Abstract
Mild cognitive impairment (MCI), often a prodromal phase of Alzheimer’s disease (AD), is frequently considered to be good target for early diagnosis and therapeutic interventions of AD. Recent emergence of reliable network characterization techniques has made it possible to understand neurological disorders at a whole-brain connectivity level. Accordingly, we propose an effective network-based multivariate classification algorithm, using a collection of measures derived from white-matter (WM) connectivity networks, to accurately identify MCI patients from normal controls. An enriched description of WM connections, utilizing six physiological parameters, i.e., fiber count, fractional anisotropy (FA), mean diffusivity (MD), and principal diffusivities (λ1, λ2, λ3), results in six connectivity networks for each subject to account for the connection topology and the biophysical properties of the connections. Upon parcellating the brain into 90 regions-of-interest (ROIs), these properties can be quantified for each pair of regions with common traversing fibers. For building an MCI classifier, clustering coefficient of each ROI in relation to the remaining ROIs is extracted as feature for classification. These features are then ranked according to their Pearson correlation with respect to the clinical labels, and are further sieved to select the most discriminant subset of features using a SVM-based feature selection algorithm. Finally, support vector machines (SVMs) are trained using the selected subset of features. Classification accuracy was evaluated via leave-one-out cross-validation to ensure generalization of performance. The classification accuracy given by our enriched description of WM connections is 88.9%, which is an increase of at least 14.8% from that using simple WM connectivity description with any single physiological parameter. A cross-validation estimation of the generalization performance shows an area of 0.929 under the receiver operating characteristic (ROC) curve, indicating excellent diagnostic power. It was also found, based on the selected features, that portions of the prefrontal cortex, orbitofrontal cortex, parietal lobe and insula regions provided the most discriminant features for classification, in line with results reported in previous studies. Our MCI classification framework, especially the enriched description of WM connections, allows accurate early detection of brain abnormalities, which is of paramount importance for treatment management of potential AD patients.
Keywords: Enriched connectivity, white-matter (WM), connectivity network, mild cognitive impairment (MCI), Alzheimer’s disease (AD), support vector machines (SVMs)
1. Introduction
Mild cognitive impairment (MCI) is commonly defined as subtle but measurable memory disorder - a stage between normal forgetfulness (due to aging) and dementia. Studies suggest that MCI patients tend to progress to probable Alzheimer’s disease (AD) at a rate of approximately 10% to 15% per year (Misra et al., 2009; Grundman et al., 2004; Peterson et al., 2001) compared with healthy controls who develop dementia at a rate of 1% to 2% per year (Bischkopf. et al., 2002). MCI is difficult to diagnose due to the subtlety of cognitive impairment, especially in high functioning individuals who are able to maintain a positive public or professional profile without showing obvious cognitive impairment. It is hence crucial to develop algorithms that can identify subtle diagnostic biomarkers for early detection of MCI patients, in order to provide possible early treatment and thus delay the transition from MCI to AD.
Models of whole-brain connectivity, which comprise networks of brain regions connected either by anatomical tracts or functional associations, have drawn a great deal of interest recently due to the increasing reliability of network characterization through neurobiologically meaningful and computationally efficient measures (Bassett and Bullmore, 2006; Hagmann et al., 2008; Sporns and Zwi, 2004). Characterization of the global architecture or topological property of anatomical connectivity patterns in the human brain can provide new and valuable insights into the association between brain functional deficits and the underlying structural disruption related to brain disorders (Sporns et al., 2005). Recent applications of whole-brain connectivity networks include exploring the anatomical and functional connectivity relationship between brain regions (Honey et al., 2009; Zhou et al., 2006) and revealing connectivity abnormalities in neurological and psychiatric disorders (Basset et al., 2008; Stam et al., 2007; Wang et al., 2009).
Understanding of brain anatomical circuitry has been experiencing considerable progress due to the development of diffusion tensor imaging (DTI), which is capable of delineating white-matter (WM) fiber bundles through the characterization of the underlying water molecule diffusion (Gong et al., 2009). WM tracts between pairs of brain regions are routinely observed in vivo using diffusion tractography (also called fiber tracking) to model a global connectivity network of macroscopic polysynaptic fiber bundles in the brain (Gong et al., 2009; Hagmann et al., 2007; Iturria-Medina et al., 2007). Derived parameters such as fractional anisotropy (FA) and mean diffusivity (MD) are widely used in statistical analyses to localize brain changes related to growth, aging and neurodegenerative disease (Dineen et al., 2009; Kochunov et al., 2007; Rose et al., 2007; Zhang et al., 2007). Nevertheless, connectivity based solely on the changes of a single physiological parameter might not be sufficient for identifying subtle differences between groups. An enriched description of WM connections, which is more sensitive to WM microstructural changes, is needed.
In this study, we propose an effective structural connectivity network-based multivariate classification framework to accurately identify MCI patients from subjects undergoing normal aging. The key of our approach involves an enriched description of WM connections via utilizing six physiological parameters, i.e., fiber count, FA, MD, and principal diffusivities (λ1, λ2, λ3), resulting in six types of connectivity networks for each subject to account for the connection topology and the biophysical properties of connection. The current study is the first attempt to characterize brain WM integrity using an enriched connectivity description quantified by DTI fiber tractography for the purpose of identification of patients affected by brain disease.
Network statistics computed from the above-mentioned networks are fed as features into a feature selection mechanism to select the most discriminant subset of features for training of support vector machine (SVM) based classifiers. Classification accuracy in this study was evaluated via leave-one-out cross-validation to ensure performance generalization. The classification accuracy obtained by the proposed method is 88.9%, which is at least 14.8% higher than that using a simple WM connection description with any single physiological parameter. Specifically, we note that the area under ROC curve (AUC) is 0.929, indicating excellent diagnostic power of the proposed framework.
The rest of the paper is organized as follows: Section 2 furnishes information on the image dataset, image acquisition protocols, and the post-processing pipeline, followed by a comprehensive description of the proposed classification framework. Advantages of the proposed MCI classification framework are evaluated in Section 3. Findings, methodological issues and limitations of our proposed framework are discussed extensively in Section 4. Section 5 concludes this paper.
2. Method and Materials
2.1. Data Acquisition and Post-Processing
The present study involved 27 participants (10 MCI patients and 17 socio-demographically matched healthy controls) who were recruited by the Duke-UNC Brain Imaging and Analysis Center, North Carolina, USA. Informed consent was obtained from all participants, and the experimental protocols were approved by the institutional ethics board. Confirmation of diagnosis for all subjects was made via expert consensus panels at the Joseph and Kathleen Bryan Alzheimer’s Disease Research Center (Bryan ADRC) and the Department of Psychiatry at Duke University Medical Center. Diagnosis was based upon available data from a general neurological examination, neuropsychological assessment evaluation, collateral and subject symptom and functional capacity reports. Furthermore, all MCI subjects met the following inclusion criteria: 1) age > 55 years and any race; 2) recent worsening of cognition, but still functioning independently; 3) score between 24 and 30 on the MMSE; 4a.) score ≤ −1.5 SD on at least two Bryan ADRC cognitive battery memory tests for single-domain amnestic MCI; or 4b.) score ≤ −1.5 SD on at least one of the formal memory tests and score ≤ −1.5 SD on at least one other cognitive domain task (e.g., language, visuospatial-processing, or judgment/executive function) for multi-domain MCI; 5) does not meet the NINCDS-ADRDA or DSM-IV-TR criteria for dementia; 6) baseline Hachinski score of 4 or lower; 7) no psychological symptoms or history of depression; 8) capacity to give informed consent and follow study procedures. All healthy controls met the following criteria: 1) age > 55 years and any race; 2) adequate visual and auditory acuity to properly complete neuropsychological testing; 3) no self-report of neurological or depressive illness; 4) normal score on a non-focal neurological examination; 5) shows no evidence of depression based on the Diagnostic Interview Schedule port based on the Diagnostic Interview Schedule portion of the Duke Depression Evaluation Schedule; 6) a score > −1 SD on any formal memory tests and a score > −1 SD on any formal executive function or other cognitive test; 7) demonstrates a capacity to give informed consent and follow study procedures. Subjects were excluded from the study if they have: 1) any of the traditional MRI contraindications, such as foreign metallic implants or pacemakers; 2) documentation of other Axis I psychiatric disorders; 3) a past head injury or neurological disorder associated with MRI abnormalities, including dementia, brain tumors, epilepsy, Parkinson’s disease, demyelinating diseases, etc.; 4) any physical or intellectual disability affecting completion of assessments; 5) any prescription medication (or nonprescription drugs) with known neurological effects.
Data acquisition was performed using a 3.0 Tesla scanner (GE Signa EXCITE, GE Healthcare). Diffusion-weighted images of each participant were acquired axially parallel to the anterior and posterior commissures (ACPC) line with twenty-five-direction diffusion-weighted whole-brain volumes using diffusion weighting values, b = 0 and 1000 s/mm2, flip angle = 90°, repetition time (TR) = 17 s and echo time (TE) = 78 ms. The imaging matrix was 128 × 128 with a rectangular FOV of 256 × 256 mm2, resulting in a voxel dimension of 2 × 2 × 2 mm3 reconstructed resolution. A total of 72 contiguous slices were acquired. Demographic information of the participants involved in this study are shown in Table 1.
Table 1.
Demographic information of the participants involved in this study.
| Group | MCI | Normal |
|---|---|---|
| No. of subjects | 10 | 17 |
| No. of males | 5 | 8 |
| Age (mean ± SD) | 74.2 ± 8.6 | 72.1 ± 8.2 |
| Years of education (mean ± SD) | 17.7 ± 4.2 | 16.3 ± 2.4 |
| MMSE (mean ± SD) | 28.4 ± 1.5 | 29.4 ± 0.9 |
2.2. Method
2.2.1. Overview of Methodology
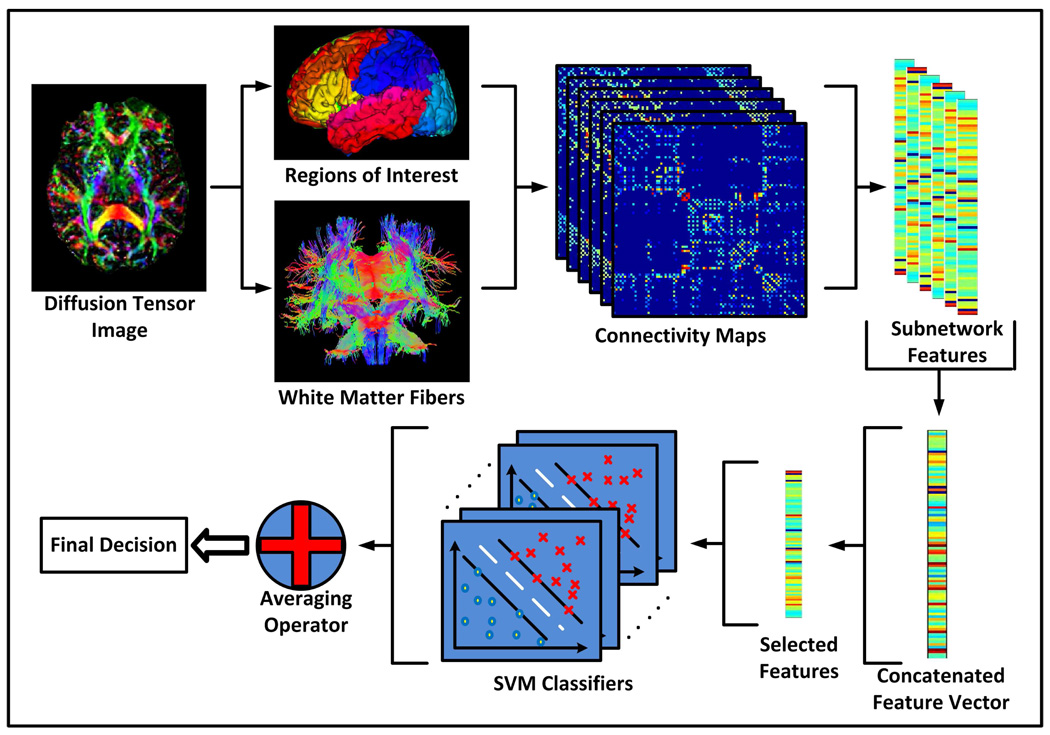
The key of the proposed classification framework involves an enriched description of WM connections utilizing six physiological parameters, i.e., fiber count, FA, MD, and principal diffusivities (λ1, λ2, λ3), resulting in six connectivity networks for each subject. The proposed MCI classification framework is shown graphically in Figure 1.
Figure 1.
Classification based on enriched description of WM connections.
Each brain image was first parcellated into 90 regions (45 for each hemisphere) by propagating the automated anatomical labeling (AAL) ROIs (Tzourio-Mazoyer et al., 2002) to each image using an efficient deformable DTI registration algorithm called F-TIMER (Yap et al., 2009, 2010) with tensor orientation corrected using the method described in (Xu et al., 2003). In F-TIMER, registration is achieved by utilizing a set of automatically determined structural landmarks via solving a soft correspondence problem. These structural landmarks are selected based on the tensors regional statistical and boundary edge information, which is grouped into an attribute vector, for each voxel, in a multiscale fashion. Upon establishing landmark correspondences, thin-plate splines are utilized to interpolate and generate a smooth, topology preserving, and dense transformation. As the registration progresses, an increasing number of voxels are permitted to participate in refining the correspondence matching. Additionally, registration in a multiscale fashion ensures that the transformation is robust to image noise and helps to alleviate the problem of local minima besides reduction in computation cost. F-TIMER is found to yield state-of-the-art performance when compared to popular methods such as DTI-TK.
Whole-brain streamline fiber tractography was then performed on each image using ExploreDTI (Leemans et al., 2009), with minimal seed point FA of 0.45, minimal allowed FA of 0.25, minimal fiber length of 20 mm, and maximal fiber length of 400 mm. The reason for choosing a relatively high FA threshold value was to extract the major matured WM fibers during the fiber tracking process. During tractography, the number of fibers passing through each pair of regions was counted. Two regions were considered as anatomically connected if fibers passing through their respective masks were present. Considering the connection between every possible pair of regions gives us the connection topology of the network. On top of the fiber count based connectivity network, averages of on-fiber FA, MD and principal diffusivity values were derived at the same time to form another five connectivity networks. These five connectivity networks shared identical connection topology as defined by the fiber count network, but conveyed different biophysical properties.
From each network, the clustering coefficient between each ROI and the remaining ROIs were extracted to form a feature vector of 90 elements. For each subject, the feature vectors of all networks were then combined to form a long concatenated feature vector. The elements in this long description vector were first ranked according to their Pearson correlation with respect to the clinical labels, and were then further sieved to select the most discriminant subset of features using the SVM-RFE algorithm (Rakotomamonjy, 2003; Guyon et al., 2004). Finally, SVMs were trained using the selected subset of features. The training process was repeated on the whole dataset in a leave-one-out fashion. Given an unseen testing sample, the final decision was determined by averaging the outcomes from all learned SVM classifiers.
2.2.2. Enriched Description of WM Connectivity
Progressive degenerative neurological disease such as Alzheimer’s disease and similar dementias exhibit subtle, spatially and temporally diffuse pathology, where the brain is damaged in a large-scale, highly connected network, rather than in one single isolated region. In view of this, designing an enriched description of interregional connections, which might be more sensitive in conveying the pathological information, is necessary for accurate diagnosis of neurological diseases. The proposed enriched description of WM connections via diffusion tractography is achieved via a collection of physiological parameters which convey rich information related to the topological and biophysical properties of the connection. This is in contrast to a simple connectivity description using a single physiological parameter that affords only limited information.
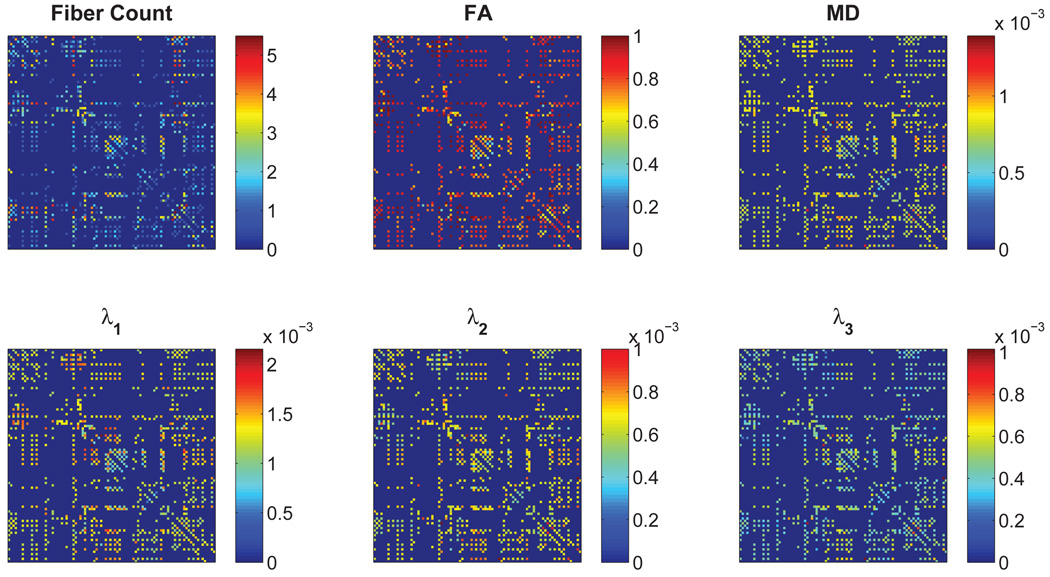
The six on-fiber physiological parameters used in this study included:
Fiber count: Number of fibers connecting a pair of regions;
FA: The average degree of anisotropy along the fibers;
MD: The average diffusivity along the fibers;
λ1: On-fiber average axial diffusivity (also called the longitudinal diffusivity);
λ2 and λ3: On-fiber average radial diffusivities (diffusivities in directions perpendicular to the axonal direction).
An example of the six connectivity networks for one subject is provided in Figure 2.
Figure 2.
Connectivity networks constructed with different physiological parameters.
2.2.3. Feature Extraction
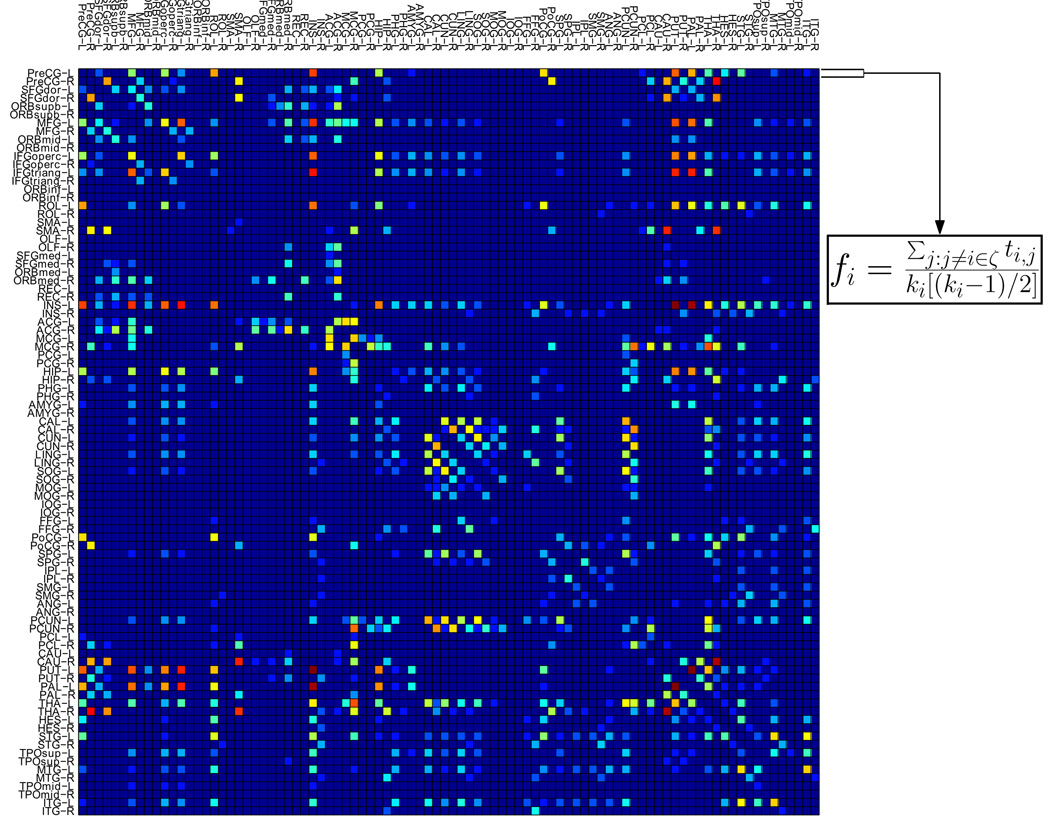
Measures in network analysis typically quantify connectivity profiles associated with the nodes and reflect the way in which these nodes are embedded in the network. Clustering coefficient (Rubinov and Sporns, 2009; Watts and Strogatz, 1998), one of the common and simple segregation measures in network analysis, quantifies the degree to which nodes in a network tend to cluster together. In this study, we utilize the weighted local clustering coefficients to extract information from the constructed networks. For each network, the weighted local clustering coefficients between each ROI and the remaining ROIs is computed as
| (1) |
where ki is the number of ROIs that are connected to the i-th ROI, ζ is the subnetwork comprising nodes directly connected to the i-th ROI, and ti,j is the parameter value between the i-th ROI and j-th ROI. Hence, for each network, a total of 90 features can be extracted (see Figure 3). For each subject, features of all networks are concatenated to form a long description vector.
Figure 3.
Extraction of clustering coefficients from a connectivity network.
2.2.4. Feature Selection
Feature selection is required for choosing an optimal subset of features from a larger pool of features in order to improve generalization performance. This is due to the fact that some features are less sensitive, irrelevant or redundant for classification, compared to others. By using the optimal subset of features, the performance of the finally constructed classifier can be improved. In this current study, we utilized a hybrid feature selection method which comprises Pearson correlation-based feature ranking and SVM-based feature selection.
The discriminative power of a feature can be qualitatively measured by its relevance to classification as well as its generalizability. The relevance of a feature to classification can be measured through its correlation with clinical labels (Fan et al., 2007b). Pearson correlation coefficient, a widely used linear correlation measure in machine learning, is used here to rank features. The larger the absolute value of the Pearson correlation coefficient, the more relevant the feature is to classification. Specifically, the Pearson correlation coefficient between a feature, f, and a clinical class label y is defined as
| (2) |
where j denotes the j-th sample in the training dataset, fj is the feature value of the j-th sample and f̅ is the mean of fj over all samples. Similarly, yj is the clinical class label (normal −1 or abnormal +1) of the j-th sample and ȳ is the mean of yj over all samples.
The generalizability of a feature can be evaluated via leave-one-out cross-validation when measuring the correlation of the feature with respect to the clinical labels via Pearson correlation coefficient (Fan et al., 2007a). For n training samples, we conservatively select the worst absolute Pearson correlation coefficient resulting from n leave-one-out correlation measurement as the effective correlation coefficient. This approach is particularly important when examining a very large number of features to minimize the effect of outliers. The formulation of this conservative principle as the generalizability of a feature f is given as (Fan et al., 2007a)
| (3) |
where g(f) is the worst absolute Pearson correlation coefficient, pj(f) is a Pearson correlation coefficient between the feature f and the clinical label y, obtained from the j-th leave-one-out case where the j-th sample is excluded.
Nevertheless, the correlation or ranking score is computed independently for each feature, without considering the correlation with other features. This inevitably causes some redundant features to be selected and eventually affects the classification performance. In order to minimize this effect, we employ a feature subset selection method, which jointly considers the discriminative power among features.
A well-known and effective wrapper-based feature selection method, namely the SVM-RFE algorithm (Rakotomamonjy, 2003; Guyon et al., 2004), is utilized in this study. In this algorithm, SVM (Vapnik, 1999) is used to evaluate the discriminative power of the selected subset of features. The SVM kernel used in this study is a Gaussian radial basic function (RBF) kernel defined as
| (4) |
where x1 and x2 are two feature vectors and σ controls the width of Gaussian kernel. The goal of SVM-RFE is to find a subset of size m among d features (m < d) which optimizes the performance of the SVM classifier. The basic principle of SVM-RFE is to ensure that the removal of a particular feature will make the classification error smallest, compared to removing other features. Note that SVM-RFE is performed via a leave-one-out procedure to estimate the generalization error with respect to the number of features and to minimize this error in order to choose the optimal combination of features.
2.2.5. Evaluation Via Cross-Validation and Bagging
In this current study, the classification performance of the proposed WM connectivity description method is evaluated using a full leave-one-out bagging cross-validation strategy to ensure a relatively unbiased estimate of the generalization power of the classifiers to new subjects. Bagging is essentially an ensemble method which improves the predictive performance of a learning algorithm based on the aggregation of a certain number of prediction models generated from bootstrap samples of the available training set (Breiman, 1996). This is accomplished by having a second stage cross-validation in each leave-one-out case to optimize the parameters used for classification. The best number of features used for classification is automatically determined by evaluating the generalization performance of the classifier through bagging cross-validation.
In each leave-one-out case, one subject is first left out as the testing subject, and the remaining subjects are used for feature extraction, feature selection and classifier training. Bagging strategy is then applied for a second round of cross-validation within the training set, to build an ensemble classifier whose parameters are automatically optimized. Specifically, for n total number of subjects involved in the study, one is left out for testing, and the remaining n − 1 are used for training. From these n − 1 samples, n − 1 different training subsets are formed by each time leaving one more sample out, giving us n − 2 subjects (bootstrap samples) in each training subset. For each subset, a bootstrap classifier is built with its performance evaluated using the second left out subject. This procedure is repeated for n − 1 times, once for each training subset. This procedure allows us to select parameters which maximize the AUC. When the completely unseen (totally left out during the entire training and parameter optimization process) test sample is to be classified, all n − 1 classifiers are used, and their outcomes are combined using an averaging operator to provide the final classification decision. This process is repeated n times, each time leaving out a different subject, finally leading to an overall bagging cross-validation classification accuracy.
2.3. Summary of Methodology
Our framework is summarized as follows.
Based on the 90-region AAL atlas (Tzourio-Mazoyer et al., 2002), whole-brain streamline tractography was performed to construct the WM connectivity network based on the number of fibers passing through each pair of regions.
Based on the common fibers connecting each region pair, on-fiber average FA, MD, and principal diffusivities were derived to construct another five connectivity networks.
For each network, the clustering coefficient for each pair of regions were determined to compute a total of 90 features. Feature vectors from all networks of each subject were concatenated to form a long feature vector to provide an enriched description of WM connections.
The elements in the concatenated vector were first ranked based on their relevance to classification using Pearson correlation coefficient, via a leave-one-out cross-validation strategy. An optimal subset of features, which was most discriminant to classification, was then selected using the SVM-RFE algorithm (Rakotomamonjy, 2003; Guyon et al., 2004).
Nonlinear SVM classifiers with Gaussian RBF kernel were constructed using the selected subset of features. The classification decision in each leave-one-out case was determined by averaging the outcomes from all the respective bootstrap classifiers. From all leave-one-out cases, a final bagging cross-validation accuracy was obtained.
3. Results
3.1. Results From Evaluation Via Cross-Validation and Bagging
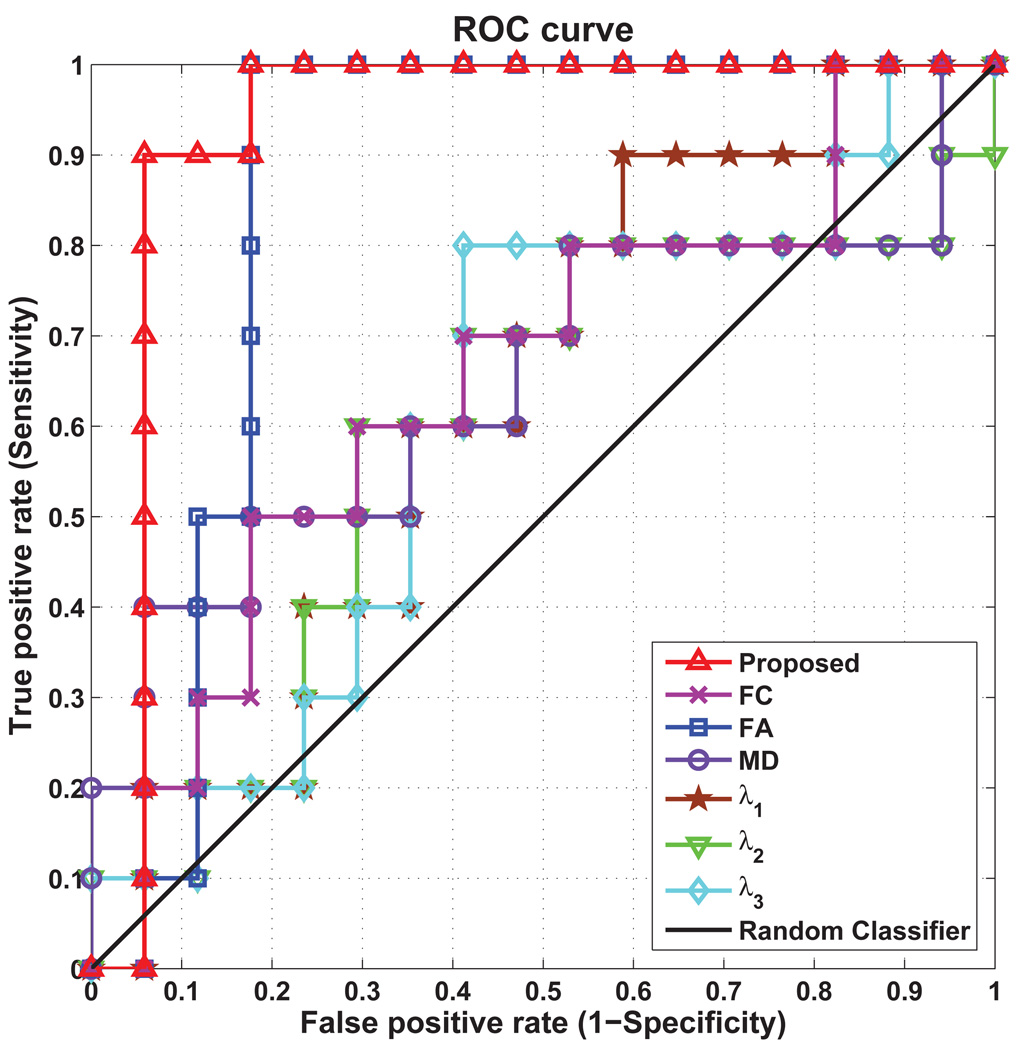
A priori knowledge of the number of features that should be used for classification is not available and this number is automatically determined as part of the bagging process. Although it generally yields slightly lower classification performance, bagging cross-validation provides a better indicator of the generalizability of a classifier. The classification accuracy by our enriched description of WM connections (with six parameters) is 88.9%, which is at least an 14.8% increment from that using any single physiological parameter. The classification performance of the enriched and simple connectivity descriptions for bagging cross-validation is summarized in Table 2.
Table 2.
Classification performance and AUC values for enriched versus simple connectivity descriptions.
| Description Parameter | Accuracy (%) | AUC |
|---|---|---|
| Proposed | 88.89 | 0.929 |
| Fiber count | 70.37 | 0.653 |
| FA | 74.07 | 0.859 |
| MD | 59.26 | 0.647 |
| λ1 | 59.26 | 0.629 |
| λ2 | 55.56 | 0.594 |
| λ3 | 59.26 | 0.612 |
The performance in terms of ROC curve is shown in Figure 4. The AUC of the proposed method is 0.929, which indicates its excellent diagnostic power. We note especially that simple connectivity description, in most of the cases, is unable to provide good generalization power, as indicated by the much smaller AUC values.
Figure 4.
ROC curves of enriched and simple connectivity descriptions evaluated via cross-validation.
An experiment was conducted to compare the classification performance of the clustering coefficient in comparison to two centrality measures, i.e., degree and betweenness centrality. In the experiment, we replaced the clustering coefficient in the feature extraction component of our method with the degree and betweenness centrality respectively, and performed the same leave-one-out training and testing procedures to compare their classification performance. The obtained classification results are summarized in Table 3. The results indicate that the clustering coefficient performed best among all compared network measures. This might be due to the fact that the weighted local clustering coefficient takes into consideration all edge weight information (including connection topology and biophysical properties) with no discretization at arbitrary cutoff level. Preserving and employing of this information might be conducive to increasing the discriminative power of the SVM classifiers in distinguishing MCI and healthy controls.
Table 3.
Classification performance and AUC values for different network measures.
| Network Measure | Accuracy (%) | AUC |
|---|---|---|
| Degree | 51.85 | 0.582 |
| Betweenness centrality | 55.56 | 0.606 |
| Clustering coefficient | 88.89 | 0.929 |
3.2. Comparison With PCA-based Feature Selection Method
In this study, performance of the proposed hybrid feature selection method was compared with a PCA-based feature selection method (Malhi and Gao, 2004). This PCA-based method finds the eigenvectors that maximize the variance of the data in the projection space (eigenspace) and the relationship between principal components and original features (Malhi and Gao, 2004). The projected data arranged based on descending eigenvalues, gives the smallest error in representation, equivalent to the largest variance. Based on this concept, PCA is used to choose the most sensitive features from the original feature set. Specifically, the eigenvector corresponding to the largest eigenvalue is chosen. Principal components with respect to this largest eigenvector are subsequently ranked based on their magnitude in descending order. Hence, features in the original space can be ranked accordingly based on the magnitude of their corresponding principal components in projected space. A subset of features is selected from the top ranked features within the original space using this PCA-based feature selection method.
Similar training and testing procedures have been applied to both hybrid and PCA-based feature selection methods, and their classification performance is summarized in Table 4.
Table 4.
Classification performance and AUC values for PCA-based and hybrid feature selection methods.
| Method | Accuracy (%) | AUC |
|---|---|---|
| PCA | 66.67 | 0.682 |
| Hybrid | 88.89 | 0.929 |
As shown in Table 4, the hybrid method performs significantly better than the PCA-based method. Specifically, the hybrid feature selection method yields a much larger AUC value than the PCA-based method. The mediocre performance of the PCA-based method is due to the fact that PCA simply performs a coordinate rotation that aligns the transformed axes with the directions of maximum variance during the feature selection process. However, there is no guarantee that these directions of maximum variance are related to the discriminative power of the features. Hence, the selected subset of features might still contain some redundant features which will deteriorate the classification performance.
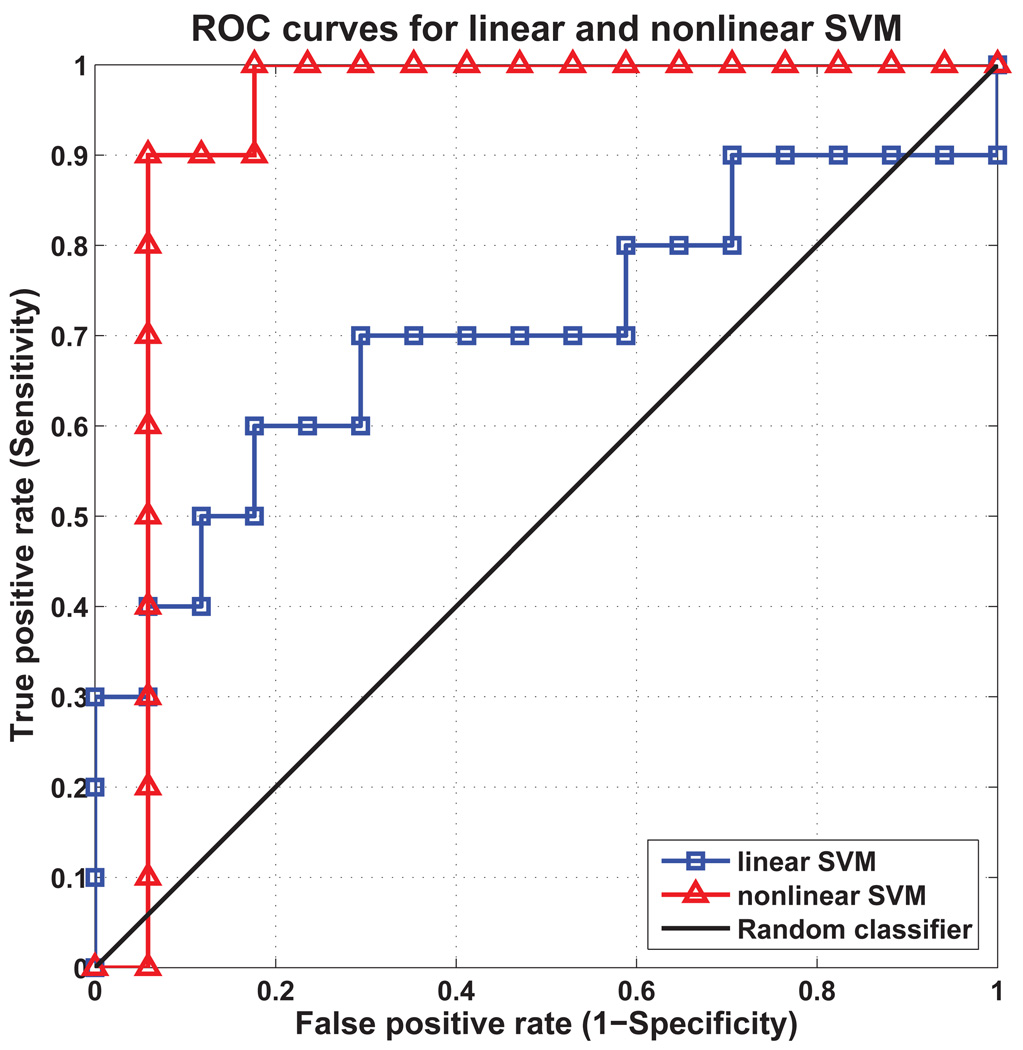
3.3. Comparison Between Linear and Nonlinear SVM Classifiers
In order to understand the effect of using nonlinear SVM classifiers in the proposed framework, linear SVM classifiers were trained and tested with the same leave-one-out procedure used in the proposed method. The classification results for the linear and nonlinear classifiers are summarized in Table 5.
Table 5.
Classification performance and AUC values for linear and nonlinear SVM classifiers.
| SVM Classifier | Accuracy (%) | AUC |
|---|---|---|
| Linear | 66.67 | 0.706 |
| Nonlinear | 88.89 | 0.929 |
The classification results obtained indicate that nonlinear SVM classifiers perform better than linear SVM classifier in terms of classification accuracy and AUC value. Nonlinear SVM classifiers, which map the input data onto a higher-dimensional feature space in a nonlinear fashion and the seek of an optimal separating hyperplane in the feature space, are useful especially for linearly inseparable data.
3.4. The Most Discriminant Regions
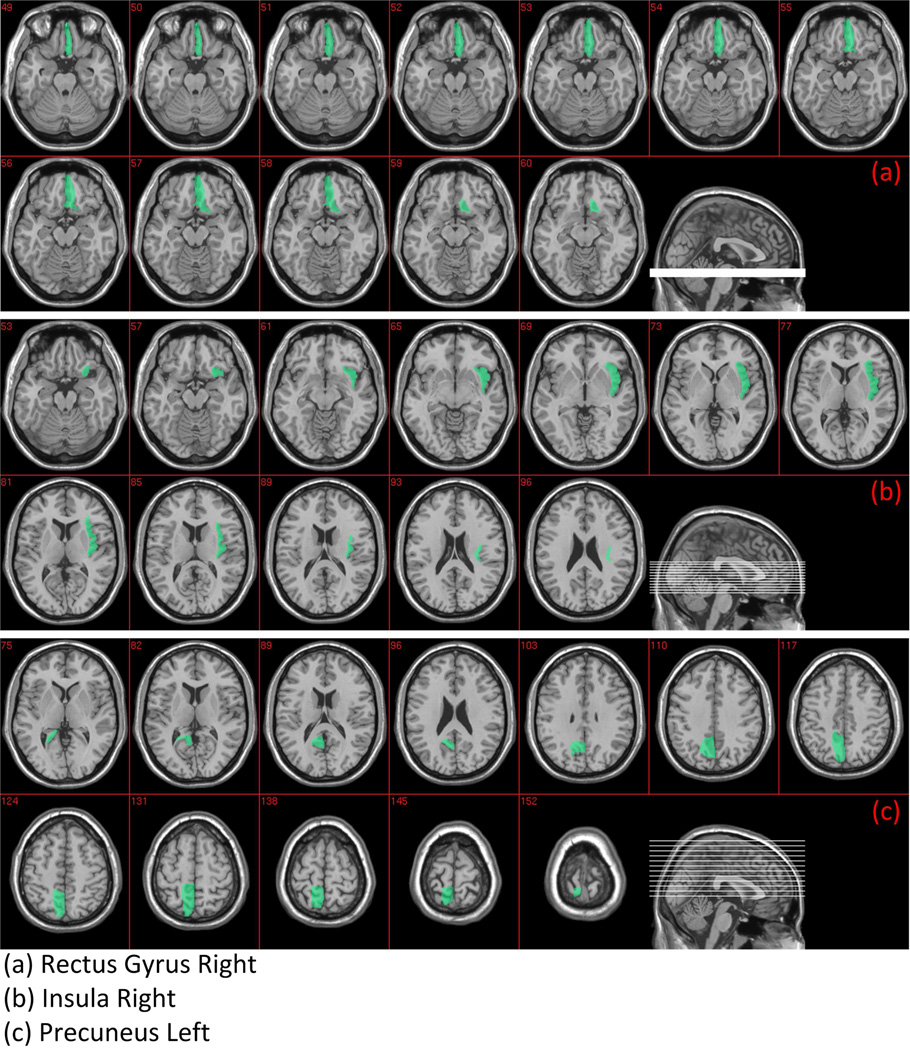
The SVM-RFE algorithm was performed to minimize the classification error in a backward sequential fashion by removing one feature at a time. The end result was a subset of most discriminant features that yields the best classification performance based on the training set. Each feature corresponds to a ROI, and a region, by capturing discriminative information, is indicative that it might be affected by the disease. Since the selected subset of features might be different for each leave-one-out case, we defined the most significant ROIs as the regions which were selected the most in all leave-one-out cases. The most discriminant regions that were selected for classification included 1) rectus gyrus, which is located on the orbital surface of the frontal lobe; 2) insula, which is located within lateral fissure between the temporal lobe and the frontal lobe; and 3) precuneus, which is a part of the superior parietal lobe hidden in the medial longitudinal fissure between the two cerebral hemispheres. The selected most discriminant regions are displayed in Figure 6.
Figure 6.
The most discriminant ROIs.
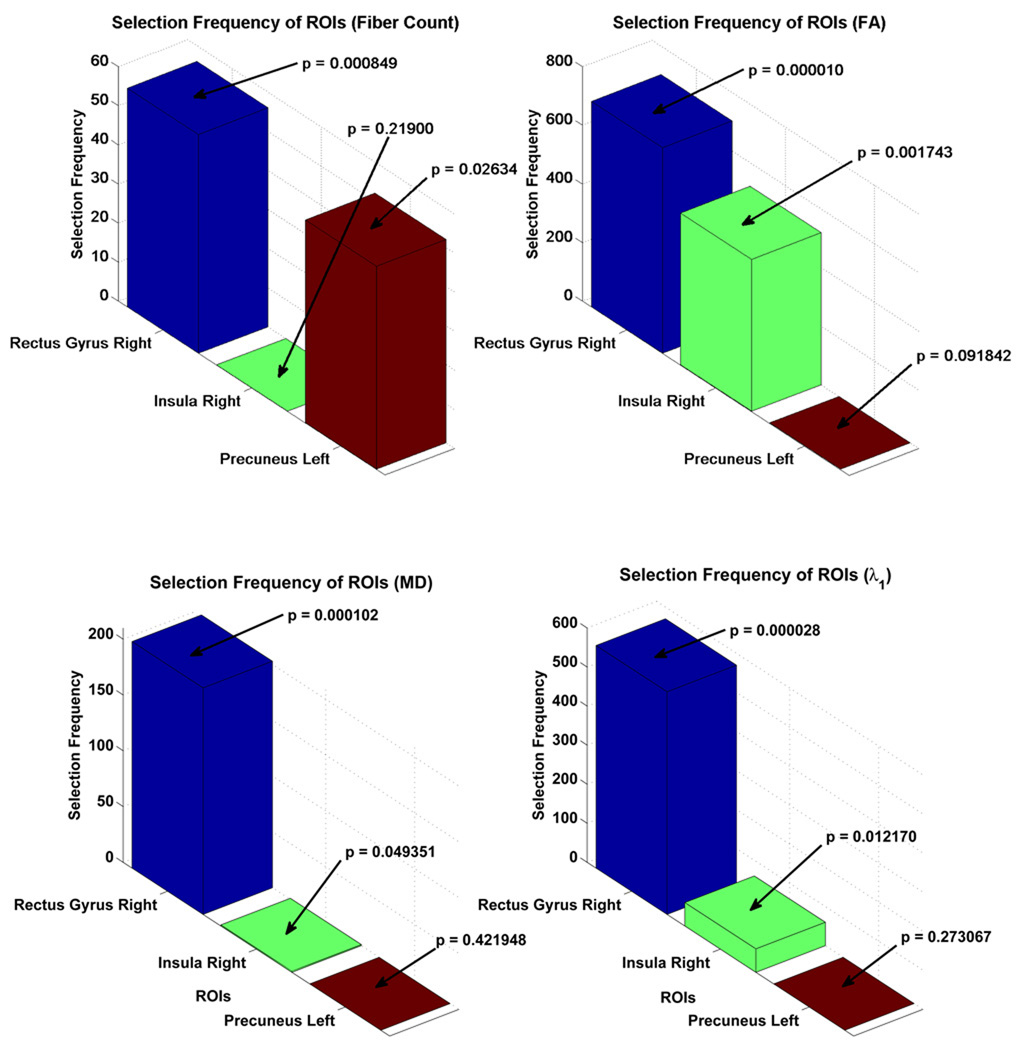
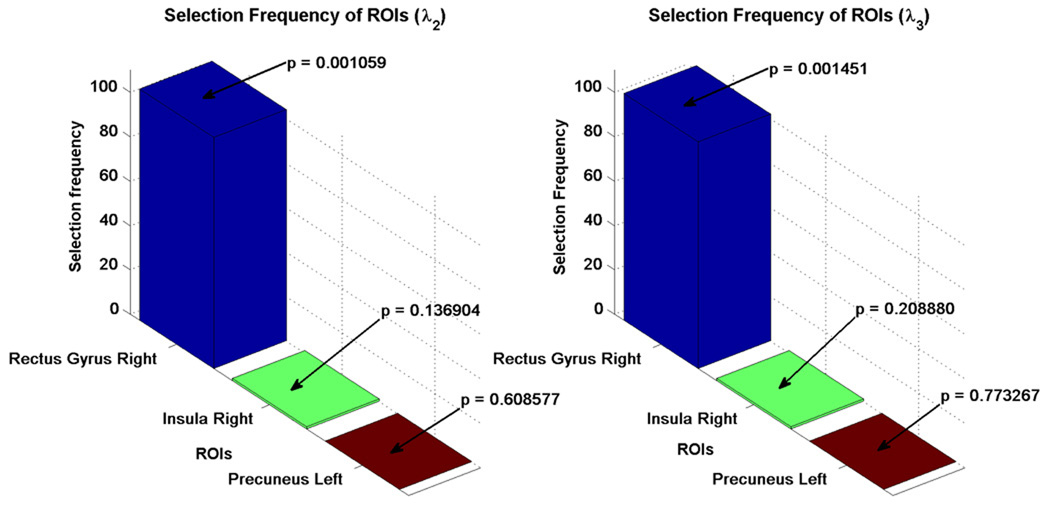
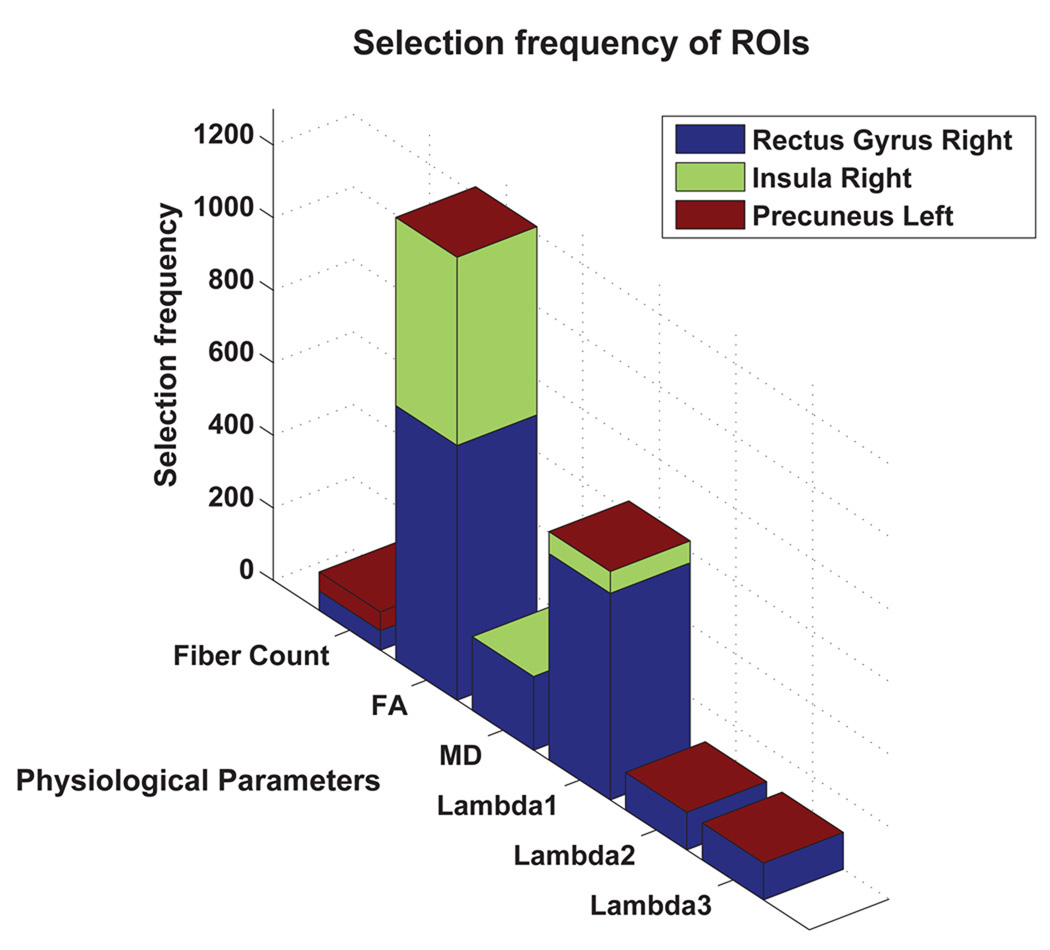
Bar charts, showing the number of times (selection frequency) the most discriminant regions were selected in all leave-one-out cases for the six physiological parameters are shown in Figure 7. The p-values, obtained by performing a two-sample t-test for each ROI, are also provided to indicate the significance of difference in connectivity between MCI and healthy controls in relation to a particular region. The p-values is in agreement with the final selected most discriminant features. The stacked bar chart showing the over-all selection frequency of the most discriminant ROIs is provided in Figure 8.
Figure 7.
Selection frequencies of the most discriminant regions.
Figure 8.
Overall selection frequencies for the most discriminant regions.
Table 6 summaries the selection frequency and corresponding p-value of each selected feature.
Table 6.
Selected features with their selection frequencies and p-values.
| Feature | Selection Frequency | p-value |
|---|---|---|
| FA-Rectus Gyrus Right | 702 | 0.000010 |
| λ1-Rectus Gyrus Right | 572 | 0.000028 |
| FA-Insula Right | 519 | 0.001743 |
| MD-Rectus Gyrus Right | 203 | 0.000102 |
| λ2-Rectus Gyrus Right | 104 | 0.001059 |
| λ3-Rectus Gyrus Right | 102 | 0.001451 |
| λ1-Insula Right | 60 | 0.012170 |
| FC-Rectus Gyrus Right | 54 | 0.000849 |
| FC-Precuneus Left | 52 | 0.026335 |
In order to understand how the selected ROIs contribute to classification, we conducted an experiment to evaluate the contribution of each selected ROI by excluding them one at a time with replacement in the training and testing processes. Initially, ROI with the highest selection frequency was excluded from the constructed connectivity networks. Then, the identical leave-one-out cross-validation procedure as in the proposed classification framework was applied to these “ROI-excluded” connectivity networks. This procedure was repeated for the other selected ROIs. The classification results are summarized in Table 7.
Table 7.
Classification performance and AUC value for each left-out ROI.
| Left-out ROI | Accuracy (%) | AUC |
|---|---|---|
| Rectus Gyrus Right | 70.37 | 0.629 |
| Insula Right | 77.78 | 0.882 |
| Precuneus Left | 85.19 | 0.894 |
| None | 88.89 | 0.929 |
It can be observed that the right rectus gyrus and right insula contribute significantly to the discriminative power, since removal of either one of them causes a significant decline in classification accuracy. Exclusion of the left precuneus however shows less prominent decline of classification accuracy and AUC value.
4. Discussion
4.1. Significance of Results
This study investigated the diagnostic value of WM connectivity networks, obtained via DTI fiber tractography, for identification of cognitively normal individuals from individuals with mild cognitive impairment. The proposed classification framework employs an enriched description of WM connectivity for more effective identification of MCI patients. Classification accuracy was evaluated via leave-one-out cross-validation to ensure performance generalization. The classification accuracy given by the proposed enriched description, utilizing six physiological parameters derived from the common fiber bundles transversing a pair of regions, is 88.9%, which is significantly higher than that using any single physiological parameter. The AUC value of the proposed method is 0.929, indicating its excellent diagnostic power, especially when in view of the relatively limited sample size available in this study. Simple description of WM connections using any single parameter can only afford limited biophysical information for distinguishing MCI patients from normal controls, as indicated by the much smaller AUC values. The enriched description, which accounts not only for the connection topology but also the biophysical properties of the connection, is more effective in conveying relevant and subtle information, particularly for the purpose of classification.
The brain regions that are selected for accurate detection of individuals with MCI includes portions of the prefrontal cortex, orbitofrontal cortex, parietal lobe and insula regions, which have already been extensively reported in previous studies. These included: parts of orbitofrontal region as reported in (Davatzikos et al., 2008), parts of the prefrontal, orbitofrontal and parietal regions as reported in (Fan et al., 2008b), parts of the orbitofrontal cortex, precuneus and insula as reported in (Misra et al., 2009), and the insula and precuneus as reported in (Fan et al., 2008a).
Our findings suggest that regions in the right hemisphere of the brain contribute most to the classification, implying that the right hemisphere sustains higher magnitude of alteration of WM connections within the cohort scanned in this study. This asymmetric alteration of WM connections is consistent with the patterns found in previous studies (Fan et al., 2008a; Wang et al., 2006) although it is also contradictory to what was reported in another study (Karas et al., 2004). The differences in these findings might be due to methodological differences in image analysis and patient selection. Nevertheless, the right-more-than-left pattern that can be observed in this study is consistent with the observation that patients who reported to clinic with memory impairments are more likely when they have language problems (Fan et al., 2008a). This implies that a smaller degree of WM alteration on the left hemisphere would trigger individuals to visit the clinic, looking for professional opinions or treatments.
In the proposed framework, the enriched connectivity description is used in conjunction with a hybrid feature selection method. The experimental results show that the PCA-based feature selection has relatively inferior classification performance, indicating that it is unable to effectively capture the prominent features for a dataset with a relatively small number of samples. Furthermore, the selection of an optimal subset of features that is most relevant to classification via the PCA-based method does not take into account information regarding class separability, and the direction of maximum variance does not necessarily correspond to the direction of maximum separability. On the other hand, the hybrid feature selection method used in the proposed framework is an efficient wrapper feature selection approach and its superior performance over the PCA-based method should be attributed to the inclusion of class separability information in both ranking-based and SVM-based feature selection processes.
In addition, we also found that nonlinear SVM classifier performs better than its linear counterpart with a relatively large margin. This hints that the dataset that we acquired and analyzed in this study is more nonlinearly, than linearly, separable.
It is noteworthy that the framework proposed herein is based on the assumption that the set of brain measurements that optimally differentiate between MCI patients and cognitively normal individuals cannot be known a priori, but can only be determined from the data. The leave-one-out cross-validation used here fundamentally guards against data overfitting, a persistent problem in high dimensionality analyses of datasets with relatively small sample size.
4.2. Methodological Issues/Limitations
DTI, while providing a convenient way of probing into the brain microstructures, has the known limitation of not being able to encode multi-directional diffusion information. Imaging techniques such as High Angular Resolution Diffusion Imaging (HARDI) (Tuch et al., 1999) should be used to provide more precise and informative description of WM connectivity.
In practice, a priori knowledge of regions of brain abnormalities is not always available. Even when good a priori hypotheses can be made about specific ROIs, a region of abnormality might be only a fraction of a whole ROI, or might span over multiple ROIs, thereby potentially reducing significantly the statistical power of the morphological analysis.
Another limitation of our current study is the relatively limited sample size, compared to the dimensionality of the connectivity measurements. Although the leave-one-out cross-validation accuracy obtained may be optimistic, the limited sample size did not allow us to explore other cross-validation techniques, since the nonlinear SVM classifier used might be undertrained. Our dataset was quite diverse, and it includes both sexes and all ages between 55 to 84 for MCI patients and 55 to 88 for normal controls. However, our results have to be evaluated in the future with larger datasets.
5. Conclusion
A novel technique has been proposed to describe the complex WM connectivity patterns for identifying individuals with MCI from normal controls. The promising results indicate that the proposed classification framework can provide an alternative and complementary approach for clinical diagnosis of alterations in brain structure associated with cognitive impairment.
Future work involves exploring how the prodromal stage of AD can be detected via functional measurements. Metabolic deficits or reduction in certain brain regions has been successfully applied for distinguishing AD from healthy aging (Greicius et al., 2004). Another interesting extension is to incorporate both physiological and functional measurements simultaneously in constructing diagnostic tools with higher sensitivity and specificity for more effective analysis of brain diseases.
Figure 5.
ROC curves of the linear and nonlinear SVM classifiers.
Acknowledgments
The authors wish to thank the anonymous reviewers for their valuable comments. This work was supported in part by NIH grants EB006733, EB008760, MH076970, EB009634, NIA L30-AG029001, P30 AG028377-02 and K23-AG028982.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basset DS, Bullmore E, Verchinski BA, Matty VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E. Small-world brain networks. The Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- Bischkopf J, Busse A, Angermeyer MC. Mild cognitive impairment - a reviews of prevalence, incidence and outcome according to current approaches. Acta Psychiatr Scand. 2002;106:403–414. doi: 10.1034/j.1600-0447.2002.01417.x. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, Bohl J, Bratzke H. Evolution of alzheimer’s disease related cortical lesions. Journal of neural transmission. 1998 Supplementum 54:97–106. doi: 10.1007/978-3-7091-7508-8_9. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Machine Learning. 1996;24:123–140. [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiology of Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen RA, Vilisaar J, Hlinka J, Bradshaw CM, Morgan PS, Constantinescu CS, Auer DP. Disconnection as a mechanism for cognitive dysfunction in multiple sclerosis. Brain. 2009;132(Pt 1):239–249. doi: 10.1093/brain/awn275. [DOI] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C the Alzheimer’s Disease Neuroimaging Initiative. Spatial patterns of brain atrophy in mci patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage. 2008a;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Rao H, Hurt H, Giannetta J, Korczykowski M, Shera D, Avants BB, Gee JC, Wang J, Shen D. Multivariate examination of brain abnormality using both structural and functional MRI. NeuroImage. 2007a;36:1189–1199. doi: 10.1016/j.neuroimage.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Fan Y, Resnick SM, Wu X, Davatzikos C. Structural and functional biomarkers of prodromal Alzheimer’s disease: A high-dimensional pattern classification study. NeuroImage. 2008b;41:277–285. doi: 10.1016/j.neuroimage.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shen D, Gur RC, Gur RE, Davazikos C. Compare: Classification of morphological patterns using adaptive regional elements. IEEE Transaction on Medical Imaging. 2007b;26:93–105. doi: 10.1109/TMI.2006.886812. [DOI] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, Beaulieu C. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cerebral Cortex. 2009;19:524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. PNAS. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA, et al. Mild cognitive impairment can be distinguished from alzheimer disease and normal aging for clinical trials. Arch. Neurol. 2004;61:59–66. doi: 10.1001/archneur.61.1.59. [DOI] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2004;46:389–422. [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Computational Biology. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Kurant M, Gigandet X, Thiran P, Wedeen VJ, Meuli R, Thiran JP. Mapping human whole-brain structural networks with diffusion MRI. PLoS ONE. 2007;2:e597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Commoun L, Gigandet X, Thairan JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. PNAS. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Canales-Rodríguez EJ, Melie-García L, Valdés-Hernández PA, Martínez-Montes E, Alemán-Gómez Y, Sánchez-Bornot JM. Characterizing brain anatomical connections using diffusion weighted MRI and graph theory. NeuroImage. 2007;36:645–660. doi: 10.1016/j.neuroimage.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Karas G, Scheltens P, Rombouts S, Visser P, van Schijndel R, Fox N, Barkhof F. Global and local gray matter loss in mild cognitive impairment and Alzheimer’s disease. NeuroImage. 2004;23:708–716. doi: 10.1016/j.neuroimage.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Thompson PM, Lancaster JL, Bartzokis G, Smith S, Royall TCDR, P. T. Fox AL. Relationship between white matter fractional anisotropy and other indices of cerebral health in normal aging: Tract-based spatial statistics study of aging. NeuroImage. 2007;35:478–487. doi: 10.1016/j.neuroimage.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Leemans A, Jeurissen B, Sijbers J, Jones DK. ExploreDTI: A graphical toolbox for processing, analyzing, and visualizing diffusion MR data. 17th Annual Meeting of Intl Soc Mag Reson Med. 2009:3537. [Google Scholar]
- Malhi A, Gao RX. Pca-based feature selection scheme for machine defect classifiaction. IEEE Transactions on Instrument and Measurement. 2004;53:1517–1525. [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: Results from ADNI. NeuroImage. 2009;44:1414–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennanen C, Testa C, Laakso MP, Hallikainen M, Helkala EL, Hanninen T, Kivipelto M, Kononen M, Nissinen A, Tervo S, Vanhanen M, Vanninen R, Frisoni GB, Soininen H. A voxel based morphometry study on mild cognitive impairment. The Journal of Neurology, Neurosurgery, and Psychiatry. 2005;76:11–14. doi: 10.1136/jnnp.2004.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch. Neurology. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Rakotomamonjy A. Variable selection using svm based criteria. Journal of Machine Learning Research: Special issue on special feature. 2003;3:1357–1370. [Google Scholar]
- Rose SE, Janke AL, Chalk JB. Gray and white matter changes in alzheimer’s disease: A diffusion tensor imaging study. Journal of Magnetic Resonance Imaging. 2007;27:20–26. doi: 10.1002/jmri.21231. [DOI] [PubMed] [Google Scholar]
- Rose SE, McMahon KL, Janke LA, ODowd B, de Zubicaray G, Strudwick MW, Chalk JB. Diffusion indices on magnetic resonance imaging and neuropsychological performance in amnestic mild cognitive impairment. Journal of Neurology, Neurosurgery & Psychiatry. 2006;77:1122–1128. doi: 10.1136/jnnp.2005.074336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex networks measures of brain connectivity: Uses and interpretations. NeuroImage Article. 2009 doi: 10.1016/j.neuroimage.2009.10.003. in Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: a structural description of human brain. PLoS Computational Biology. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. The small world of the cerebral cortex. Neuroinformatics. 2004;2:145–161. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. Small-world networks and functional connectivity in Alzheimer’s diease. Cerebral Cortex. 2007;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- Tuch D, Weisskoff R, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging of the human brain. ISMRM’ 1999. 1999 [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. The nature of statistical learning theory (Statistics for Engineering and Information Science) Springer-Verlag; 1999. [Google Scholar]
- Wang L, Miller JP, Gado MH, McKeel DW, Rothermich M, Miller MI, Morris JC, Csernansky JG. Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. NeuroImage. 2006;30:52–60. doi: 10.1016/j.neuroimage.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhu C, He Y, Zhang Y, Cao Q, Zhang H, Zhang Q, Wang Y. Altered small-world brain functional networks in childrean with attention-deficit/hyperactivity disorder. Human Brain Mapping. 2009;30:638–649. doi: 10.1002/hbm.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Xu D, Mori S, Shen D, van Zijl PCM, Davatzikos C. Spatial normalization of diffusion tensor fields. Magnetic Resonance in Medicine. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- Yap PT, Wu G, Zhu H, Lin W, Shen D. Fast tensor image morphing for elastic registration. MICCAI 2009 LNCS. 2009;5761:721–729. doi: 10.1007/978-3-642-04268-3_89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap PT, Wu G, Zhu H, Lin W, Shen D. F-TIMER: Fast tensor image morphing for elastic registration. IEEE Transactions on Medical Imaging. 2010;29:1192–1203. doi: 10.1109/TMI.2010.2043680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schuff N, Jahng GH, Bayne W, Mori S, Schad L, Mueller S, Du AT, Kramer J, Yaffe K, Chui H, Jagust W, Miller B, Weiner M. Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and alzheimer disease. Neurology. 2007;68:13–19. doi: 10.1212/01.wnl.0000250326.77323.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zemanova L, Zamora G, Hilgetag CC, Kurths J. Hierarchical organization unveiled by functional connectivity in complex brain networks. Physical Review Letter. 2006;97:238103. doi: 10.1103/PhysRevLett.97.238103. [DOI] [PubMed] [Google Scholar]