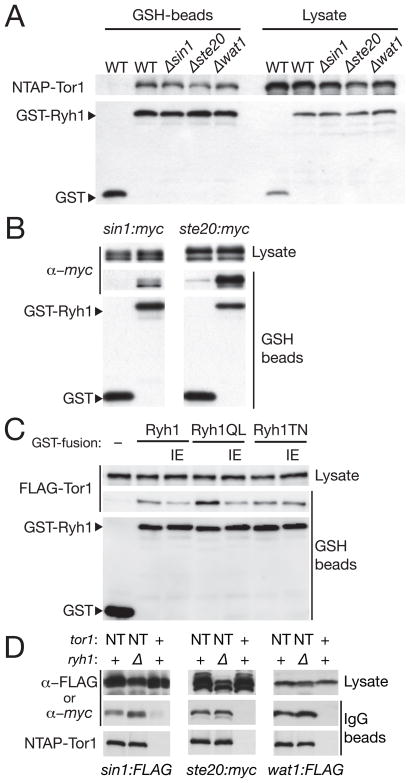
Figure 4. Physical interaction of Ryh1 GTPase with TORC2.
(A) Interaction of Ryh1 with Tor1 in the presence and absence of other TORC2 subunits. In the NTAP:tor1 strain, expression of GST or GST-Ryh1 was induced from the nmt1 promoter in EMM medium without thiamine, and the proteins were affinity-purified onto glutathione (GSH)-Sepharose beads, followed by immunoblotting. The experiment was performed in the wild-type, Δsin1, Δste20 and Δwat1 backgrounds. (B) Co-purification of TORC2 subunits with GST-Ryh1. GST or GST-Ryh1 proteins were expressed in sin1:myc and ste20:myc strains, followed by purification with GSH-beads. Co-purification was detected by immunoblotting with anti-myc antibodies. (C) The I44E mutation to the effector domain of Ryh1 partially compromises the interaction between Ryh1 and Tor1. GST alone or GST-fused to the wild-type or mutant Ryh1 proteins were expressed in the FLAG:tor1 strain and affinity-purified by GSH-beads. Crude cell lysate and proteins bound to the beads were analyzed by immunoblotting. The Ryh1 mutations used were I44E (“IE”), GTP-locked Q70L (“QL”), GDP-locked T25N (“TN”) as well as Q70L I44E and T25N I44E double mutations. (D) The Δryh1 mutation does not impair the integrity of TORC2. TORC2 was affinity-purified onto IgG-Sepharose beads from tor1+ (“+”) and NTAP:tor1 (“NT”) strains carrying the sin1:FLAG, ste20:myc or wat1:FLAG in both ryh1+ (“+”) and Δryh1 (“Δ”) backgrounds. Proteins bound to the beads and those in the cell lysate were detected by immunoblotting.
The Tor1 fragment composed of only the N-terminal HEAT repeat region (amino acid residues 1-1360; 1360Δ C) was found to co-precipitate with GST-Ryh1 (Figure S4), However, unlike the full-length Tor1 (Figure 4C), the amount of the co-precipitated 1360ΔC fragment was not reduced by the Ryh1I44E mutation. We also tested whether the Ryh1 GTPase affects the integrity of TORC2, by examining association between Tor1 and the other TORC2 subunits in wild-type and Δryh1 mutant strains. The amounts of Sin1, Ste20 and Wat1 associated with Tor1 were comparable between the two strains (Figure 4D), suggesting that Ryh1 is not required for stable TORC2 formation.
Ryh1-dependent regulation of the TORC2–Gad8 pathway
The autophosphorylation assay and the in vitro Tor1 kinase assay using PHAS-I as substrate [31] showed that Tor1 isolated from wild-type, Δryh1, Δsat1 and Δsat4 cells had comparable catalytic activity (Figure 5A). Thus, it is likely that Ryh1 stimulates TORC2–Gad8 signaling without increasing the intrinsic kinase activity of Tor1. Since the Ryh1 GTPase is known to localize on Golgi/endosomes to play a role in intracellular protein transport [17,21], we set out to determine by live-cell imaging whether Ryh1 affects the cellular localization of TORC2. Because of the difficulty in GFP-tagging Tor1 [8], TORC2-specific subunits, Ste20 and Bit61 [8], were fused to three copies of GFP and expressed from their chromosomal loci. No TORC2-defective phenotype was observed with the resultant strains (data not shown), reflecting the functionality of the GFP-fusion proteins. Ste20-3GFP (Figure 5B) and Bit61-3GFP (Figure S5C) showed very similar punctate signals throughout the cell surface in interphase cells, while more intense signals were at the cell division site (Figure S5A). This characteristic localization of Ste20 and Bit61 was dependent on Tor1 (Figure 5C and Figure S5C) and therefore, the observed Ste20- and Bit61-3GFP signals are likely to represent the localization of TORC2. Similar cortical distribution of TORC2 has been reported also in budding yeast [32,33]. We found that the Ste20-3GFP and Bit61-3GFP localization was not noticeably disturbed by the Δryh1, Δsat1 and Δsat4 mutations (Figure 5D; Figure S5B, C), indicating that the Ryh1 GTPase is not required for the normal cellular localization of TORC2. Although the Sin1 ortholog in budding yeast, Avo1, was proposed to be essential for targeting TORC2 to the plasma membrane [33], the Δsin1 mutation did not impair the cortical localization of S. pombe TORC2 (Figure 5C). However, TORC2 appeared more concentrated to cell tips in Δsin1 strains, as well as in strains carrying the catalytically inactive tor1-D2137A or Δgad8 mutations. Thus, TORC2-activated Gad8 kinase might promote uniform distribution of TORC2 foci throughout the cell surface.
Figure 5. Ryh1-dependent regulation of the TORC2–Gad8 pathway.
(A) The catalytic activity of Tor1 kinase is not altered in strains lacking the Ryh1 GTPase. Wild type, Δryh1, Δsat1 and Δsat4 strains carrying the NTAP:tor1+ allele were grown in YES medium, and the NTAP-Tor1 protein was affinity-purified onto IgG-Sepharose beads, followed by an in vitro kinase assay in the presence of [©-32P]ATP and PHAS-I [31]. Phosphorylated PHAS-I (32P-PHAS-I) and NTAP-Tor1 (32P-NTAP-Tor1) are presented together with immunoblotting of NTAP-Tor1 (NTAP-Tor1). The NTAP:tor1D2137A strain expressing catalytically inactive Tor1 kinase was used as a negative control (“tor1DA”). (B) Cortical localization of Ste20 in S. pombe. Z-axial images of wild-type cells carrying the ste20:3GFP allele were deconvolved, and mid- and top-section images are shown. (C) Cortical localization of Ste20 is dependent on the Tor1 protein. Δ tor1, tor1D2137A (“tor1DA”), Δ sin1 and Δ gad8 mutants carrying the ste20:3GFP allele were observed by fluorescence microscopy. Mid-section images are presented after deconvolution. (D) Cellular localization of TORC2 is not affected by the Δ ryh1, Δ sat1 and Δ sat4 mutations. Deconvolved mid-section images of Δ ryh1 ste20:3GFP, Δ sat1 ste20:3GFP and Δ sat4 ste20:3GFP mutants are shown. See also Figure S5. (E) Gad8 is localized throughout the cytoplasm in wild type and the sat mutants. Wild-type, Δ ryh1, Δ sat1 and Δ sat4 strains carrying the gad8:3GFP allele were observed by fluorescence microscopy. Mid-section images are presented after deconvolution. Exclusion of Gad8-3GFP from vacuoles is apparent in wild-type cells, but not in the Δ ryh1, Δ sat1 and Δ sat4 cells that have much smaller, fragmented vacuoles (see Figure 6A). (F) Expression of GTP-locked Ryh1 promotes physical interaction of TORC2 with Gad8. Unfused GST (“-“), GST-Ryh1Q70L (“QL”), GST-Ryh1T25N (“TN”), GST-Ryh1Q70L/I44E or GST-Ryh1T25N/I44E proteins were expressed using the nmt1 promoter in a gad8:FLAG sin1:myc strain grown in the absence of thiamine. Immunoprecipitation was performed with anti-FLAG affinity gel and co-purified Sin1myc was detected with anti-myc antibodies. Scale bars, 5 μm.
We also examined the cellular localization of Gad8 by fusing the GFP sequence to the chromosomal gad8+ gene in wild-type, Δryh1, Δsat1 and Δsat4 strains. The Gad8-3GFP signal was diffused throughout the cytoplasm but excluded from the nucleus and vacuoles in both wild-type and mutant strains tested (Figure 5E). Together, these results indicate that the Ryh1 GTPase does not significantly affect the cellular localization of TORC2 and Gad8. Therefore, regulation of their cellular localization does not appear to be part of the mechanism by which Ryh1 modulates TORC2–Gad8 signaling.
It is possible that Ryh1 promotes the TORC2-dependent phosphorylation of Gad8 by enhancing the physical interaction between TORC2 and Gad8. Association between TORC2 and Gad8 was hardly detectable in wild-type cells, probably because their interaction was too weak to be captured by co-precipitation experiments. However, after the ectopic expression of GTP-locked Ryh1Q70L, the TORC2-specific Sin1 subunit was detected with immunoprecipitated Gad8 kinase (Figure 5F). Such interaction was not observed after expression of GDP-locked Ryh1T25N, or Ryh1Q70L with the I44E effector domain mutation, conforming to their inability to stimulate the TORC2-dependent Gad8 phosphorylation (Figure 3A). These results suggest that the activation mechanism of TORC2 signaling by Ryh1 involves enhanced interaction between TORC2 and its substrate Gad8.
Ryh1 GTPase and the TORC2–Gad8 pathway are important for vacuolar integrity
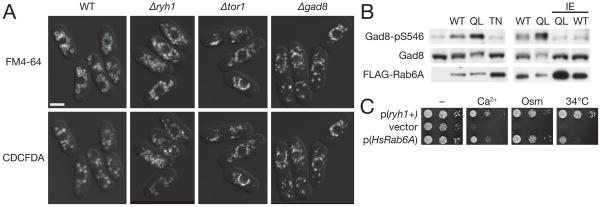
When stained by the membrane dye FM4-64 and the CDCFDA dye that fluoresces in acidic vacuolar compartments, smaller fragmented vacuoles were observed in Δryh1 cells in comparison with those in wild-type cells (Figure 6A). As expected, Δsat1 and Δsat4 mutant cells exhibited similar vacuolar defects (data not shown). We further discovered that vacuoles were fragmented and smaller also in cells lacking the functional TORC2 or Gad8 kinase, though to a lesser extent than in Δryh1 cells (Figure 6A). Thus, both Ryh1 GTPase and the TORC2–Gad8 pathway appear to be implicated in a common cellular function, vacuolar integrity, consistent with their regulatory interactions described above. Yeast vacuoles play important roles in cellular resistance to high osmolarity and extracellular Ca2+ [34,35], and the vacuolar defects in mutants without functional Ryh1, TORC2 or Gad8 kinase may be responsible for their hyper-sensitivity to Ca2+ and osmostress (Figure 1B).
Figure 6. Vacuolar defects in ryh1, TORC2 and gad8 mutants.
(A) Cells lacking functional Ryh1 GTPase, TORC2 and Gad8 have smaller, fragmented vacuoles. Vacuolar membrane was visualized in wild type, Δ ryh1, Δ tor1 and Δ gad8 by the fluorescent dye FM4-64, together with the CDCFDA dye that generates fluorescence in acidic vacuolar compartments. Z-axial images were deconvolved, and mid-section images are shown. Scale bar, 5 μm (B) Human Rab6 GTPase can stimulate TORC2–Gad8 signaling in S. pombe. The Δ ryh1 strain was transformed by the empty pREP1 vector, or by the plasmid carrying wild-type Rab6A, GTP-locked Rab6A(Q72L), GDP-locked Rab6A(T27N), effector domain-defective Rab6A(I46E) or Rab6A(Q72L/I46E). The transformants were grown in thiamine-free EMM medium to induce the plasmid genes, and their lysate was analyzed by anti-phospho-Ser546 and anti-Gad8 immunoblotting. (C) Human Rab6 can substitute the Ryh1 function in S. pombe. Serial dilutions of Δ ryh1 cells carrying the empty pREP1 vector, pREP1-ryh1+, or pREP1-HsRab6A were spotted onto a YES plate at 25°C (“-”), 34°C, or those containing either 50 mM CaCl2 or 0.4 M KCl at 25°C.
Human Rab6 can activate TORC2–Gad8 signaling in fission yeast
Because of the high sequence homology of Ryh1 to human Rab6 [17], we tested whether human Rab6 can stimulate TORC2–Gad8 signaling in S. pombe. As shown in Figure 6B, Gad8 Ser-546 phosphorylation was increased by expression of human Rab6A, and even more by the GTP-locked Rab6 mutant, Rab6A(Q72L) (Figure S2; [36]). In contrast, the GDP-bound Rab6A(T27N) mutant failed to increase the Gad8 phosphorylation level. Moreover, the I46E mutation to the effector domain (Figure S2; [29,30]) prevented Rab6A and Rab6A(Q72L) from stimulating Gad8 Ser-546 phosphorylation (Figure 6B, right panel). These observations suggest that the ability to induce TORC2 signaling appears to be conserved between Ryh1 and human Rab6. In addition, the human Rab6A gene complemented the temperature-sensitive growth [17] and other stress-sensitivity (Figure 6C) of the Δryh1 mutant, demonstrating the evolutionary conservation of the cellular function of Rab6.
Discussion
A Ras superfamily GTPase, Rheb, is a key regulator of TORC1 both in mammals [37,38] and fission yeast [39], though the molecular mechanism of TORC1 activation by Rheb remains to be understood [5–7]. More recently, a set of Ras-family Rag GTPases have been identified as mediators of nutrient signaling to TORC1 through its Raptor subunit [40,41]. Our findings reported here strongly suggest that another GTPase in the Ras superfamily, Ryh1/Rab6, interacts with TORC2 and modulates TORC2-dependent activation of the AGC-family kinase Gad8 in fission yeast. Thus, regulation by Ras-family GTPases may be a common theme in the control of the TOR kinase, and the distinct subunit compositions of TORC1 and TORC2 may determine the specificity of the regulation by different GTPases.
The following genetic, biochemical and cell-biological observations consistently indicate that the Rab GTPase Ryh1 has a role as a positive regulator of TORC2–Gad8 signaling in S. pombe. First, mutations to Ryh1 or its putative GEF, Sat1-Sat4, significantly reduce the TORC2-dependent phosphorylation of the hydrophobic motif in Gad8 kinase. The residual Gad8 phosphorylation detected in the absence of Ryh1 implies an additional TORC2 activator, possibly other Rab-family GTPases, though the Δypt4, Δypt7 and Δypt71 mutants showed no defect in TORC2 signaling (Table S4). Second, expression of GTP-locked Ryh1Q70L is sufficient to induce the Gad8 phosphorylation, while GDP-locked Ryh1T25N has a dominant-negative effect on TORC2–Gad8 signaling. Third, Ryh1 GTPase and TORC2 show physical interaction, which appears to be partly dependent on the effector domain of Ryh1. The effector domain is essential for GTP-bound Ryh1 to stimulate TORC2–Gad8 signaling. However, association of Tor1 with GDP-locked Ryh1T25N was also detected, suggesting that Ryh1 does not need to bind GTP to interact with Tor1. Interestingly, nucleotide-independent interaction of the Rheb GTPase with TOR kinase has also been reported [37,38]. Fourth, in a strain expressing Ryh1Q70L, increased interaction between Gad8 and the TORC2 subunit Sin1 is detectable, implying that GTP-bound Ryh1 promotes physical interaction between TORC2 and Gad8. A recent report proposes that the Rheb GTPase regulates TORC1 activation also by promoting the interaction between TORC1 and its substrate, 4E-BP1 [7]. Fifth, mutational inactivation of Ryh1 or the TORC2–Gad8 pathway results in similar phenotypes, such as stress sensitivity and vacuolar defects, corroborating that Ryh1, TORC2 and Gad8 are involved in a common cellular process. Lastly, TORC2 is mostly localized to the cellular cortex (Figure 5B), where a fraction of GTP-locked Ryh1QL has also been detected [21]; therefore, we speculate that activation of TORC2 by GTP-bound Ryh1 takes place at the plasma membrane. It is conceivable that certain plasma membrane lipid is also involved in TORC2 activation, as TORC2 in higher eukaryotes appears to be under the regulation of the phosphatidylinositol 3-kinase [3,42]. Our current effort focuses on reconstituting the Ryh1-dependent activation of TORC2 in vitro.
We have noticed that some of the Δryh1 phenotypes are more severe than those of the TORC2 and gad8 mutants, such as temperature-sensitivity [12,21](data not shown) and vacuolar fragmentation. In addition, overexpression of the Ykt6 SNARE partially complements the temperature-sensitive growth of Δryh1 cells [21] but not their defect in TORC2–Gad8 signaling (data not shown). The simplest interpretation of these observations is that, in addition to TORC2 signaling, Ryh1 GTPase has an additional function in intracellular membrane traffic, such as a role in retrograde traffic from endosome to the Golgi [21]. Other observations also suggest that the mechanism of TORC2 regulation by Ryh1 is not through its function in intracellular trafficking. First, control of the intracellular membrane traffic by Rab GTPases is dependent on their perpetual cycling between the GTP- and GDP-bound states [27]. However, activation of TORC2 signaling is not dependent on the nucleotide cycle of Ryh1, because activation of TORC2–Gad8 signaling is induced in cells expressing only GTP-locked Ryh1Q70L. Second, the cellular localization of TORC2 and Gad8 is not altered in the Δryh1 mutant, indicating that Ryh1 does not affect transport and/or localization of TORC2 and Gad8. Third, only Δryh1 and Δsat1 were isolated in our screen of the null mutant library, which include additional 163 genes implicated in intracellular membrane/protein transport (Table S4). For example, TORC2-Gad8 signaling was not altered in the mutants of the retromer subunits [43] and the Tlg2 SNARE [44] that mediate the retrograde traffic from endosome to the Golgi, the process dependent also on Ryh1 [21]. Thus, TORC2–Gad8 signaling is not readily affected by mutations in various steps of intracellular trafficking, except ypt3-i5 that alters the cellular localization of Ryh1 [21].
In mammals, insulin-activated TORC2 and its downstream AGC-family kinase, Akt, promote intracellular traffic of vesicles carrying the GLUT4 glucose transporter [3]. Recent identification of a Golgi protein, GOLPH3, as an activator of mTOR signaling in cancers also implies an association of TOR with vesicular trafficking [45]. The Rab GTPase-dependent regulation of the TORC2–Gad8 pathway in S. pombe is also indicative of a functional link between TORC2 signaling and intracellular membrane trafficking. Moreover, Ryh1 is highly homologous to human Rab6, and indeed, human Rab6 can substitute Ryh1 in S. pombe TORC2 signaling. Considering the evolutionary conservation of TOR signaling from yeast to humans, Rab GTPases may also be involved in the regulation of mammalian TORC2 to modulate vesicular transport processes during cellular responses to insulin and growth factors.
Experimental Procedures
Fission yeast strains used in this study are listed in Supplemental Table S2. Details of the S. pombe deletion library screen, immunological methods, protein interaction assays and microscopy are described in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to D. Richter, J. Del Rosario, M. Shiozaki, B. You, R. Maximo and N. Tahara for technical assistance. T. Toda kindly provided sat1-11, sat4-86 and sat7-110 mutants. K. Gould, T. Kuno, K. Nakayama, R. Sugiura, K. Takegawa, M. Yamamoto, M. Yanagida, and the National BioResource Yeast Project of Japan also provided essential reagents. We thank K. Nakashima and T. Takizawa for their help in microscopy, T. Powers for discussion, and H. Takagi and H. Maki for encouragement. This work was supported by grants from NIH (GM059788) and the UC Cancer Research Coordinating Committee to K.S., and Grant-in-Aid for Scientific Research (C)21570198 awarded to S. Morigasaki from the Japan Society for the Promotion of Science. S. Morigasaki is a International Research Fellow of the Nara Institute of Science and Technology Global COE Program funded by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Jacinto E, Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem J. 2008;410:19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- 3.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2008;283:30482–30492. doi: 10.1074/jbc.M803348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi T, Hatanaka M, Nagao K, Nakaseko Y, Kanoh J, Kokubu A, Ebe M, Yanagida M. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12:1357–1370. doi: 10.1111/j.1365-2443.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- 9.Pearce LR, Huang X, Boudeau J, Pawłowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jenö P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuo T, Kubo Y, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 2003;22:3073–3083. doi: 10.1093/emboj/cdg298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda K, Morigasaki S, Tatebe H, Tamanoi F, Shiozaki K. Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle. 2008;7:358–364. doi: 10.4161/cc.7.3.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilti N, Baumann D, Schweingruber AM, Bigler P, Schweingruber ME. Gene ste20 controls amiloride sensitivity and fertility in Schizosaccharomyces pombe. Curr Genet. 1999;35:585–592. doi: 10.1007/s002940050456. [DOI] [PubMed] [Google Scholar]
- 14.Kawai M, Nakashima A, Ueno M, Ushimaru T, Aiba K, Doi H, Uritani M. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr Genet. 2001;39:166–174. doi: 10.1007/s002940100198. [DOI] [PubMed] [Google Scholar]
- 15.Weisman R, Choder M. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J Biol Chem. 2001;276:7027–7032. doi: 10.1074/jbc.M010446200. [DOI] [PubMed] [Google Scholar]
- 16.Kominami K, Seth-Smith H, Toda T. Apc10 and Ste9/Srw1, two regulators of the APC-cyclosome, as well as the CDK inhibitor Rum1 are required for G1 cell-cycle arrest in fission yeast. EMBO J. 1998;17:5388–5399. doi: 10.1093/emboj/17.18.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengst L, Lehmeier T, Gallwitz D. The ryh1 gene in the fission yeast Schizosaccharomyces pombe encoding a GTP-binding protein related to ras, rho and ypt: structure, expression and identification of its human homologue. EMBO J. 1990;9:1949–1955. doi: 10.1002/j.1460-2075.1990.tb08322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 19.Aiba H, Kawaura R, Yamamoto E, Yamada H, Takegawa K, Mizuno T. Isolation and characterization of high-osmolarity-sensitive mutants of fission yeast. J Bacteriol. 1998;180:5038–5043. doi: 10.1128/jb.180.19.5038-5043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Sugiura R, Ma Y, Kita A, Deng L, Takegawa K, Matsuoka K, Shuntoh H, Kuno T. Genetic and functional interaction between Ryh1 and Ypt3: two Rab GTPases that function in S. pombe secretory pathway. Genes Cells. 2006;11:207–221. doi: 10.1111/j.1365-2443.2006.00935.x. [DOI] [PubMed] [Google Scholar]
- 22.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 23.Shinozaki-Yabana S, Watanabe Y, Yamamoto M. Novel WD-repeat protein Mip1p facilitates function of the meiotic regulator Mei2p in fission yeast. Mol Cell Biol. 2000;20:1234–1242. doi: 10.1128/mcb.20.4.1234-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27:3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakashima A, Sato T, Tamanoi F. Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J Cell Sci. 2010;123:777–786. doi: 10.1242/jcs.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H, Sugiura R, Wu W, Fujita M, Lu Y, Sio SO, Kawai R, Takegawa K, Shuntoh H, Kuno T. Role of the Rab GTP-binding protein Ypt3 in the fission yeast exocytic pathway and its connection to calcineurin function. Mol Biol Cell. 2002;13:2963–2976. doi: 10.1091/mbc.01-09-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick F, Wittinghofer A. Interactions between Ras proteins and their effectors. Curr Opin Biotechnol. 1996;7:449–456. doi: 10.1016/s0958-1669(96)80123-6. [DOI] [PubMed] [Google Scholar]
- 29.Echard A, Jollivet F, Martinez O, Lacapère JJ, Rousselet A, Janoueix-Lerosey I, Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 30.Recacha R, Boulet A, Jollivet F, Monier S, Houdusse A, Goud B, Khan AR. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 2009;17:21–30. doi: 10.1016/j.str.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Petersen J, Nurse P. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol. 2007;9:1263–1272. doi: 10.1038/ncb1646. [DOI] [PubMed] [Google Scholar]
- 32.Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7:1819–1830. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009;20:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham KW, Fink GR. Ca2+ transport in Saccharomyces cerevisiae. J Exp Biol. 1994;196:157–166. doi: 10.1242/jeb.196.1.157. [DOI] [PubMed] [Google Scholar]
- 36.Yamane J, Kubo A, Nakayama K, Yuba-Kubo A, Katsuno T, Tsukita S, Tsukita S. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp Cell Res. 2007;313:3472–3485. doi: 10.1016/j.yexcr.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 38.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 39.Urano J, Comiso MJ, Guo L, Aspuria PJ, Deniskin R, Tabancay AP, Kato-Stankiewicz J, Tamanoi F. Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol Microbiol. 2005;58:1074–1086. doi: 10.1111/j.1365-2958.2005.04877.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.