Summary
DNA·RNA hybrid duplexes are functionally important structures in gene expression that are underutilized as potential drug targets. Several tools are described here for the discovery and characterization of small molecules capable of the selective recognition of DNA·RNA hybrid structures. Competition dialysis and thermal denaturation of mixtures of polynucleotide structures can be used to identify small molecules that bind selectively to DNA·RNA hybrids. An assay that measures small molecule inhibition of RNase H can be used to measure a functional response to these ligands.
Keywords: DNA·RNA hybrid, RNase H, dialysis, thermal denaturation, enzyme inhibition, UV spectrophotometry
1. Introduction
The DNA·RNA hybrid helix was first discovered in 1960, only seven years after the discovery of the famed DNA double helix (1, 2). The hybrid structure was immediately recognized as a key component of genetic information transfer, even though mRNA and tRNA had yet to be discovered (2). After nearly half a century, DNA·RNA hybrid duplexes have yet to be exploited as a target in drug design, despite their unquestioned functional significance. This stands in contrast to other non-canonical nucleic acid structures (e.g. DNA triplexes and quadruplexes, Z-DNA, RNA secondary structures), all of which have been recognized as potential therapeutic targets. The reasons underlying the failure to target the DNA·RNA hybrid are most likely related to uncertainties and misunderstandings about the precise nature of the hybrid duplex structure, along with the lack of available lead ligands (whether of natural or synthetic origin) and, finally, the dearth of convenient assays to assess structural-selective binding to the hybrid.
Structure- and sequence-selective ligands for DNA·RNA hybrids have a variety of potential pharmaceutical applications. For instance, the first-formed product of HIV reverse transcriptase is a DNA·RNA hybrid duplex. Generation of the viral DNA duplex and its integration into the host genome are dependent on digestion of the RNA strand of this hybrid by RNase H (3). Loading DNA·RNA hybrid duplexes with small-molecule ligands inhibits RNase H (4, 5) and thus provides a new route to the control of HIV infection. The tumour-specific enzyme, telomerase is a specialized reverse transcriptase, with protein structure domains closely-related to the HIV enzyme (6). A DNA·RNA hybrid duplex is generated in the course of the telomerase reaction, so telomerase is also amenable to such a small-molecule targeting strategy (7, 8). Indeed, the thermodynamic stability of the DNA-RNA hybrid appears to regulate telomerase activity (9). A long-term goal of nucleic acid-directed drug discovery is the control of gene expression. This may potentially be achieved through sequence-selective intervention in the RNA polymerase products of normal gene expression or binding to the Okazaki fragment RNA·DNA hybrid formed at the initiation of DNA replication (10).
The consensus view of hybrid duplex structures in solution is that the overall helix conformation lies somewhere between the global B-form adopted by pure DNA and the A-form of pure RNA duplexes. The conformation of each strand is driven by the sugar puckers, which in turn are determined by the β-effect of the 3' and 2' substituents of the furanose rings. Thus in the DNA strand, “North” puckers predominate, driving that strand towards a B-conformation. In contrast, for the RNA strand, more heterogeneous sugar puckers are detected (11) resulting in an overall A-like structure. The duplex formed on combination of such diverse strands differs significantly from both pure DNA·DNA and RNA·RNA duplexes (12). The structures of different DNA·RNA hybrids can differ, depending on whether the DNA strand is purine or pyrimidine (13, 14). DNA·RNA hybrids of differing sequence may thus present different backbone conformations and groove geometries, distinctive features that may be selectively targeted by small molecules.
Intuitive attempts to identify ligands for DNA·RNA hybrids using established DNA duplex groove binders or intercalators are perhaps doomed. The grooves of the hybrid are not recognized by the classic minor groove binders distamycin (for examples, see data in Figure 3). Amongst intercalators, matching the planar surface shape and size of a ligand to the cross-sectional dimensions of triplex and quadruplex DNA proved a fruitful approach to the identification of structure-selective binding (15). However, the search for structure selective intercalators of DNA·RNA hybrids is constrained by the similarity of the Watson-Crick base-pairs, and hence intercalation site dimensions, to those of the pure DNA or RNA duplexes. None-the-less some success has been achieved in identifying ethidium as possessing a marked binding preference for the poly(A)·poly(dT) hybrid duplex (16) and re-designing actinomycin to enhance hybrid selectivity (17, 18). The few hybrid binding ligands so far reported are dominated by ethidium and aminoglycoside structures (4, 8, 19, 20) and have recently been the subject of a comprehensive review (19). All of these investigations focus on measuring binding phenomena using spectrophotometric, fluorimetric and calorimetric techniques but offer little rationalization of the inter-molecular interactions that drive the recognition event.
Figure 3.

Melting of mixtures data for “classic” duplex DNA minor groove binders (0.25 µM) that imply lack of binding to structures other than the DNA duplex.
Herein we describe three assays, used in our laboratories, that are useful in exploring the structure and sequence binding preferences of small molecules that target DNA·RNA hybrid duplexes. Competition equilibrium dialysis (21) delivers a detailed and thermodynamically-rigorous profile of the binding strengths and preferences of an investigational ligand across a broad range of up to 48 different nucleic acid structures and sequences (22–25). The assay in its current form typically includes both poly(dA)·poly(U) and poly(A)·poly(dT) hybrids alongside the counterpart pure DNA and pure RNA duplexes. The melting of mixtures assay (26) is an extension of the well-established DNA thermal denaturation assay (27, 28) that detects the effects of added ligand on thermal denaturation of four duplexes simultaneously. It is valuable for rapidly assessing binding preferences amongst DNA, RNA and hybrid duplexes; for the hybrid systems it also provides evidence of selectivity between dA·U and A·dT sequences. Finally a RNase H assay is included that provides a pharmaceutically-relevant biological endpoint for studies of ligands for DNA·RNA hybrids. Various protocols for the detection and quantitation of telomerase inhibition are described in the literature (29–31).
All three assays described here are UV spectrophotometric methods, directly detecting either ligand or nucleic acid without the use of radio-labels or fluorescent tags. This latter is particularly important since the presence of large aromatic groups, typical of fluorophores and quenchers, will itself alter the nucleic acid structures and thereby ligand binding events.
1.1 Overview of Methods and Illustrative Results
1.1.1 Competition Dialysis
The competition dialysis assay is a simple extension of equilibrium dialysis binding assays (32, 33). In the competition dialysis assay, an array of nucleic acid structures is dialyzed against a common test ligand ("drug") solution. Each structure in the array is at an identical concentration and is contained within its own dialysis unit, isolated from all other structures. All structures are in contact with the same ligand solution. At equilibrium, the free ligand concentration is identical for all structures in the array, but each structure will bind ligand according to its affinity for the ligand. The difference in the amount of ligand bound to each structure is proportional to the association constant of the ligand for that structure. A simple bar graph reveals at a glance the sequence or structural preferences of the ligand. Figure 1 shows a representative result for the DNA intercalator ethidium. In this case, binding to an array of 13 nucleic acid structures reveals that ethidium binds with a slight, but significance, preference to a DNA·RNA hybrid structure.
Figure 1.

Competition dialysis data for the interaction of ethidium with an array containing 13 nucleic acid structures. The data show preferential binding to the DNA·RNA hybrid poly(rA)·poly(dT) over all other structures and sequences..
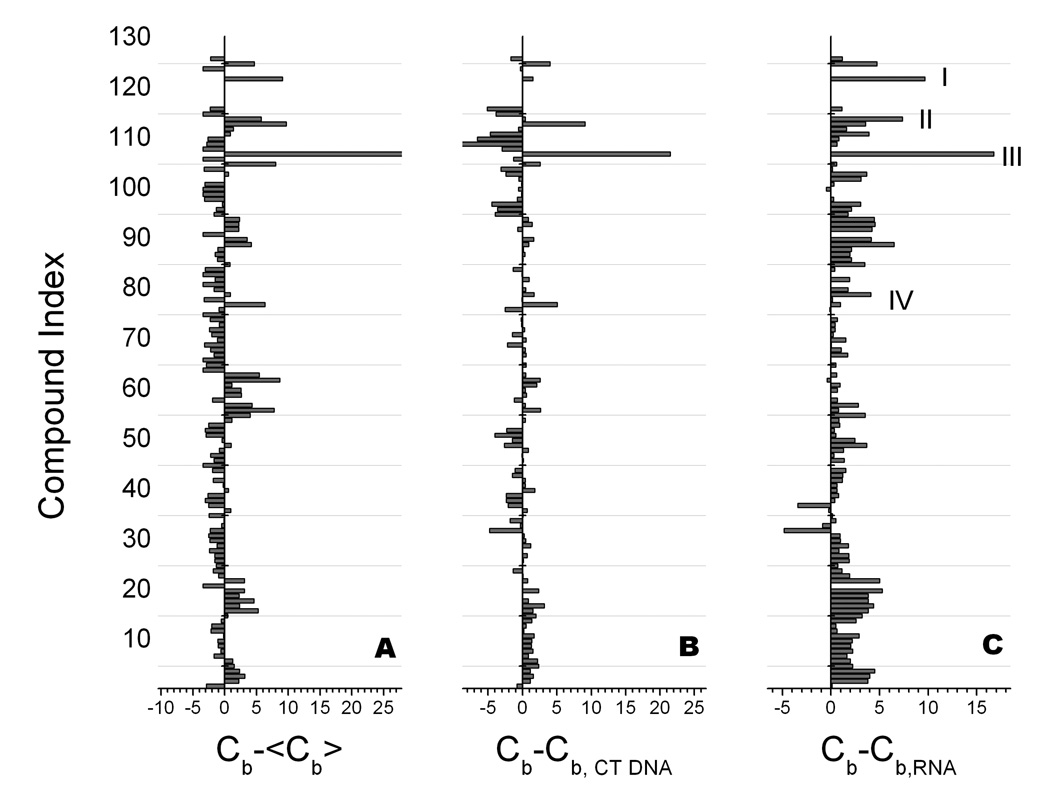
Competition dialysis is particularly powerful when examining a library of small molecules. In such a case, competition dialysis difference plots can be used to highlight those ligands that bind with high affinity and selectivity to a particular structure or sequence of interest (34, 35). Figure 2 shows an example in which ligands that bind to a DNA·RNA hybrid structure were sought. In this case, the binding of a library of 126 nucleic acid binding compounds to an array of 13 nucleic acid structures and sequences was studied. Figure 2 shows three difference plots that emphasize difference binding to a DNA·RNA hybrid structure, to a standard DNA duplex, and to an RNA duplex. Four compounds stand out as having higher than average affinity for the DNA·RNA hybrid, while showing clear preferential binding to the hybrid over duplex DNA or RNA. These compounds are ellipticine (III), the topoisomerase poison TAS103 (I), thiazole orange (II), and ethidium (IV). These compounds are all DNA intercalators, and while they show modest preferential binding to the DNA·RNA hybrid, the intercalator motif may not provide optimal lead compounds for more stringent targeting of hybrid structures and sequences. The primary point of Figure 2 is to illustrate the power of competition dialysis for screening libraries to reveal binding preferences.
Figure 2.
Differential binding plots derived from competition dialysis studies of 126 compounds with 13 nucleic acids structures. (A). Difference in the amount bound to poly(rA)·poly(dT) relative to the average amount bound <Cb>. Positive values indicate higher than average affinity. (B) Difference in the amount bound to poly(rA)·poly(dT) relative to duplex calf thymus DNA. Positive values indicate preferential binding to the hybrid form over this standard duplex DNA. (C) Difference in the amount bound to poly(rA)·poly(dT) relative to poly(rA)·poly(U). Positive values indicate preferential binding to the hybrid form over RNA. Four compounds are highlighted (I – IV) that bind with higher than average affinity to the hybrid structure while showing preferential binding to the hybrid form over DNA or RNA. The construction and use of differential binding plots is fully described in (34, 35, 44)
Detailed, step-by-step protocols for the competition dialysis assay and for the analysis of competition dialysis data have been presented in several publications (21, 22, 25, 34, 35). These protocols will not be repeated here, and the interested investigator should consult the published protocols. Competition dialysis is an important element of the toolbox for the discovery of DNA·RNA hybrid binders, as this brief discussion indicates.
1.1.2 Melting of Mixtures
Thermal denaturation is a powerful tool for monitoring binding to nucleic acids (27, 28, 36, 37). Ligands that bind to structured nucleic acids stabilize the ordered structure and elevate the melting temperature for the denaturation of that structure. The magnitude of the change in melting temperature is a complex function of binding affinity and stoichometry (28). Qualitatively, an increase in the melting temperature for a nucleic acids structure provides unambiguous evidence for binding. A recent extension of the thermal denaturation assay resulted from the simple expediency of making a mixture of different nucleic acid structures that could be distinguished by their melting temperatures(26). Addition of low binding ratios of a test ligand altered the melting temperature of the nucleic acid structure to which it bound most avidly, providing a simple, rapid evaluation of its structural preference. A mixture containing DNA, RNA, and two DNA·RNA hybrid forms was described and tested (26), and a protocol for that preparation and use of that mixture will be described here.
Example melting of mixtures data are presented for established DNA minor groove binders (Figure 3) and a family of analogues of the biarylpyrimidine 1a (Figure 4, panel A.) These demonstrate the value of this assay in rapidly and simultaneously assessing thermal stabilization and selectivity amongst the four nucleic acid structures. Thus although analogue 1d showed the greatest ΔTm for the poly(dA)·poly(U) duplex it was the analogue 1b that showed the most selectivity between the structures, with minimal shifting of the DNA duplex.
Figure 4.


Nucleic acid melting and RNase H inhibition data for a family of analogues of biarylpyrimidine 1a; assays performed according to the protocols described in the text. Panel A (top), melting mixtures assay, 4 × 10 µM bp nucleic acids, 0.25 µM ligand; inset shows an expansion of the poly(dA)·poly(U) melting curves. Panel B (bottom), inhibition of RNase H digestion of poly(dA)·poly(U) hybrid duplex; nucleic acid 10 µM bp; inhibitors, 3 µM.
The data of Figure 4 also show a direct correlation between the ligand-induced ΔTms of the poly(dA)·poly(U) duplex (panel A, inset) and the potency of RNase H inhibition (panel B). The rank order of ΔTms is the same as the rank order as inhibitors of RNase H digestion of that hybrid duplex. Thus there is a strong correlation between strength of the ligand–hybrid interaction and RNase H inhibitor potency; this means that the relatively quick and inexpensive mixed melting assay may be used as a surrogate for the more expensive and time-consuming RNase H assay.
1.1.3 Ribonuclease H Assay
This spectrophotometric assay is based on that previously described by Rasche et al (38). The release of nucleotides from the hybrid duplex substrate is followed as an increase in A260. The overall ΔA260 change detected for complete digestion of the RNA strand of a hybrid duplex approaches that observed following loss of the duplex secondary structure in the UV thermal melting experiment. This is a direct assay that monitors disappearance of substrate: the signal is independent of the inhibitor under investigation. The scale on which the reaction is performed (enzyme and substrate concentrations) is determined by the sensitivity of the UV spectrophotometer (Note 1) and there is no need for either radioactive labelling or the introduction of fluorescent tags.
Solutions of the hybrids 10 µM bp were found to give a final ΔA260 ≈ 0.06, appropriate for monitoring the reaction. Typical time courses for the digestion of poly(dA)·poly(rU) and poly(A)·poly(dT) are shown in Figure 5 panel A. A burst phase is apparent at the start of the reaction which is typical of “ping-pong” kinetics involving a covalently-linked enzyme-substrate intermediate. This phase was usually flowed by a linear phase from which the Vinital was estimated as the gradient of a best fit straight line; at high inhibitor concentrations, curvature of the line could complicate estimation of this gradient.
Figure 5.
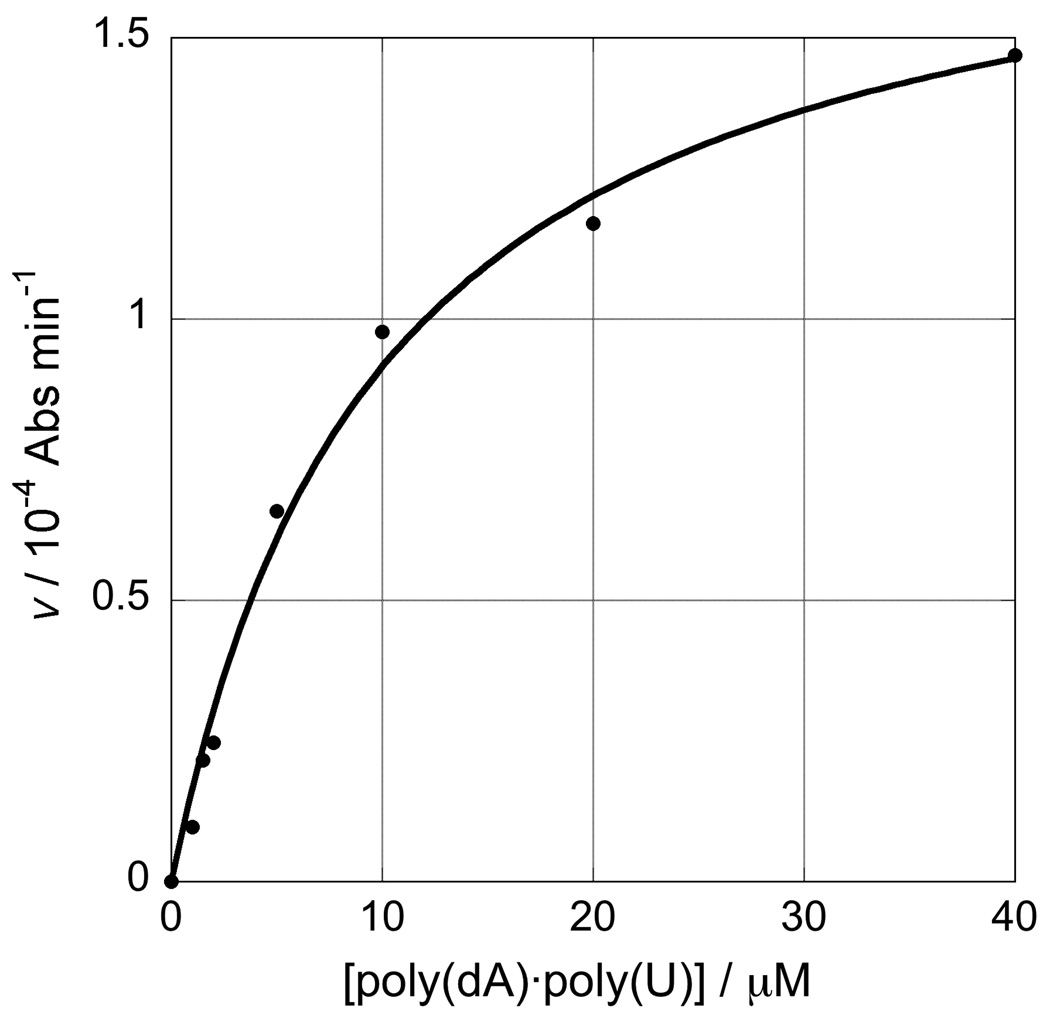
Kinetics of the RNase H reaction. Panel A, A260 change during the course of reaction for the RNase H degradation of poly(dA)·poly(rU) (10 and 20 µM bp) and poly(rA)·poly(dT) (10 µM bp) hybrid duplexes showing the initial burst phase followed by the linear section of the curve used to estimate Vmax; panel B, dependence of V on the concentration of poly(dA)·poly(rU) substrate from which KM was determined (KM= 9.99±1.39 µM, R = 0.996); panel C, effect of varying concentration of inhibitor 1d when [poly(dA)·poly(rU)] = 40 µM, from which IC50 = 6.53±0.03 µM (R = 0.997).
As with any enzyme reaction, it was first necessary to determine the KM of the enzyme for the specific substrate under investigation. Figure 5 panel B shows the dependence of Vmax on [poly(dA)·poly(rU)] from which KM was determined (KM= 9.99±1.39 µM for poly(dA)·poly(rU)). Subsequent kinetic runs were performed with [S] = 4KM to ensure maximal velocity (39). Formally, the inhibitor strength should be measured as the Ki. However, this requires repetitions of the assay at ranges of [I] and [S] which may become expensive in both time and the quantity of enzyme consumed; for most purposes it is adequate to measure the IC50. Data obtained for a range of concentrations of the most potent inhibitor, compound 1d, and the curve-fit for the determination of the IC50 (6.53±0.03 µM) are shown in Figure 5 panel C.
The RNase H assay can be used in a survey mode to screen series of compounds at a fixed concentration in order to select the most potent inhibitors for detailed investigation. Figure 4, Panel B shows data for a range of analogues of biarylpyrimidine 1a inhibiting digestion of the poly(dA)·poly(U) hybrid. Significantly, the rank order of potency of inhibition thus detected followed the magnitude of the ΔTm shifts detected in the in the melting of mixtures assay (Figure 4, panel A) even though this melting assay was performed at non-saturating concentrations of ligand using a different buffer system.
2. Materials
2.1 Competition Dialysis
Materials for the competition dialysis assay in a variety of different formats containing arrays of 13, 19 and 48 nucleic acid structures have been presented in several publications (21, 22, 25, 34, 35)
Dialysis Units. The first generation assay used 1.0 or 0.5 mL DispoDialyzer® units (Spectrum Medical Industries, Inc.; www.spectrumlabs.com) of the desired molecular weight cutoff, usually 3500 MWCO. Subsequently, a less expensive and overall more suitable alternative was found, Pierce Slide-A-Lyzer® MINI Dialyzer units (Pierce Chemical Company; www.piercenet.com). Dialysis units may be reused several times after careful rinsing with buffer solution.
Nucleic acids. The nucleic acid structures used in one version of the competition dialysis assay are listed in Table 1. All samples are dissolved in BPES buffer, consisting of 6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 0.185 M NaCl, pH 7.0. Samples of DNA from Clostridium perfringens calf thymus and Micrococus lysodeikticus were purchased from Sigma Chemical Co. (St. Louis, MO), and were sonicated, phenol extracted, and purified as previously described (40). Poly(dA), poly(dTpoly(U),poly(dC), poly(dA)-poly(dT), poly(dAdT), and poly(dGdC)) were purchased from Pharmacia Biotech, Inc. (Piscataway, NJ). Poly(rA) and poly(A)-poly(U) were purchased from Sigma Chemical Co. Deoxyoligonucleotides 5’T2G20T2, 5’G10T4G10 and 5’AG3(TTAG3)3 were purchased from Research Genetics (Huntsville, AL). Synthetic single-stranded and duplex polynucleotides were used without further purification. The poly(rA):poly(dT) DNA-RNA hybrid was prepared by mixing poly rA and poly dT in a 1:1 molar ration, heating to 90°C, and slow cooling to room temperature. The DNA and RNA triplex forms prepared by mixing poly(dA)-poly(dT) with poly(dT) (or poly(A)-poly(U) with poly(U)) in a 1:1 mole ratio, heating to 90 °C, and slowing cooling to room temperature. Guadruplex DNA and i-motif DNA were prepared by heating the oligonucleotides (5’T2G20T2), (5’G10T4G10), (5’AG3(T2AG3)3) or poly(dC) to 90°C for 2 min, slowly cooling to room temperatue, and then equilibrating for 48 h at 4°C before use. Left-handed Z DNA was prepared by bromination of poly(dGdC) as previously described (41).
Concentration Determinations. Concentration of nucleic acid samples were determined by UV absorbance measurements using the extinction coefficients and absorption maxima listed in Table I. Stock solutions of nucleic acids were prepared and maintained at 75 µM concentration, using the monomeric unit of each polynucleotide as the concentration standard. That means nucleotides (nt) for single stranded forms, base pairs (bp) for duplex forms, triplets for triplex forms and quartets for quadruplex forms. The exinction coefficients listed in Table 1 all refer to these concentration standards.
Table 1.
Nucleic acid conformation and samples used in competition dialysis
| Conformation | DNA/oligonucleotide | λ (nm) |
ε (M−1cm−1) |
|---|---|---|---|
| Single strand purine | poly dA | 257 | 8600 |
| polyA | 258 | 9800 | |
| Single-strand pyrimidine | poly dT | 264 | 8520 |
| polyU | 260 | 9350 | |
| Duplex DNA | C. perfringens (31%GC) | 260 | 12476 |
| calf thymus (42%GC) | 260 | 12824 | |
| M. lysodeikticus (72%GC) | 260 | 13846 | |
| poly dA: poly dT | 260 | 12000 | |
| poly (dAdT) | 262 | 13200 | |
| poly (dGdC) | 254 | 16800 | |
| DNA-RNA hybrid | poly rA: poly dT | 260 | 12460 |
| Duplex RNA | poly A: poly U | 260 | 14280 |
| Z DNAa | Br- poly (dGdC) | 254 | 16060 |
| Triplex DNA | poly dA: (poly dT)2 | 260 | 17200 |
| Triplex RNA | PolyA(polyU)2 | 260 | 17840 |
| Quadruplex DNA 1 | (5’T2G20T2)4 | 260 | 39267 |
| Quadruplex DNA 2 | 5’AG3(TTAG3)3 | 260 | 73000 |
| Quadruplex DNA 3 | (5’G10T4G10)x | 260 | 39400 |
| i-motif | (poly dC)4 | 274 | 27200 |
Key: λ: Wavelength; ε: molar extinction coefficient
2.2 Melting of Mixtures
Polynucleotides poly(rA), poly(dA), poly(rU), poly(dT) were purchased from Sigma Aldrich (Pool, UK) and used without further treatment.
Assay buffer ¼(BPES (1.5 mM Na2HPO4, 0.5 mM NaH2PO4, 0.25 mM, Na2EDTA, 46.25 mM NaCl, pH = 7.0). All materials Sigma-Aldrich molecular biology grade.
2.3 Ribonuclease H Assay
RNase H was obtained from Promega UK Ltd. (Southampton, UK) in HEPES buffer–glycerol (1.5 U µl−1) and used directly, Note 2.
Reaction buffer: 50 mM Tris pH 8.0 (Note 3), 50 mM NaCl, 10 mM MgCl2. All materials Sigma-Aldrich molecular biology grade.
Polynucleotides poly(dA), poly(dT), poly(U), poly(rA) were used as supplied by Sigma.
3. Methods
3.1 Competition Dialysis
Detailed, step-by-step protocols for the competition dialysis assay in different formats have been presented in several publications (21, 22, 25, 34, 35).
For each competition dialysis assay, place 400ml of the dialysate solution containing 1µM test ligand concentration into a beaker.
Pipette 180 uL (at 75µM monomeric unit) of each of the DNA samples listed in Table 1 into a separate Slide-A-Lyzer® MINI dialysis unit with 7000 molecular weight cutoff membrane. Place all 19 dialysis units were placed in a MINI dialysis flotation device (Pierce Chemical Company) and then place the whole unit in the beaker containing the dialysate solution.
Cover the beaker with parafilm, wrap the beaker in foil, and equilibrate with continuous stirring for 24 h at room temperature (20–22°C).
At the end of the equilibration period, carefully collect DNA samples in the corner of the mini dialysis unit and transfer to microfuge tubes. Add 10% sodium dodecyl sulfate (SDS) to DNA samples to a final concentration of 1% (w/v) (SDS).
The total concentration of drug (Cf) with each mini dialysis unit is then determined spectrophotometrically using wavelengths and extinction appropriate for each ligand. The free ligand concentration (Cf) is determined spectrophotometrically using an aliquot of the dialysate solution, although its concentration usually does not vary appreciable from the initial 1 µM concentration.
The amount of bound drug is determined by difference, Cb=Ct-Cf. Plot as a bar graph using Orgin software (Version 5.1, Micalcal, Inc., Northampton, MA) or any other available graphics package suitable for plotting and analysis.
3.2 Melting of Mixtures
Commercial polynucleotides were dissolved in a small volume of assay buffer to give stock solutions approximately 5 mg or 10 U per ml. The concentrations (in phosphate group) were then determined spectrophotometrically using the data of Table 2.
-
Preparation of duplexes
poly(dA)·poly(T)
poly(A)·poly(dT)
poly(dA)·poly(U)
poly(A)·poly(U)
Working solutions (40 µM bp) of each duplex were prepared in 100 ml volumetric flasks by mixing the required volume (Note 4) of each polynucleotide single strand solution and then making up to the line with buffer. Solutions were transferred to Sterilin screw-cap jars and annealed by heating at 100 °C for 5 min in a water bath followed by slow cooling overnight. All four working solutions were stable in the refrigerator over several months, Note 5.
Test compounds (approx 1 mg weighed accurately) were dissolved in water (0.5 ml) and diluted 10-fold, if necessary, to give volumes convenient for handling.
In a typical assay (Figure 4A) solutions of each duplex (1 ml) were mixed in a polythene vial and the requisite amount of test compound (2–5 µl in water) added to give a final mixture 10 µM in each duplex, 0.25 µM in test compound. This solution was mixed gently and allowed to equilibrate for at least 12 h at 4 °C before use.
Data were acquired at 260 nm using the “Thermal” module within the Cary WinUV software to provide the melting curve and 1st differential. In a typical run, equilibration for 30 min at 19 °C was followed by a ramp from 20–80 °C at 1 °C min−1.
Table 1.
Data for the Spectrophotometric quantitation of polynucleotides (42).
| Polynucleotide | λ / nm | ε / M−1 cm−1 |
|---|---|---|
| poly(dA) | 257 | 8600 |
| poly(dT) | 264 | 8520 |
| poly(A) | 258 | 9800 |
| poly(U) | 260 | 9350 |
3.3 Ribonuclease H Assay
Concentrated stock solutions of poly(dA)·poly(rU) and poly(dT)·poly(rA) (100 µM bp) were prepared in reaction buffer. Formation of the duplexes was checked by thermal melting (Tm = 73.0 °C, ΔA260 ≈ 0.06, for poly(dA)·poly(rU) 10 µM bp), the solutions were stored at 0–4 °C and were stable over several months at 4 °C.
Test compounds dissolved in water (approx. 2 mg ml−1) were added to portions of diluted nucleic acid stock solution in glass or polythene vials to achieve the desired experimental concentrations, then equilibrated at 0–4 °C for at least 12 h before use.
In a typical assay, the nucleic acid (or test compound–nucleic acid) solution was equilibrated at 37 °C; 1.2 ml was transferred to a cuvette, 2 µl of enzyme preparation added directly and the reaction mixture mixed by gentle inversion 12 times. The sample was placed in the spectrophotometer block at 37 °C and A260 monitored for 1–3 h following an initial 2 min delay. Cuvettes were cleaned thoroughly before re-use Note 6.
Data were transferred to software packages for analysis. Typical plots of dA260 vs. time are shown in Figure 5, panel A. The initial burst phase was followed by a region of steady reaction with approximately linear gradient. Reaction rate, V, was determined as the gradient of this linear region: typically over a period of 20 min during the first 1.5 h of reaction, although in the presence of the more potent inhibitors, this extended up to 90 min. The linear region was identified graphically and the gradient calculated using a software-fitted a straight line (R>0.996). Concentration and velocity (Note 7) data were transferred to the KaleidaGraph (43) package for calculation of KM, Vmax and IC50 using the integral curve-fitting routines of the program (see Figure 5 panels B, C).
Acknowledgments
This work supported by award number R01GM077422 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official vies of the NIGMS or NIH.
Footnotes
The assays described herein were performed using a CARY400Bio spectrophotometer equipped with a multi cell (6+6) block and Peltier heating control. A matched par of Hellma masked quartz cuvettes (114-QS) was used. The test solution cuvette was placed in the front position and buffer in the back (or nothing where relative absorbance data were required); sample volumes were 1.2 ml. The temperature was monitored using a temperature probe placed in a third cuvette, containing buffer, placed in the position adjacent to the sample cuvette in the block.
Attempts at dilution of the enzyme stock into reaction buffer prior to use resulted in loss of activity. The enzyme preparation is sufficiently fluid at room temperature to be pipetted into the reaction cuvette. Thorough mixing by gentle inversion 12 times initiates the reaction.
Tris buffer was used as made up from the supplied solid; the pH was not checked or adjusted.
Precision digital pipettes were used for all test compound and nucleic acid measurements.
Before use, each of the four duplexes should be melted individually for quality assurance. The absorbance change observed for each duplex is about the same.
It is critical that the cuvettes are cleaned thoroughly: during the KM study, cuvettes were soaked for 3–6 h in 2% Hellmanex solution between reactions.
From the total absorbance change at “infinity” (about 6.5 h for poly(dA)·poly(U)), and assuming complete release of rU nucleotides from the hybrid, the absorbance data could be scaled to nmoles of rU released (1 Abs unit ≡ 287.08 nmoles rU) and hence rate data converted to pmol(rU) s−1.
References
- 1.Rich A. A hybrid helix containing both deoxyribose and ribose polynucleotides and its relation to the transfer of information between the nucleic acids. Proc. Nat. Acad. Sci. USA. 1960;46:1044–1053. doi: 10.1073/pnas.46.8.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rich A. Discovery of the hybrid helix and the first DNA-RNA hybridization. J Biol Chem. 2006;281:7693–7696. doi: 10.1074/JBC.X600003200. [DOI] [PubMed] [Google Scholar]
- 3.Williams DA, Lemke TL. Foye's Principles of Medicinal Chemistry. Baltimore, MD: USA: Lippincott, Williams & Wilkins; 2002. [Google Scholar]
- 4.Ren J, Qu X, Dattagupta N, Chaires JB. Molecular recognition of a RNA:DNA hybrid structure. J Am Chem Soc. 2001;123:6742–6743. doi: 10.1021/ja015649y. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri CM, Li TK, Guo S, et al. Aminoglycoside Complexation with a DNA·RNA Hybrid Duplex: the Thermodynamics of Recognition and Inhibition of RNA Processing Enzymes. J. Am. Chem. Soc. 2003;125:6469–6477. doi: 10.1021/ja021371d. [DOI] [PubMed] [Google Scholar]
- 6.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum Telomerase Catalytic Subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 7.Francis R, West C, Friedman SH. Targeting telomerase via its key RNA/DNA heteroduplex. Bioorganic Chemistry. 2001;29:107–117. doi: 10.1006/bioo.2000.1196. [DOI] [PubMed] [Google Scholar]
- 8.Rangarajan S, Friedman SH. Design, synthesis, and evaluation of phenanthridine derivatives targeting the telomerase RNA/DNA heteroduplex. Bioorg Med Chem Lett. 2007;17:2267–2273. doi: 10.1016/j.bmcl.2007.01.070. [DOI] [PubMed] [Google Scholar]
- 9.Yu HQ, Zhang DH, Gu XB, Miyoshi D, Sugimoto N. Regulation of telomerase activity by the thermodynamic stability of a DNA ×RNA hybrid. Angew Chem Int Ed Engl. 2008;47:9034–9038. doi: 10.1002/anie.200803577. [DOI] [PubMed] [Google Scholar]
- 10.Gmeiner WH, Cui W, Konerding DE, et al. Shape-Selective Recognition of a Model Okazaki Fragment by Geometrically-Constrained Bis-Distamycins. J. Biomol. Struct. Dyn. 1999;17:507–518. doi: 10.1080/07391102.1999.10508381. [DOI] [PubMed] [Google Scholar]
- 11.Saenger W. Principles of Nucleic Acid Structure. New York: Springer Verlag; 1984. [Google Scholar]
- 12.Noy A, Perez A, Marquez M, Luque FJ, Orozco M. Structure, recognition properties, and flexibility of the DNA.RNA hybrid. J Am Chem Soc. 2005;127:4910–4920. doi: 10.1021/ja043293v. [DOI] [PubMed] [Google Scholar]
- 13.Gyi JI, Conn GL, Lane AN, Brown T. Comparison of the thermodynamic stabilities and solution conformations of DNA.RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry. 1996;35:12538–12548. doi: 10.1021/bi960948z. [DOI] [PubMed] [Google Scholar]
- 14.Gyi JI, Lane AN, Conn GL, Brown T. Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine- rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry. 1998;37:73–80. doi: 10.1021/bi9719713. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins TC. Targeting multi-stranded DNA structures. Current Medicinal Chemistry. 2000;7:99–115. doi: 10.2174/0929867003375551. [DOI] [PubMed] [Google Scholar]
- 16.Alberti P, Ren J, Teulade-Fichou MP, et al. Interaction of an acridine dimer with DNA quadruplex structures. J Biomol Struct Dyn. 2001;19:505–513. doi: 10.1080/07391102.2001.10506758. [DOI] [PubMed] [Google Scholar]
- 17.Shinomiya M, Chu WH, Carlson RG, Weaver RF, Takusagawa F. Structural, Physical, and Biological Characteristics of RNA·DNA Binding-Agent N8-Actinomycin-D. Biochemistry. 1995;34:8481–8491. [PubMed] [Google Scholar]
- 18.Takusagawa F, Takusagawa KT, Carlson RG, Weaver RF. Selectivity of F8-actinomycin D for RNA·DNA Hybrids and its Anti-leukemia Activity. Bioorg. Med. Chem. 1997;5:1197–1207. doi: 10.1016/s0968-0896(97)00062-x. [DOI] [PubMed] [Google Scholar]
- 19.Shaw NN, Arya DP. Recognition of the Unique Structure of DNA:RNA Hybrids. Biochimie. 2008;90:1026–1039. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Shaw NN, Xi H, Arya DP. Molecular Recognition of a DNA:RNA Hybrid: Sub-nanomolar Binding by a Neomycin-methidium Conjugate. Bioorg. Med. Chem. Lett. 2008;18:4142–4145. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 21.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 22.Chaires JB. A competition dialysis assay for the study of structure-selective ligand binding to nucleic acids. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Current Protocols in Nucleic Acid Chemistry. Vol. 1. New York: John Wiley & Sons, Inc.; 2002. pp. 8.3.1–8.3.8. [DOI] [PubMed] [Google Scholar]
- 23.Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007;43:313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragazzon PA, Garbett NC, Chaires JB. Competition dialysis: a method for the study of structural selective nucleic acid binding. Methods. 2007;42:173–182. doi: 10.1016/j.ymeth.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Chaires JB. Rapid screening of structurally selective ligand binding to nucleic acids. Methods Enzymol. 2001;340:99–108. doi: 10.1016/s0076-6879(01)40419-8. [DOI] [PubMed] [Google Scholar]
- 26.Shi X, Chaires JB. Sequence- and structural-selective nucleic acid binding revealed by the melting of mixtures. Nucleic Acids Res. 2006;34:e14. doi: 10.1093/nar/gnj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson WD, Tanious F, Fernades-Saiz M, Rigl CT. Evaluation of Drug-Nucleic Acid Interactions by Thermal Melting Curves. In: Fox KR, editor. Drug-DNA Interaction Protocols. Vol. 90. Totowa, NJ: Humana Press; 1997. pp. 219–240. [DOI] [PubMed] [Google Scholar]
- 28.Shi X, Chaires JB. Thermal Denaturation of Drug-DNA Complexes: Tools and Tricks. In: Waring M, editor. Sequence-specific DNA Binding Agents. Cambridge: RSC Publishing; 2006. pp. 130–151. [Google Scholar]
- 29.Sun D, Hurley LH, Von Hoff DD. Telomerase Assay Using Biotinylated-primer Extension and Magnetic Separation of the Products. Biotechniques. 1998;25:1046–1051. doi: 10.2144/98256cr03. [DOI] [PubMed] [Google Scholar]
- 30.Kim NW, Wu F. Advances in Quantification and Characterization of Telomerase Activity by the Telomeric Repeat Amplification Protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francis R, Friedman SH. An interference-free fluorescent assay of telomerase for the high-throughput analysis of inhibitors. Anal Biochem. 2003;323:65–73. doi: 10.1016/j.ab.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Craig LC, King TP. Dialysis. Methods Biochem Anal. 1962;10:175–199. doi: 10.1002/9780470110270.ch6. [DOI] [PubMed] [Google Scholar]
- 33.Muller W, Crothers DM. Interactions of heteroaromatic compounds with nucleic acids. 1. The influence of heteroatoms and polarizability on the base specificity of intercalating ligands. Eur J Biochem. 1975;54:267–277. doi: 10.1111/j.1432-1033.1975.tb04137.x. [DOI] [PubMed] [Google Scholar]
- 34.Chaires JB. Competition dialysis: an assay to measure the structural selectivity of drug-nucleic acid interactions. Curr Med Chem Anti-Canc Agents. 2005;5:339–352. doi: 10.2174/1568011054222292. [DOI] [PubMed] [Google Scholar]
- 35.Chaires JB. Structural Selectivity of Drug-Nucleic Acid Interactions Probed by Competition Dialysis. In: Waring MJ, Chaires JB, editors. DNA Binders and Related Subjects. Vol. 253. Berlin: Springer-Verl; 2005. pp. 33–54. [Google Scholar]
- 36.Crothers DM. Calculation of melting curves for DNA. Biopolymers. 1968;6:1391–1404. doi: 10.1002/bip.1968.360061003. [DOI] [PubMed] [Google Scholar]
- 37.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 38.Raschke TM, Kho J, Marqusee S. Confirmation of the Hierarchical Folding of RNase H: a Protein Engineering Study. Nature Struct. Biol. 1999;6:825–830. doi: 10.1038/12277. [DOI] [PubMed] [Google Scholar]
- 39.Tipton KF. Principles of Enzyme Assay and Kinetic Studies. In: J Danson MJ, editor. Enzyme Assays a Practical Approach. Oxford, UK: Oxford University Press; 1992. [Google Scholar]
- 40.Chaires JB, Dattagupta N, Crothers DM. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry. 1982;21:3933–3940. doi: 10.1021/bi00260a005. [DOI] [PubMed] [Google Scholar]
- 41.Moller A, Nordheim A, Kozlowski SA, Patel DJ, Rich A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry. 1984;23:54–62. doi: 10.1021/bi00296a009. [DOI] [PubMed] [Google Scholar]
- 42.Shi XC, Chaires JB. Sequence- and Structural-selective Nucleic Acid Binding Revealed by the Melting of Mixtures. Nucleic Acids Res. 2006;34:e14. doi: 10.1093/nar/gnj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.KaleidaGraph: Synergy Software. 2004 [Google Scholar]
- 44.Cai L, Chen L, Raghavan S, Ratliff R, Moyzis R, Rich A. Intercalated cytosine motif and novel adenine clusters in the crystal structure of the Tetrahymena telomere. Nucleic Acids Res. 1998;26:4696–4705. doi: 10.1093/nar/26.20.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]