Abstract
Repair and integrity of DNA ends at breaks, replication forks and telomeres are essential for life; yet, paradoxically, these responses are, in many cases, controlled by a single protein complex, Mre11-Rad50-Nbs1 (MRN). The MRN complex consists of dimers of each subunit and this heterohexamer controls key sensing, signaling, regulation, and effector responses to DNA double-strand breaks including ATM activation, homologous recombinational repair, microhomology-mediated end joining and, in some organisms, non-homologous end joining. We propose that this is possible because each MRN subunit can exist in three or more distinct states; thus, the trimer of MRN dimers can exist in a stunning 63 or 216 states, a number that can be expanded further when post-translational modifications are taken into account. MRN can therefore be considered as a molecular computer that effectively assesses optimal responses and pathway choice based upon its states as set by cell status and the nature of the DNA damage. This extreme multi-state concept demands a paradigm shift from striving to understand DNA damage responses in separate terms of signaling, checkpoint, and effector proteins: we must now endeavor to characterize conformational and assembly states of MRN and other DNA repair machines that couple, coordinate, and control biological outcomes. Addressing the emerging challenge of gaining a detailed molecular understanding of MRN and other multi-state dynamic DNA repair machines promises to provide opportunities to develop master keys to controlling cell biology with probable impacts on therapeutic interventions.
Introduction
The Mre11-Rad50-Nbs1 (MRN) complex is a multifaceted molecular machine, critical for biological processes that detect and repair double strand breaks (DSBs) [1,2]. DSBs arise from exposure to ionizing radiation (IR) and genotoxic chemicals, as well as from cellular metabolism during replication, meiosis, and V(D)J recombination [3–7]. The ends of chromosomes can also be considered as forms of DSBs if they are not correctly protected as telomeres. Failure to expeditiously repair DSBs or protect DNA ends at either replication forks or telomeres can lead to chromosomal rearrangements, loss of chromosome arms or aneuploidy. Thus, DSBs present one of the most deleterious forms of DNA damage and failure to appropriately process them can ultimately result in human disorders including developmental, immunological and neurological disease, and especially cancer [8,9]. Protection against these disease phenotypes therefore requires efficient recognition of DSBs, precise activation of cell cycle checkpoints, and coordinated repair via one of several pathways including non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ, also referred to as Alt-NHEJ) or homologous recombinational repair (HRR) [10,11].
The MRN complex has been implicated in all aspects of DSB processing: from initial detection, to triggering signaling pathways, and facilitating repair through the pathways outlined above (Fig. 1). Furthermore, MRN is present at replication forks and telomeres and plays key roles in preventing DSBs from forming at these sensitive locations. Thus, MRN has emerged as a critical regulator of DSB biology as emphasized by human disorders associated with misregulation or inherited mutations in any one of the subunits. Mutations in Nbs1 were described in patients suffering from Nijmegan breakage syndrome (NBS) [12], with subsequent mutations in Mre11 found to cause ataxia-telangiectasia-like disorder (ATLD) [13] and one mutation so far described for Rad50 in NBS-like disorder [14].
Figure 1.
The MRN complex acts as a sensor, signaler and effector to protect DNA ends and process DSBs. MRN senses DSBs and, in collaboration with CtIP, processes DNA ends before channeling into one of 3 distinct DNA repair pathways (HRR, MMEJ or NHEJ). ATM (red) is a negative regulator of MMEJ. MRN’s MMEJ and NHEJ functions are also important for immunological roles during V(D)J and class-switch recombination. In addition, MRN is found at the telomere where it interacts with TRF2 and is involved in telomere maintenance. MRN is also associated with the replication fork and functions to stabilize forks through it’s DNA bridging activity and is involved in replication restart pathways. MRN signals both DSBs, through ATM interactions, and collapsed replication forks, through ATR and RPA interactions. Red stars indicate DNA damage.
Diverse functions of the MRN machine
The MRN complex can be considered a flexible scaffold that acts as a combined sensor, signaling and effector complex via dynamic states that control biological outcomes to DSBs. MRN imparts three key functions critical for its diverse roles: 1) DNA binding and processing, 2) DNA tethering to bridge DNA over short and long distances, and 3) activation of DSB response and checkpoint signaling pathways. MRN structural architecture, separated into distinct “head”, “coil”, “hook” and flexibly attached adapter regions, underlies these roles (Fig. 2A).
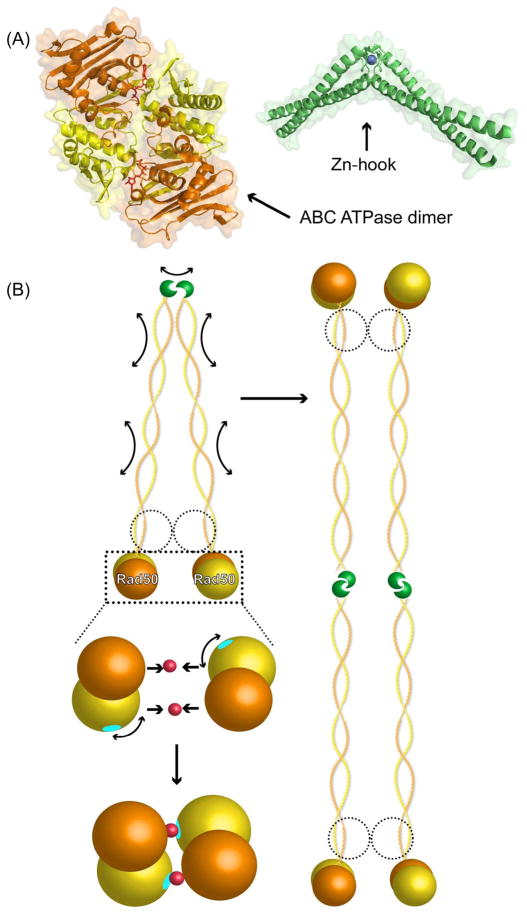
Figure 2.
Overall MRN assembly and key domains. (A) MRN can assemble as a heterohexamer and consists of 4 key regions: the processing “head”, formed by the Mre11 dimer and two Rad50 ABC ATPase domains (indicated by dotted circle), the “coil” and “hook” encoded by the region of Rad50 separating the N- and C-terminal ABC ATPase halves, and the Nbs1 “flexible adapter” (indicated by dotted circle) that provides the key link to signaling functions. (B) Schematic representations of the MRN subunits Mre11, Rad50 and Nbs1 showing key domains, colored as in other figures. The approximate location of reported methylation sites are indicated by M and DNA damage inducible phosphorylation sites by P (see text for details). The major sites corresponding to inherited human disorders associated with each gene are indicated by a red triangle, with amino acid substitutions labeled for Mre11 and Rad50 (X is a stop codon) and 657del5 representing the major Nbs1 mutation responsible for >90% of NBS cases.
The head region comprises two Rad50 ABC-ATPase domains and two Mre11 nucleases bound to the base of the Rad50 coiled-coils (Fig. 2A). This M2R2 head is the DNA binding and processing core of MRN. Mre11, which contains an N-terminal phosphodiesterase and C-terminal DNA binding domains (Fig. 2B), provides ssDNA endonuclease and 3’-5’ dsDNA exonuclease activities [15]. Rad50 has regulatory ATPase and adenylate kinase activities [16,17], and can also bind to DNA. The importance of the Mre11-Rad50 core complex is underscored by its conservation in bacteria, archaea and throughout the eukarya [18,19].
The Rad50 ATPase domain of the MRN head is formed from two halves encoded at either end of the primary sequence that come together from the collapse of the intervening sequence, which forms a long (~500 Å) extended anti-parallel coiled-coil [20,21]. This coil region of MRN flexibly extends from the M2R2 head and forms a Zn-hook domain at the apex of the Rad50 coiled-coil (Fig. 2A and 2B).
In eukaryotes, Nbs1 (Xrs2 in S. cerevisiae) forms the flexible adapter domain of MRN (Fig. 2A), acting as a regulatory and protein recruitment module. Nbs1 consists of an N-terminal phosphoprotein-binding core that is flexibly attached to the MR core complex through an Mre11 interaction motif towards the end of the unfolded C-terminal half (Fig. 2B). Nbs1 provides the MRN complex with its signaling role through interactions with, and ultimately activation of, either ATM (ataxia-telangiectasia mutated) or ATR (ATM- and Rad3-related) protein kinases in response to DSBs or replication fork stalling, respectively. Nbs1 is also essential for localization of MRN to the nucleus, and works in tandem with Rad50 ATP binding and hydrolysis to regulate Mre11 nuclease activities.
MRN functions in a diverse set of cellular contexts including repair of DSBs by NHEJ, MMEJ, and HRR, signaling of DSBs within chromatin, at sites of V(D)J and class-switch recombination, at replication forks, and at telomeres (Fig. 1). These different cellular environments require different functionalities of MRN to protect against inappropriate end processing or signaling. For example, MRN endonuclease activity is essential for initiating HRR [22,23], whereas its exonuclease activity is required for MMEJ [24,25] and inappropriate nuclease activity at dysfunctional telomeres results in chromosome fusions [26]. Further, initiation of the DSB response through ATM activation is likely to be necessary only when DSBs cannot be efficiently repaired, such as after high levels of damage [27], and activation of this pathway at undamaged replication forks or telomeres would be severely detrimental to the cell [28].
Major questions in the field have therefore centered around how MRN activities and cellular locations are controlled to provide its diverse functions. More simply put; how does MRN know what to do, where to do it and when? Progress towards answering these questions has been made by employing a wide range of experimental techniques and model organisms; results indicate that MRN is assembled into larger macromolecular complexes that contain different protein components and that this defines its multiple functions in the DNA damage response. These MRN assemblies and states are further impacted by post-translational modifications (PTMs), providing additional layers of complexity and regulation. Thus, MRN states in terms of shape, conformation, and interactions are information controlling its functions and biological outcomes.
The emerging picture underlying MRN assembly and its control of biological outcomes can therefore be considered in terms of three key elements that control biologically-relevant states: 1) macromolecular shape and conformation, 2) availability of interaction sites and 3) allosteric regulation from interactions or PTMs that impact on its shape and function. Structural biology plays a critical role in addressing these questions as structures define macromolecular shape, conformation, and assembly; give detailed information of interfaces; and, in combination with biochemical assays, determine catalytic mechanisms. Further, structural methods allow changes in conformation and dynamics to be visualized upon interactions and in response to PTMs. Therefore, united with biochemistry and genetics, structural insights into macromolecular assemblies provide the basis from which to predict biological outcomes, a necessary step to allow designed intervention of these pathways for therapies. Here, we consider how MRN conformations, assemblies, interactions, and functions are controlled to provide the desired biological activity in a given cellular context. Emerging results indicate that distinct MRN states define different outcomes in cell biology.
Mre11 complexes and interactions
Recent results show that MRN acts as a functional dimer. Small-angle X-ray solution (SAXS) envelopes show that in solution and in a DNA-free state Mre11 forms a lop-sided U-shaped dimer [23]. A hydrophobic 4-helix bundle forms the dimer interface between the two phosphodiesterase domains that form the base of the U-shape. The Mre11 DNA binding motifs extend away from this base to form capping domains, with the groove of the U-shape forming the DNA binding site. Analysis of mutations that disrupt the Mre11 dimer show that it is clearly important for MRN functions, as the resulting monomeric form has reduced DNA affinity and, unlike wild type Mre11, does not restore resistance to clastogenic agents in S pombe.
Crystal structures of Mre11 bound to DNA substrate mimics of both a 2-ended DSB and 1-ended replication fork reveal that the DNA-bound state is also dimeric, forming the same U-shaped dimer observed by SAXS [23] (Fig. 3A). However, this DNA-bound state of Mre11 can be further divided into at least two distinct conformations. When bound to the 2-ended DSB substrate, the Mre11 dimer synapses the two DNA ends and is symmetric. In contrast, when bound to the 1-ended DSB substrate, the Mre11 dimer binds a single DNA molecule and adopts an asymmetric conformation. This asymmetry arises from rotation of the Mre11 monomer that is not engaging DNA, which sterically blocks the binding of a second DNA substrate (Fig. 3B).
Figure 3.
The Mre11 dimer adopts different conformational states at a 2-ended Vs 1-ended DSB. (A) Crystal structures of Mre11 in complex with a 2-ended DSB (top) or a 1-ended DSB replication fork mimic (bottom) revealed Mre11 binds these DNA substrates as symmetric and asymmetric dimers respectively. (B) Cartoon of Mre11 bound to DNA substrates as in (A) with phosphodiesterase and capping domains colored blue and gray, respectively, as in Fig. 1B. Arrows highlight the Mre11 phosphodiesterase and DNA capping domain rotations required to move between 2-ended and 1-ended bound states. ATLD sites that reduce Nbs1 binding (N117S and W210C, green and yellow respectively) are mapped onto the surface and the dotted outline of the symmetric Mre11 dimer is overlaid on the asymmetric dimer to highlight rearrangements between the two states.
The MRN complex initiates different damage and checkpoint signaling responses for 2-ended DSBs, which generally arise in chromatin and result in ATM activation, compared to 1-ended breaks, which arise from collapse of replication forks and lead to ATR activation (Fig. 1). Therefore, we propose that the distinct conformations of Mre11 bound to DNA represent a critical signal within the cell to mediate downstream signaling pathways, with symmetric Mre11 activating ATM signaling and asymmetric Mre11 initiating ATR signaling. This provides an elegant means by which Mre11 can control pathway choices at a DSB, with ATM activation important for DSB repair by HRR and ATR activation leading to replication fork rescue.
Rad50 complexes and interactions
Rad50 contains a globular ABC-ATPase domain at one end of the folded protein and, at the other end, 500 Å away, a Zn-hook, separated by an anti-parallel coiled-coil (Fig. 2A, 2B). Rad50’s unique architecture has led to challenges in understanding the structural basis for its function. However, hybrid structural approaches have defined several distinct and functionally important conformational states.
Within the MRN complex two Rad50 ABC-ATPase domains are juxtaposed with the Mre11 dimer at the M2R2 head (Fig. 2A), and ATP binding regulates at least two distinct states. 1) Upon ATP and Mg2+ binding the two Rad50 ABC-ATPase domains come together to form a head-to-tail dimer, sandwiching two molecules of ATP at the dimer interface (Fig. 4A). Contributions to the active site are made by conserved motifs from both the N-terminal ABC-ATPase half of one monomer and the C-terminal ABC-ATPase half from the other monomer. 2) Upon hydrolysis and release of ATP, the Rad50 ABC-ATPase domains monomerize, forming an open M2R2 head conformation [21,29]. In switching between these closed and open conformations there is a significant ~35º rotation of the N-terminal ABC-ATPase half with respect to the C-terminal half (Fig. 4B). As Rad50 is a prototype for other ABC-ATPases, including the mismatch repair protein MutS, the ATP-driven conformational states of Rad50 have broad implications for ABC-ATPase functions [30–32]
Figure 4.
Rad50 states include ATP-dependent association of ABC ATPase domains, and Zn-hook mediated inter-and intramolecular dimers. (A) Structures of the Rad50 ABC ATPase dimer, with N- and C-terminal ABC ATPase halves colored orange and yellow respectively, bound to ATP (in red) and the Zn-hook dimerization domain (green). (B) Cartoon showing several possible Rad50 states. The Rad50 Zn-hook can either intermolecularly dimerize Rad50 within a single MRN complex, which connects the Rad50 ABC ATPase domains present within an M2R2 head (top left), or intramolecularly connect two MRN complexes to form a dumbbell-like structure with M2R2 heads at either end (right). In these cartoons Mre11 is outlined as a dotted circle to show that it can bridge Rad50 molecules in the absence of direct Rad50 dimerization through Zn-hook or ATP-mediated connections. ATP-induced dimerization brings together two Rad50 ABC ATPase domains (bottom left), inducing an ~35º subdomain rotation of the C-terminal subdomain (yellow) with respect to the N-terminal subdomain (orange). ATP is indicated in red. The evolutionarily conserved ABC signature motif is in cyan to facilitate visualization of this rotation.
The ATP-induced conformational states are important for MRN functions, with ATP binding and hydrolysis by Rad50 affecting Mre11 nuclease activities in an Nbs1-dependent manner in eukaryotes [17]. Mutations that disrupt Rad50 ATPase activity result in sensitization of cells to DSB causing agents [33]. At the other end of Rad50, the Zn-hook acts as a dimerization domain. It can either intramolecularly link two Rad50 monomers that are part of a single M2R2 core of MRN, or intermolecularly link two MRN complexes with M2R2 heads on either end (Fig. 4B). This latter architecture allows bridging of DNA up to 1,000 Å away, providing the basis for long-range tethering, an essential MRN function [20,34].
Other states of Rad50 have also been observed. Rad50, in the absence of Mre11, can exist in DNA bound and free states [21]. Additionally, atomic force microscopy experiments have shown that juxtaposed coiled-coils from adjacent Rad50 molecules can interact, which has been proposed to act in conjunction with intermolecular Zn-hook dimerization to tether DNA ends together [34]. In the context of assembled MRN, DNA and ATP binding at the M2R2 head regulates Rad50 states, acting to straighten the Rad50 coiled-coils and this in turn favors intermolecular Zn-hook bridging dimerization and interactions between adjacent coiled-coils.
Nbs1 interaction states
The recent crystal structures of the folded core of Nbs1 reveals that it consists of dual phosphopeptide binding domains encoded within an FHA domain and a structurally apposed tandem BRCT repeat domain (Fig. 5A) [35,36]. This arrangement of domains suggests that Nbs1 acts as a multimodal adapter, linking MR to host of proteins that are phosphorylated as part of the DNA damage response (Fig. 5B). Potential interaction states include the unbound form and states bound to phosphoproteins through connections with the FHA domain only, the BRCT repeat only, or simultaneously to both domains. This latter state can further be divided into simultaneous interactions to an individual protein, containing two compatible phosphorylated regions, or to two different proteins. Proteins shown to interact with the FHA/BRCT region of Nbs1 include CtIP [35,36], MDC1 [35,37,38], ATR [39] and the WRN helicase [40]. Phosphoproteomics studies have revealed that exposure of human cells to IR or UV radiation induces close to a thousand phosphorylation events on several hundred different proteins [41–43] and it seems likely that other phosphoproteins will interact with Nbs1 through its FHA/BRCT domains.
Figure 5.
The Nbs1 flexible adapter. (A) The crystal structure of the Nbs1 N-terminal phosphopeptide binding domain containing the structurally apposed FHA (blue), BRCT1 (yellow) and BRCT2 (red) domains. (B) Cartoon showing interaction states mediated by full length Nbs1. The N-terminal domain mediates interactions with phosphoproteins through binding to phosphothreonine (FHA domain) and/or phosphoserine (BRCT repeat domain) consensus motifs. The extended C-terminus, revealed by SAXS, contains adjacent interaction motifs for Mre11 (dark blue) and ATM (green).
The N-terminal Nbs1 phosphoprotein-binding core is connected through a flexible linker to adjacent Mre11 and ATM interaction motifs at the C-terminus (Fig. 5B). The Nbs1 interaction with Mre11 is critical for MRN localization to the nucleus and DSBs [12,44] and the interaction with ATM is critical for MRN-dependent activation of ATM following DSBs [45,46]. Additional factors are also important for efficient MRN-dependent ATM signaling and DSB repair. These include 53BP1, which interacts with MRN through Rad50 [47] and can act to amplify the accumulation of MRN and ATM at DSBs, in particular in the context of heterochromatin [48], and BRCA1 that interacts with MRN indirectly through CtIP [49,50].
While Nbs1 is almost always associated with MR in the cell, of the interaction with ATM is dynamic, occurring in response to DSBs. Thus, phosphoprotein bound states of Nbs1 may possibly exist in ATM bound and free states, doubling the possible number of states.
States are transmitted within the MRN complex to effect biological outcomes
In the above sections we have detailed key states of the MRN subcomponents. In the context of the assembled MRN complex, emerging results reveal that integration and communication between these different states, as well as interactions with additional proteins, are key for fine-tuning MRN functions for its diverse roles in the cell (Fig. 6).
Figure 6.
The MRN complex assembles as a flexible sensor, signaler and effector for DSB responses. Model of the MRN complex combining structures of Mre11, Rad50 and Nbs1 domains with known regions of conformational change and/or flexibility (curved arrows) and key interactions (straight arrows) as discussed in the text and figures 3–5. CtIP binding to Nbs1 (shown on the left) induces a conformational change within the phosphopeptide binding core that is suitable to signal to Mre11 through a pull on the flexible tether.
Different Rad50 nucleotide states have clear roles in regulating MRN activities, with ATP binding and hydrolysis regulating Mre11 nuclease activities, resulting in a switch from exonuclease to endonuclease activity [17,51]. In eukaryotes, this switch is dependent on Nbs1, and Rad50 active site mutations that disrupt ATPase activity sensitize cells to DSB inducing agents [33]. Further, ATP affects DNA end binding specificity of the M2R2 head [52], with DNA binding inducing straightening of the Rad50 coiled-coils that favors intermolecular Zn-hook dimers [34] (Fig. 4B). Thus, Rad50 conformational states can be communicated throughout the protein to effect biological outcomes.
Mre11 appears to be a key signal to distinguish between MRN binding to 1-ended breaks, found at replication forks, vs 2-ended breaks formed throughout the genome, largely from exogenous sources of DNA damage. Crystal structures show that Mre11 is symmetric when bound to two DNA ends, but asymmetric when bound to the replication fork mimic [23] (Fig 3A). Intriguingly, residues mutated in ATLD that reduce Nbs1 binding map across the Mre11 dimer interface and their relative geometric positioning alters between symmetric and asymmetric Mre11 states [23] (Fig. 3B). This suggests that Nbs1 senses Mre11 dimer shape to effect different biological outcomes at a replication fork vs a 2-ended DSB. The juxtaposition of Mre11 and ATM interaction motifs on Nbs1 suggest a possible mechanism for eliciting these different responses. When bound to DSBs, the ATM interaction site must be available as ATM is recruited and activated [46]. In contrast, at a 1-ended break, i.e. a replication fork, ATM is no longer directly recruited but ATR is activated, through interactions dependent on the N-terminal phosphoprotein core [39]. This suggests that the Mre11 conformational change to an asymmetric dimer at the replication fork may reposition or sterically block the ATM interaction site on Nbs1 to make it inaccessible to ATM.
Besides Mre11 mediated changes at the C-terminal end of Nbs1, important conformational changes at the N-terminal phosphopeptide-binding core are important for DSB responses. The structure of the Nbs1 core in complex with a peptide of CtIP bound to the FHA domain revealed that the close juxtaposition of FHA and BRCT repeat domains allows communication between the dual phosphopeptide binding sites [36]. CtIP phosphopeptide binding triggers a long-range conformational change that alters the interface between the BRCT repeat domains (Fig 6). This movement results in a rotation of the BRCT2 domain with respect to the BRCT1 domain, altering the phosphopeptide-binding site at the interface. This mechanical linkage provides a means of communication between binding sites to control interacting partners. Further, since Nbs1 interactions with CtIP or other damage response proteins such as MDC1 would anchor Nbs1 to DNA and chromatin, such a conformational change could act as a pull on the tether to communicate phosphopeptide bound states to the Mre11-Rad50 core complex.
Post-translational modifications of MRN
Like most eukaryotic proteins, especially those responding to environmental and endogenous stresses, components of the MRN complex are modified by PTMs in vivo. Initial studies focused on ATM-dependent phosphorylation events, such as IR-induced phosphorylation of Nbs1, controlling cell cycle checkpoint and apoptosis (reviewed in [53], see also [54]). Recent phosphoproteomics studies have revealed extensive phosphorylation of MRN components; to date 17, 9 and 25 in vivo phosphorylation sites have been reported in Mre11, Rad50 and Nbs1, respectively (http://www.phosphosite.org) [41–43,55]. Many sites do not conform to the ATM/ATR/DNA-PK SQ/TQ consensus, indicating potential roles for other serine/threonine protein kinases, including CK2 in several cases and even tyrosine kinases, in regulating MRN activity, location and/or function. DNA damage-inducible phosphorylation sites have been reported on serines 676, 678, and 681 (Mre11), serine 635 and threonine 690 (Rad50) and serines 58, 278, 341, 343, 397, 432, 611 and threonines 335 and 434 (Nbs1) [41–43]. Mre11 is also methylated on arginines located between amino acids 572 and 587, while acetylation sites have been identified in Nbs1 and on Rad50 (http://www.phosphosite.org).
While the effects of PTMs on the states of Mre11, Rad50 and Nbs1 are not well understood, PTMs are clearly involved in regulating MRN activities in the cell. Recently, it has been shown that Mre11 exonuclease dependent MMEJ is negatively regulated both by ATM [24] and by methylation in the glycine-arginine-rich region of Mre11 between 572 and 587 [25]. Phosphorylation of Nbs1 has also been shown to regulate responses to replicative stress, with serine 343 phosphorylation important for down-regulating replication following UV damage [39], and necessary for RPA hyperphosphorylation following hydroxurea exposure [56]. Thus, PTMs provide a means to tune individual subunits and overall MRN conformations and interactions in response to cell state and thereby aid MRN functions in helping to coordinate sensing, signaling, and effector functions in response to DSBs.
Synopsis and prospects
The tragic human diseases resulting from mutations of the MRN complex provide powerful opportunities for insights into the biological roles and importance of this complex molecular machine. For the XPD helicase, integrated structural, biochemical, and mutational analyses provided the basis to understand how three distinct diseases involving cancer, aging, and developmental defects could result from single site mutations in one gene [57]. Similarly, for Mre11-Rad50-Nbs1 many of the key insights have resulted from an increasingly detailed understanding of MRN subunit structures, their interfaces, conformations, and interactions. Yet, we have actually only achieved an initial understanding of these critical features. Detailed and comprehensive structure determinations of MRN are extremely challenging due to the multiple conformations, interfaces, and dynamic nature of the complex. However, achieving a more complete picture of MRN conformations and interactions promises great rewards for biological understanding of its roles in controlling cellular outcomes to DSBs and for informed interventions relevant to human diseases.
These rewards and challenges presented by characterizations of DNA repair machines have prompted the problem-driven development of powerful new methods and technologies for structural analyses of dynamic complexes. The most promising approaches to defining dynamic macromolecular complexes such as MRN come from combined methods [58,59]. SAXS has proven particularly powerful to comprehensively characterize multiple conformations and assemblies in solution due to its high-throughput and ability to provide data on most samples [60]. For example, SAXS provided the means for recent characterization of DNA-PK conformations in solution and, furthermore, of conformational changes in DNA–PKcs resulting from autophosphorylation (See Dobbs et al this issue) [61]. As SAXS is especially informative when combined with crystal and NMR structures [62], SAXS will be increasingly valuable as more structures of components are defined for macromolecular machines.
Currently, we know that the MRN heterohexamer controls key sensing, signaling, regulation, and effector responses to DSBs. We also now understand that this is possible because each MRN subunit can exist in multiple states, allowing extreme complexity despite its simple three-member composition (Fig. 6). Mre11 states include symmetric and asymmetric dimers bound to DNA ends or forks and linked to multiple Nbs1 states. The Rad50 ATPase, an archetype for the ABC-ATPase superfamily, has DNA-bound, intra-and intermolecular Zn-hook dimers, and ATP, ADP or nucleotide free states. Whereas Mre11 shows structure-specific binding to DNA ends and forks, Rad50 DNA binding has eluded characterizations. For other aspects of DNA repair, such as base excision repair, DNA binding motifs and minor groove interactions have profoundly changed our understanding of repair activities and pathway coordination [63,64], so defining Rad50 DNA binding states is likely critically important. As metal ion sites, even iron-sulfur clusters, have proven to act in positioning residues for DNA interactions as well as providing direct interactions and chemistry [65], the Zn-hook as well as the Rad50 coiled-coils may act in DNA binding and distinct Rad50 states. Like Rad50 and Mre11, Nbs1 has multiple states including bound and free FHA, BRCT and ATM domains/motifs. While PTMs of each of the MRN subunits adds an extra layer of complexity, it is clear that this plethora of biological states, which represents extreme multi-state allostery, is critical to determining biological outcomes.
MRN results to date therefore prompt a conceptual shift that entails a change from seeking separate sensor, signaling, and effector proteins to the recognition of the MRN complex as a molecular machine. In fact, it is useful to consider MRN as a type of molecular analog computer that can effectively calculate optimal responses and pathways based upon its states as set by the cell status and the nature of the bound DNA damage. Indeed, the evolution of complexes such as MRN with many conformational states underscores the ability of DNA repair machines to integrate information on cellular status and coordinate signaling, checkpoint and effector activities in different ways as needed to optimally maintain genetic integrity.
This conceptual change is critical. It means that new strategies for experimental analyses of cell biology must be developed to appropriately solve problems regarding DSB responses: information from gene knockouts and knockdowns must be better complemented by characterizations of complexes with multiple states defining functional outcomes. Interactions and PTMs that set MRN states allow the integration of information from both cell state and DNA damage, so that initial sensing, signaling, and effector functions can be coupled and thereby coordinated. Now we must employ combined structural, biochemical and genetic methods to meet the challenge of decoding these analog computers that solve DNA damage problems by determining optimal biological responses. Such combined structural and biological studies offer the opportunity to understand and even reprogram DNA repair machines: an opportunity that promises substantial rewards in all areas of biology and medicine.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Structural Cell Biology of DNA Repair Machines P01 grant CA92584. We thank T. Dobbs and Kathleen Dixon for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hopfner KP, Putnam CD, Tainer J. DNA double-strand break repair from head to tail. Current Opinion in Structural Biology. 2002;12:115–122. doi: 10.1016/s0959-440x(02)00297-x. [DOI] [PubMed] [Google Scholar]
- 2.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 3.Bierne H, Ehrlich SD, Michel B. Deletions at stalled replication forks occur by two different pathways. Embo J. 1997;16:3332–3340. doi: 10.1093/emboj/16.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. Embo J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 6.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 8.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 9.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 10.Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 11.Lieber M. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 13.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 14.Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, Schindler D, Dork Human T. RAD50 Deficiency in a Nijmegen Breakage Syndrome-like Disorder. The American Journal of Human Genetics. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 16.Bhaskara V, Dupré A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull T. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DR, Kanaar R, Wyman C. Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J Mol Biol. 2004;339:937–949. doi: 10.1016/j.jmb.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopfner K, Craig L, Moncalian G, Zinkel R, Usui T, Owen B, Karcher A, Henderson B, Bodmer J, Mcmurray C, Carney J, Petrini J, Tainer J. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 21.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 22.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahal EA, Henricksen LA, Li Y, Williams RS, Tainer J, Dixon K. ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell cycle (Georgetown, Tex) 2010;9:2866–2877. doi: 10.4161/cc.9.14.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease Function of Human Mre11 Promotes Deletional Nonhomologous End Joining. Journal of Biological Chemistry. 2009;284:30565–30573. doi: 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Chen J. MRE11-RAD50-NBS1 Complex Dictates DNA Repair Independent of H2AX. Journal of Biological Chemistry. 2010;285:1097–1104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrova N, De Lange T. Cell Cycle-Dependent Role of MRN at Dysfunctional Telomeres: ATM Signaling-Dependent Induction of Nonhomologous End Joining (NHEJ) in G1 and Resection-Mediated Inhibition of NHEJ in G2. Molecular and Cellular Biology. 2009;29:5552–5563. doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams R, Williams J, Tainer J. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 30.Hopfner KP, Tainer JA. DNA mismatch repair: the hands of a genome guardian. Structure. 2000;8:R237–241. doi: 10.1016/s0969-2126(00)00545-1. [DOI] [PubMed] [Google Scholar]
- 31.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 32.Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem. 2005;280:2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- 34.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd J, Chapman JR, Clapperton J, Haire L, Hartsuiker E, Li J, Carr AM, Jackson S, Smerdon S. A Supramodular FHA/BRCT-Repeat Architecture Mediates Nbs1 Adaptor Function in Response to DNA Damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams J, Guenther G, Classen S, Glover JM, Iwasaki H, Russell P, Tainer J. Nbs1 Flexibly Tethers Ctp1 and Mre11-Rad50 to Coordinate DNA Double-Strand Break Processing and Repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hari FJ, Spycher C, Jungmichel S, Pavic L, Stucki M. A divalent FHA/BRCT-binding mechanism couples the MRE11–RAD50–NBS1 complex to damaged chromatin. EMBO reports. 2010;11:387–392. doi: 10.1038/embor.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. The Journal of Cell Biology. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson E, Nievera CJ, Lee AY, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–22952. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi J, Okui M, Asaithamby A, Burma S, Chen BP, Tanimoto K, Matsuura S, Komatsu K, Chen D. WRN participates in translesion synthesis pathway through interaction with NBS1. Mechanisms of Ageing and Development. 2010;131:436–444. doi: 10.1016/j.mad.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Molecular & Cellular Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge S. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 43.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerosaletti KM, Desai-Mehta A, Yeo TC, Kraakman-Van Der Zwet M, Zdzienicka MZ, Concannon P. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis. 2000;15:281–286. doi: 10.1093/mutage/15.3.281. [DOI] [PubMed] [Google Scholar]
- 45.Lee J, Paull T. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH, Paull T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 47.Lee J, Goodarzi AA, Jeggo PA, Paull T. 53BP1 promotes ATM activity through direct interactions with the MRN complex. The EMBO Journal. 2010;29:574–585. doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, Ogi T, Takeda S, Taniguchi Y. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 52.de Jager M, Wyman C, van Gent DC, Kanaar R. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Research. 2002;30:4425–4431. doi: 10.1093/nar/gkf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rupnik A, Lowndes N, Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–135. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 54.Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 56.Manthey KC, Opiyo S, Glanzer JG, Dimitrova D, Elliott J, Oakley GG. NBS1 mediates ATR-dependent RPA hyperphosphorylation following replication-fork stall and collapse. J Cell Sci. 2007;120:4221–4229. doi: 10.1242/jcs.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer J. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perry JJP, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Current Opinion in Structural Biology. 2010;20:283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rambo R, Tainer J. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Current Opinion in Structural Biology. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hura GL, Menon AL, Hammel M, Rambo R, Poole FL, Tsutakawa SE, Jenney FE, Classen S, Frankel KA, Hopkins RC, Yang S, Scott JW, Dillard BD, Adams MWW, Tainer JA. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammel M, Yu Y, Mahaney B, Cai B, Ye R, Phipps B, Rambo R, Hura G, Pelikan M, So S, Abolfath R, Chen D, Lees-Miller S, Tainer J. Ku and DNA-dependent Protein Kinase Dynamic Conformations and Assembly Regulate DNA Binding and the Initial Non-homologous End Joining Complex. Journal of Biological Chemistry. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 63.Hitomi K, Iwai S, Tainer J. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 64.Huffman JL, Sundheim O, Tainer J. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 65.Kuo CF, McRee DE, Fisher CL, O’Handley SF, Cunningham RP, Tainer JA. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]