Abstract
A subset of the neutralizing anti-HIV antibodies recognize epitopes on the envelope protein gp120 of the human immunodeficiency virus. These epitopes are exposed during conformational changes when gp120 binds to its primary receptor CD4. Based on chemical modification of lysine and arginine residues followed by mass spectrometric analysis, we determined the epitope on gp120 recognized by the human monoclonal antibody 559/64-D, which was previously found to be specific for the CD4 binding domain. Twenty-four lysine and arginine residues in recombinant full-length glycosylated gp120 were characterized; the relative reactivities of two lysine residues and five arginine residues were affected by the binding of 559/64-D. The data show that the epitope is discontinuous and is located in the proximity of the CD4-binding site. Additionally, the reactivities of a residue that is located in the secondary receptor binding region and several residues distant from the CD4 binding site were also altered by Ab binding. These data suggest that binding of 559/64-D induced conformational changes which result in altered surface exposure of specific amino acids distant from the CD4-binding site. Consequently, binding of 559/64-D to gp120 affects not only the CD4-binding site, which is recognized as the epitope, but appears to have a global effect on surface exposed residues of the full-length glycosylated gp120.
Shortly after infection with the human immunodeficiency virus (HIV-1), patients develop antibodies (Abs) against the virus; however, ultimately the immune system fails to control the virus, leading to acquired immune deficiency syndrome (AIDS). The characterization of neutralizing antibodies, which are generated by most individuals infected with HIV and are a major component of host defense against the virus, will provide insight on the structural features of antigen-recognition and the development of neutralization resistance. HIV neutralizing antibodies are mainly directed against the transmembrane protein gp41 and the envelope protein gp120 (1, 2). Both gp41 and gp120 are expressed as the precursor protein gp160, which matures into the individual proteins by cleavage. The proteins remain non-covalently associated and form a trimeric oligomer (gp41/gp120)3 (3). During the initial step of infection, a non-covalent complex is formed between gp120 and its primary receptor CD4, which is located as a transmembrane glycoprotein on circulating CD4 lymphocytes and monocytes (4, 5). Subsequently, conformational changes occur in gp120 and CD4, which expose the binding site on the surface of gp120 for a coreceptor of the chemokine receptor family. After formation of the complex of gp120/CD4/coreceptor, additional conformational changes in gp41 occur, facilitating the fusion of the viral membrane with the host cell membrane and entry of the viral RNA into the cell (6, 7).
Homology analyses of the glycoprotein gp120 have identified five conserved regions and five variable regions (8). The conserved regions form the core of the protein (the core consists of Gly-Ala-Gly tripeptide substitutions for 67 V1/V2 loop residues and 32 V3 loop residues; and the removal of all sugar groups beyond the linkages between the two core N-acetylglucosamine residues and deletions of 52 and 19 residues from the N and C termini, respectively) (9), and disulfide bridges in the variable regions result in the formation of flexible loops (10). Both conserved and variable regions are extensively glycosylated with approximately 50% of the mass of the glycoprotein derived from glycans (10-12). The variable loops and the carbohydrate moieties on the surface of gp120 shield conserved regions that are functionally important and are only accessible after a sequence of conformational changes in the envelope protein. Nevertheless, several regions are targets for neutralizing antibodies, in particular the variable loops V1/V2 and V3, the conserved region C4 (which contains residues involved in CD4-binding), the CD4 binding site (CD4-BS, formed by the inner domain, the outer domain and part of the bridging sheet of gp120), and the CD4-induced binding site for the coreceptor (13-19). Mapping of epitopes on gp120, mainly through competition analyses and mutagenic analyses, resulted in the identification of a “neutralizing face” recognized by neutralizing antibodies, a “non-neutralizing face” recognized by non-neutralizing antibodies and a “silent face” which has a very poor immunogenicity due to its extensive glycosylation (15, 20). Although a significant number of neutralizing antibodies targeted against epitopes in T-cell line adapted (TCLA) HIV strains have been characterized, only a small number of antibodies are capable of neutralizing primary HIV isolates in vitro (reviewed in (1, 2, 21).
The reasons for the differences in neutralization sensitivity are not fully understood, but it appears that laboratory-adapted viruses and primary isolates adopt different configurations of the trimeric envelope complex (21). Studies also suggest that various neutralizing antibodies might recognize different conformations of gp120 that do not necessarily correspond to the conformation of the truncated deglycosylated gp120 core in complex with CD4. Analysis of the thermodynamics of gp120-CD4 association showed large enthalpy and entropy changes indicating significant flexibility of gp120 in the unbound form and conformational changes upon binding of the primary receptor (22). Based on the crystal structure, gp120 in the bound state has a narrow binding pocket for CD4 (9). Xiang et al. described a gp120 mutant with the amino acid substitution S375W that adopts a narrow conformation at the CD4-BS in the absence of CD4 (23). This S375W mutant did not bind any of the tested Abs directed against the CD4-BS, also indicating different conformations of the CD4 binding site in absence and presence of the primary receptor. More recently, Kwong and co-workers showed by crystallographic structure determinations that CD4 induces a conformational change in gp120 (24) and that the conformation of the bridging sheet in gp120 undergoes significant conformational change upon CD4 binding (25). Molecular dynamics simulations also suggest different conformations for gp120 in the bound and unbound states (26). Recently, the crystal structure of a SIV gp120-core, which represents the unliganded state of the glycoprotein, was published (27). Comparison of the structure with the liganded HIVHXBc2 gp120-core showed significant conformational differences of the inner domain. In the unliganded conformation, the constituents of the CD4-binding domain form a cavity, which is long and narrow compared to the open cavity in the CD4-bound HIV-gp120.
Here we present the mapping of the epitope of full-length, non-deglycosylated gp120 recognized by the human monoclonal antibody (MAb) 559/64-D by chemical modification of arginine (R) and lysine (K) residues and subsequent mass spectrometric analyses. Immunochemical studies suggested that the epitope of this MAb is located in the CD4-BS (28), and functional studies show that this MAb is able to neutralize several TCLA-adapted strains of HIV (29) Interestingly, this and other MAbs to the CD4-binding site have been shown to prevent proteolysis of gp120 and subsequent presentation of gp120 to CD4-T-cells (30-33) that sera with broadly cross-neutralizing Abs have anti-CD4-BS Abs which can be responsible or contribute to the breadth of neutralization (34, 35). MAb 4559/64-D may also have characteristics similar to the anti-CD4-BS MAb F105 which is often used in many studies and is also not broadly neutralizing for non-TLCA strains(36). The CD4-BS is conserved among HIV strains and is functionally required for the initial step of infection. Consequently, it is important to determine the binding site of neutralizing and non-neutralizing Abs such as the MAb 559/64-D targeting the CD4-BS to understand the relationship between gp120-structure and neutralization potency of the Abs.
In this study, gp120 bound to the antibody 559/64-D, as well as free in solution, was modified by either lysine acetylation or arginine hydroxyphenylglyoxylation, and differences in the modification pattern were used to indicate interaction sites between the antibody and its antigen. The acetylation of the ε-amino group at the N-terminus of a protein and of the α-amino group in lysine residues has been previously used to characterize the topology of proteins (37-40). The derivatization of arginine residues with hydroxyphenylglyoxal (HPG) has been established as a modification to determine the participation of arginines in the active site of proteins and in the interaction site with a binding partner (41, 42). In the present case, observed differences in reactivity of arginines and lysines are used as a measure of the accessibility of the residue to the reagent. It should be noted that this is not identical to surface accessibility as changes in reactivity resulting from, for example changes in pKa due to variations in residue reactivities (e.g., salt bridges) are eliminated by taking the ratio of reactivity in the bound and unbound states.
Experimental Procedures
Materials
The CHO-cell expressed glycoprotein gp120 from HIV strain SF2 was purchased from Austral Biologicals (San Ramon, CA). The monoclonal antibody (MAb) 559/64-D was produced as described in Karwowska et al. (29). Endoproteinases GluC, AspN and LysC were obtained from Roche Molecular Biologicals (Indianapolis, IN), trypsin-(tosyl-amido-2-phenyl) ethylchloromethyl ketone from Worthington Biochemical (Freehold, NJ) and PNGaseF from Roche Diagnostics (Mannheim, Germany) and Glyko (Novato, CA). Hydroxyphenylglyoxal was obtained from Pierce (Rockford, IL), iodoacetamide and α-cyano-4-hydroxycinnamic acid from Aldrich (Milwaukee, WI), and acetic anhydride and hexadeuteroacetic anhydride from Sigma (St. Louis, MO).
Derivatization of lysine residues
HIV-gp120 and the MAb 559/64-D were combined and incubated for 1 h at RT. A two-fold molar excess of the MAb compared to gp120 was used to assure that all gp120 was bound to the antibody based on the dissociation constant of the complex (14 × 10−9 M) [29]. The molar excesses that are necessary for the partial and the complete acetylation of gp120 were determined by acetylation of the protein with 1000, 10,000 and 200,000 fold molar excess and subsequent purification by reversed phase HPLC. The conditions were as following: Protein-C4-column (4.6 mm × 250 mm; Vydac, Hesperia, CA) with a C4-guard column (Vydac); flow rate 1 mL/min; A: 0.1% trifluoroacetic acid in water, B: 0.09% trifluoroacetic acid, 10% water in acetonitrile; gradient 10% B to 70 % B over 50 min; 1 mL-fractions. The collected fractions were lyophilized, re-dissolved in PBS, and analyzed by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS). The purified protein was digested with the endoproteinase GluC in 50 mM NH4HCO3 buffer at 37°C and analyzed by MALDI/MS. Acetylated peptides were identified by their mass shift of 42 Da. A 1000 fold and a 10,000 fold molar excess, respectively, lead to partial acetylation of gp120. With a molar excess of 200,000, complete acetylation was observed. Note that the molar excess does not include the aqueous environment which also reacts with acetic anhydride, albeit at a slower rate than does the primary amine. To ensure that the antigen-antibody complex remains intact during modification, the antibody was immobilized on cyanogen bromide-activated Sepharose beads (Pierce Chemical Co. Rockford, IL) as described previously (38), gp120 was added, and the Ab-Ag complex was formed by incubation for 1 h at RT. Subsequently, the beads were washed to remove any unbound protein and analyzed by MALDI/MS for the presence of bound gp120. The complex was then acetylated with 10,000 fold molar excess acetic anhydride and washed again. The subsequent analysis of the beads by MALDI/MS showed that the acetylated gp120 was still affinity-bound.
For differential modification of lysine residues, the antigen-antibody complex in solution was initially acetylated with a 10,000 fold molar excess of acetic anhydride. Under these conditions, the native conformation of the protein was largely retained as indicated by continued recognition by the antibody. The acetylated gp120/MAb complex was purified by reversed phase HPLC. Fractions containing the acetylated gp120 were combined and derivatized with a 200,000 fold molar excess of hexadeuteroacetic anhydride. Under these conditions, the protein became denatured, allowing exhaustive acetylation. Acetylated residues were accessible to the reagent in the native or complexed state, while deuteroacetylated residues became accessible to the reagent only after denaturing the protein, regardless of whether the protein was initially complexed or non-complexed. The acetylated/trideuteroacetylated protein was again purified by HPLC. After lyophilization, the protein was dissolved in 10 μL 50% acetonitrile/0.1% formic acid. The solution was diluted 1:5 with 50 mM ammonium bicarbonate (pH 8.3) and digested with endoproteinase GluC (enzyme:substrate ratio 1:20) for 12 h at 37°C. The solution was split into two equal parts. One part was digested further with trypsin for 4 h at 37°C (enzyme:substrate ratio 1:20). The peptides in both samples were deglycosylated with PNGaseF for 12 h at 37°C. To reduce and alkylate the disulfide bridges, DTT was added to a final concentration of 10 mM (ca. 25:1 molar ratio of DTT per disulfide bond), and the solution was incubated at 56°C for 1 h. The peptides were alkylated with 55 mM iodoacetamide for 45 min at 56°C in the dark. Subsequently, the samples were analyzed by MALDI/MS and liquid chromatography (LC) electrospray ionization (ESI) MS/MS. All reactions were performed in duplicate. The ratio of deuteroacetylated lysine containing peptide to non-deuteroacetylated lysine containing peptide was determined from the abundances of the extracted ion currents of the protonated molecules.
Derivatization of arginine residues
For the formation of a non-covalent complex, HIV-gp120 and the MAb 559/64-D were incubated for 2 h at RT in a 2:1 ratio with a final concentration of 3 pmoles gp120/μL. Arginine residues were derivatized with 10 mM HPG in 25 mM NaHCO3 for 18 h 30 min with rotation at RT in the dark. As a control, gp120 was incubated with 10 mM HPG in the absence of the MAb. After derivatization, gp120 was purified from the antibody and the excess HPG by reversed phase HPLC. The conditions were as following: Protein-C4-column (4.6 mm × 250 mm; Vydac, Hesperia, CA) with a C4-guard column (Vydac); flow rate 1 mL/min; A: 0.1% trifluoroacetic acid in water, B: 0.1% trifluoroacetic acid in acetonitrile; gradient: 2% B, 5 min; 2% B to 95 % B, 50 min; 95% B, 5 min; collection of 1 mL fractions. After lyophilization, the protein in each fraction was resuspended in 5 μL 50% acetonitrile/0.1% formic acid and analyzed by MALDI/MS. Fractions containing derivatized gp120 were combined. Multiple reactions and digestion conditions were performed at different times. Due to limited MAb availability, only single derivatizations under each set of conditions could be performed.
To determine the sites of derivatization, HPG-modified gp120 was digested with various endoproteinases. A 100 pmole aliquot of modified gp120 was diluted 10 fold to give a concentration of 5% acetonitrile and then incubated with endoproteinase LysC at a ratio of 1:20 in 25 mM Tris-HCl (pH 8.0) for 18 h at 37°C. The peptides were deglycosylated with PNGaseF for 24 h at 30°C. Additionally, an aliquot of the deglycosylated LysC digest was further digested with the endoproteinase AspN at an enzyme:substrate ratio of 1:62.5 for 18 h at 35°C. Equivalent changes in reactivity were observed for specific residues determined from different reactions and digested with different reagents.
MALDI/MS
Mass spectra were acquired on a Voyager DE-STR Super MALDI time-of-flight instrument (Applied Biosystems, Framingham, MA). α-Cyano-4-hydroxycinnamic acid was re-crystallized in hot methanol and stored in the dark. Prior to analysis of acetylated proteins and peptides, a fresh solution of saturated α-cyano-4-hydroxycinnamic acid in water/ethanol/formic acid 45/45/10 (v/v) was prepared. Matrix solution (0.5 μL) was mixed with sample (0.5 μL), pipetted onto the target and dried at RT. Mass calibration was performed using an external calibrant (Applied Biosystems, Framingham, MA).
LC-ESI/MS) and MS/MS
For the LC-ESI/MS analyses, an aliquot of the acetylated peptide sample was injected onto a Hypersil RP-18 column (75 μm i.d. × 15 cm, LC Packings, San Francisco, CA) and eluted with a linear gradient of 5 to 95% B in 30 min (A: 0.1% formic acid in water, B: 0.1% formic acid in acetonitrile). The solvents delivered from the pumps were split before the sample injector such that 200 nL/min were delivered to the mass spectrometer.
Separation of HPG-derivatized peptides from an 8 μL-aliquot, which had been incubated with 5 mM dithioerythritol for 30 min at RT prior to analysis, was achieved on a C18-PepMap column (5 μm, 75 μm i.d. × 150 mm, LC Packings, San Francisco, CA); conditions: A: 0.1% formic acid in water; B: 0.1% formic acid in acetonitrile; gradient: 0.3 μL/min; 5% B, 5 min; 5-95% B over 35 min.
For online analysis of the eluting peptides, the nano-HPLCcolumn was connected to an electrospray ionization source on a quadrupole-time-of-flight instrument (QToF-I and QToF-Micro, Micromass, Altrincham, U.K.). Instrument conditions for QTof-I: capillary 2800-3095 V, sample cone 19-21 V, source temperature 80°C, scan: MS - m/z 300-4000 over 2 s, MS/MS - m/z 50-2000 over 2 s, collision energy 30 V. Conditions on QTof-Micro for MS: capillary voltage 3000 V, cone voltage 30 V, extraction voltage 2.5 V, collision energy 10 V, source temperature 20°C, and scan: m/z 400-1800 over 0.98 s; alternatively for MS/MS: cone voltage 40 V, collision energy 30 V, scan: m/z 80-2800 over 2 s. Data were acquired using MassLynx 3.5 and deconvoluted via MaxEnt3 (Micromass, Altrincham, U.K.).
Data Analysis
The relative abundances of HPG-derivatized peptides were calculated from the sum of counts of the peptides in the various charge states, with the total for unmodified and modified peptides defining 100%. To determine the significance of the relative changes, the ratios of the relative abundance of a specific peptide derived from Ab-bound gp120 to the relative abundance of the corresponding peptide derived from unbound gp120 were calculated. In cases where only one modification state (either modified or unmodified) was observed the relative abundance of this peptide was defined as 99% whereas the relative abundance of the complimentary peptide (unmodified or modified) was defined as 1% for calculation purposes only. A ratio of 1 would indicate that the relative reactivity of an arginine residue was equal in the presence and absence of the antibody. The mean of all values was 0.87 with a standard deviation of 0.33. Data points with a variation from the mean of greater than 1 standard deviation, i.e., R480 and R476, were then omitted. The mean of the remaining data points was calculated to be 0.98 with a standard deviation of 0.14. Based on this standard deviation, ratios within 0.98 ± 0.14 indicated no significant change, changes within 2 standard deviation changes were considered moderately significant, and ratios with greater than ±0.28 deviation from the mean were termed significant.
Similarly, the relative reactivity of lysine residues was determined as a ratio of the abundance of acetylated to trideuteroacetylated peptide. To determine changes in reactivity of lysine residues, the ratios of the relative reactivity of gp120 derivatized in the presence of the MAb to the relative reactivity of gp120 in the absence of MAb were calculated. The reactivity was considered unchanged for ratios with deviations within ± 20% of 1, moderate for ratios with deviations within ± 20% to ± 40% of 1 and significant for ratios with deviations greater than ± 40%.
Residue numbering system
The sequence of the HIV-1SF2 gp120 used in this study is shown in Figure 1. The numbering system used is according to Korber et al. (43). In this system, the sequence of the protein is numbered according to the sequence of the protein in HIV HXB2CG (http://hiv-web.lanl.gov/content/hiv-db/LOCATE_SEQ/locate.html; http://www.hiv.lanl.gov/content/hiv-db/REVIEWS/HXB2.html). Insertions are indicated as xxxa=AA where the inserted amino acid is inserted after residue xxx. If more than one amino acid is inserted, the residues are indicated sequentially by their single letter designations. AA is the amino acid inserted. Deletions are indicated by HXBC region numbers with deleted amino acid numbers in parentheses, i.e., (del 460-461) or (Δ460-461) for deletion of two amino acids at positions 460 and 461.
FIGURE 1.
Alignment of the sequences of recombinant HIV-1SF2 gp120 corresponding to amino acids 31-509 of the env-precursor protein (54) and of the residues 1-511 of the reference HXBc2 env-protein (43). Arginine and lysine residues with a significantly decreased relative reactivity are shown in red, residues with a slightly decreased relative reactivity are shown in purple, residues with no change in relative reactivity in blue, and residues with an increase in relative reactivity are shown in dark-green. Lysine and arginine residues not detected are in orange. N-glycosylation sites in HIVSF2 gp120 as determined by mass spectrometry are italicized (11, 12). In the numbering system used here, the annotation ‘Δ405-407’ indicates that SF2 gp120 contains a deletion of three residues (405-407) relative to the HXBc2 reference sequence and ‘ins 459a=T’ indicates that the SF2 gp120 sequence contains an additional T residue between residues 459 and 460 in the HXBc2 reference sequence (43).
Results and Discussion
To develop a better understanding of the correlation of antigen structure and the neutralization potency of the antibody, it is important to determine the epitope recognized by the antibody using a full-length glycosylated gp120 that closely resembles the wild-type protein. In this study we used non-deglycosylated, non-truncated gp120 from recombinantly expressed HIV strain SF2gp120. To characterize the epitope recognized by the antibody 559/64-D, lysine and arginine residues in gp120 were chemically modified and their relative reactivities between gp120 bound to mAb559/64-D and unbound gp120 were determined. The recombinant HIV envelope protein gp120 of HIV strain SF2 is a major representative of the TCLA-strains and corresponds to amino acids 31-509 of the HIVSF2 env-protein. It contains 30 lysine residues and 25 arginine residues (Figure 1).
Lysine Acetylation
Two acetylation steps were performed, the first with acetic anhydride, and the second, with d6-acetic anhydride. The relative reactivities of the lysine residues for which peptides were observed was determined from the ratios of the abundance of acetylated and trideuteroacetylated peptide, respectively, within one sample. Ratios of the relative reactivity of gp120 derivatized in the presence of the MAb to the relative reactivity of gp120 in the absence of the MAb were used to determine the changes of reactivity upon binding of the MAb 559/64-D (Table 1). The calculation of the ratio of relative reactivities results in a normalization of the relative reactivities, thereby allowing the comparison of the two experimental conditions of gp120 bound to the MAb and free in solution. The ratio of 1 would indicate that the relative reactivity of a specific residue did not change upon binding of the MAb, while a ratio of 0 indicates greatly reduced access in the presence of the antibody. A ratio of greater than 1.0 indicates increased accessibility/reactivity due to the presence of the MAb.
Table 1.
Relative Reactivities of Lysine Residues
| Lysine Residuea |
Peptide Sequence (HIVSF2) | Ratio of modified to unmodified In Antibody-Bound vs. Antibody Free HIV-1 gp120:b |
Change of Reactivityc |
|---|---|---|---|
| 155 | 154IKNCSFNITTSIR166 | 1.18 ± 0.05 | - |
| 168, 171 | 167DKIQKE172 | 1.18 ± 0.1 | - |
| 282 | 270VVIRSDNFTNNAKTIIVQLNE290 | 1.11 | - |
| 305 | 305KSIYIGPGR315 Δ310-311 | 1.16 ± 0.4 | - |
| 409 | 404GTKGNDTIILPCR419 Δ405-407 | 0 | s ↓ |
| 432 |
430VGKAMYAPPIGGQISCSSNITG LLLTRDGGTNVTNDTE466 ins 459a=T |
1.56 | m ↑ |
| 485, 487, 490, 500, 502 |
483LYKYKVIKIEPLGIAPTKAKRRV VQRE509 |
1.16±.07 | - |
Residue numbers are based on HIVHXBc2 gp120 as shown in Figure 1 (denoted “HXBc2-env”;)
Ratios of relative reactivity of gp120 derivatized in the presence of the MAb to relative reactivity of gp120 in absence of MAb.
Reactivity was considered unchanged (−) for ratios with deviations within ± 20% of 1, moderate (m) for ratios with deviations within ± 20% to ± 60% of 1 and significant (s) for ratios with deviations greater than ± 60%.
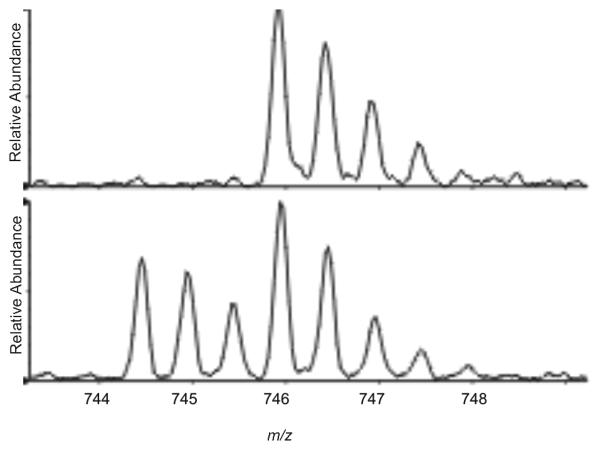
K409
Representative LC/ESI/MS spectra of the doubly-charged peptide 404GTKGNDTIILPCR419Δ405-407 after derivatization of lysine residues, deglycosylation and reductive alkylation of the cysteine residues from the Ab-Ag complex and from gp120 free in solution are shown in Figure 2A and 2B, respectively. The absence of an ion of m/z 744.49 in the spectrum of acetylated, deglycosylated and proteolyzed gp120 from the gp120:MAb complex (Figure 2A) shows that, in the presence of the antibody, derivatization of K409 did not occur during the first step but was only achieved under denaturing conditions, resulting in an ion of m/z 745.94. In contrast, approximately 57% of K409 was already seen derivatized during the first step of acetylation when the antibody was absent (Figure 2B, Supplemental Figure S-1). The relative reactivity of K409 changed from 0.75 for gp120 free in solution to 0 for gp120 bound.
FIGURE 2.
Representative LC/ESI/MS spectra (sum of five individual spectra) of the doubly charged peptide containing residues 404-419 Δ405-407 with lysine residue 409 derivatized in presence (A) and absence (B) of the MAb. These spectra indicate that K409 was protected from derivatization in the presence of the antibody.
K432
A ca. 50% increase in reactivity was observed for this residue in the presence of MAb 559-64D. This increase in reactivity of this residue may indicate that a conformational change occurs in the gp120 in the presence of the antibody that leads to an increased surface accessibility and/or reactivity.
K155, 168, 171, 282, 305, 485, 487, 490 , 500 and 502
The reactivities of these lysines were not affected, within experimental error, by the presence/absence of the antibody. This indicates that the antibody does not interact with these residues.
In summary, of 12 lysine residues that were characterized, the surface reactivity of one residue (K409) was found to be significantly reduced by the presence of 559/64-D, and the surface reactivity of one residue (K432) was found to be moderately enhanced in the presence of the antibody. Additionally, modifications of residues that lead to change of the formal charge on the residue side chain may also influence the solubility of the modified protein, leading to reduced information about the reactivity of some residues.
Arginine Hydroxyphenyl glyoxylation
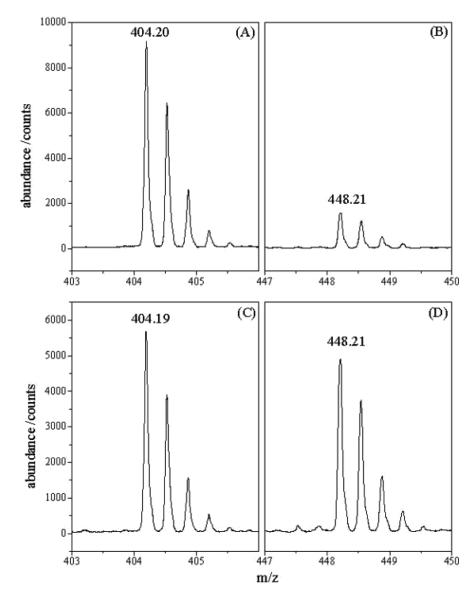
Arginine residues were modified with hydroxyphenylglyoxal in antibody-bound gp120 and gp120 free in solution. The relative abundances of HPG-derivatized peptides were calculated from the sum of counts of the peptides in the various charge states, with the total for unmodified and modified peptides defining 100%. Representative spectra from an LC/ESI/MS analysis of the triply charged peptide 477DNWRSELYK485 are shown in Figure 3. R476 was found in peptide 463-476 with one missed cleavage arising from combined trypsin and GluC protolysis. This peptide also contained R469. R469 was also observed by itself in peptide 463-473, in which no change in extent of modification to R469 was observed between bound and unbound gp120. Thus, changes in the relative extent of modification between bound and unbound gp120 peptide 463-476 was attributed to changes in the extent of modification to R476. Peptides containing 13 of 25 arginine residues were observed whose sequences were confirmed by MS/MS.
FIGURE 3.
Representative LC/ESI/MS spectra of the triply charged peptide containing residues 477-485 with arginine residue 480 modified in the presence (A: without label, B: with label) and absence (C: without label, D: with label) of the MAb. Note that after derivatization with hydroxyphenylglyoxal the retention time of the peptide increases compared to the underivatized peptide.
R456, R476 and R480
The reactivity of these residues were significantly reduced (by > 1 standard deviation) in the presence of 559/64-D, with R476 and 480 being most strongly affected. The reactivity of R456 is less strongly affected by the presence of the Ab (Figure 3, Supplemental Figure S-2). These residues are in the protein core of gp120 (Figure 4A). It should be noted that R456 was observed in several peptides after PNGase F treatment, 448-456, 448-459 ins 459a=T (one missed cleavage site) and 448-462 ins 459a=T (two missed cleavage sites). The unmodified peptide dominated in all cases. Peptide 448-462 ins 459a=T contains a glycosylation site at N460 that is only partially glycosylated (11). During deglycosylation with PNGaseF the glycosylated asparagine residue will be transformed to an aspartic acid residue, while the asparagine residue of the non-glycosylated peptide will not be affected. Peptide 448-462 ins 459a=T with N460 and peptide 448-462 ins 459a=T with D460 could be separated by 0.23 min by reversed phase chromatography. The sequence of these peptides was confirmed by MS/MS (data not shown). For all peptides, the ratios of modified to unmodified peptides in the bound and unbound forms showed moderate protection.
FIGURE 4.
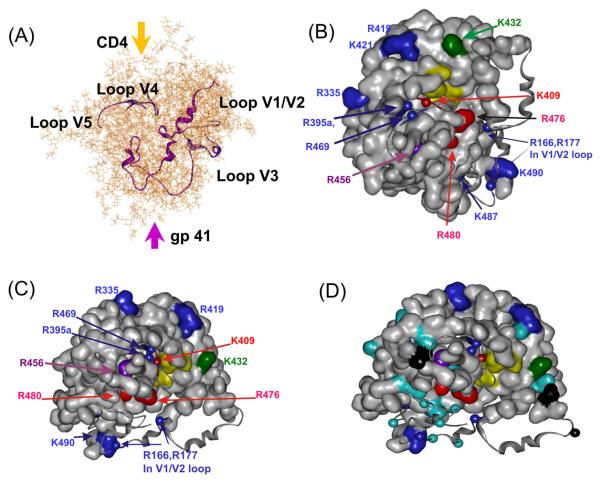
(A) The structural model of glycosylated full-length gp120 of HIV strain SF2 as determined by Zhu et al. (12) was used to depict the protein core (gray ribbon), the variable loops (blue and purple ribbons) and the glycan chains (stick models). The model is oriented with the gp41-interaction site on the bottom of gp120 (purple arrow) and CD4 entering its binding site from the top (gold arrow). (B) The amino acids characterized in this study were mapped to HIVSF2 gp120 with the protein core as a surface model and the loops as ribbons. Residues with a significantly decreased relative reactivity are shown in red, residues with a slightly decreased relative reactivity are shown in purple, residues with no change in relative reactivity in blue, and residues with an increase in relative reactivity are shown in dark-green. Amino acids in light-yellow reside within the CD4 binding area as found in the X-ray structure of HIVHXBc2 gp120 (9). The orientation is the same as in (A). (C) The gp120-model was tilted 90° forwards such that an interacting CD4-molecule would stick out of the paper plane and gp41 would be located “behind” gp120. Color code is as in (B). (D) Same view as in (C) with all lysines and arginines which were not verifiably detected in these experiments colored in cyan and black, respectively
R59
The reactivity of R59 is moderately increased in the presence of the Ab. The position of R59 in the gp120 structure was not determined in the solved structure of the construct, which did not include the first 52 N-terminal residues. It was also omitted from our computational model of HIVSF2 gp120. (9, 12). Our results, however, indicate that this residue is slightly more accessible in the presence of MAb 559/64-D.
R252
This arginine exhibited slightly lower reactivity when bound to the antibody. R252 is located spatially close to A287 of the V3 loop. The slightly increased protection of this residue may be due to movement of the V3 loop and/or glycans upon Ab binding.
R166, 177, 273, 298, 335, 395a, 419, 469
The relative reactivities of these arginines did not vary significantly from the mean. Thus, the locations of these residues are such that the reactivities are unaffected by the presence of the antibody.
Peptides for the remaining 12 arginine residues were not observed. One possible reason for this is the size of the peptides formed during proteolysis may be too small or too large to be analyzed. Blocked cleavage at acetylated lysine can lead to large peptides. For example, a complete digest of gp120 with LysC/AspN would result in arginine-containing peptides with a mass smaller than 600 Da, and trypsin/Glu-C digestion can still lead to peptides with a mass of over 4000 Da containing two arginine residues. In summary, 13 of 25 arginine residues were characterized, and 5 of these were affected by the binding of 559/64-D.
Structural implications - localization in the three-dimensional structure of gp120
Based on the high sequence homology between HIV strains SF2 and HXBc2 (see Figure 1), the structural data of the gp120 core of HIVHXBc2 (9), and the characterization of the glycan chains of recombinantly expressed HIVSF2 gp120, a molecular model of glycosylated full-length gp120 was developed (Figure 4A) (12). The model is depicted to illustrate the extensive shielding of the gp120 core by the carbohydrate moieties and the position of the variable loops V1/V2, V3, V4, and V5. Using this model, localization of the residues characterized in this study is shown in Figure 4B (same orientation as in Figure 4A) and in Figure 4C (gp120 tilted forwards by 90°), omitting the glycans for clarity. The amino acids for which the relative reactivity was significantly reduced by the presence of the MAb form a cluster around the CD4-binding site, predominantly on one side of the CD4-BS cleft, and are indicative of the epitope recognized by the MAb 559/64-D. The MAb-shielded residues R456, R476 and R480 are located in the core of gp120 (as modeled as the surface described in the caption of Figure 4), whereas K409 is within the flexible loop V4, but within 8.8 Å of the significantly affected R456 ((Cα-atoms).
The position of residues R59, K500 and K502 could not be determined because 52 residues at the N-terminus and 19 residues at the C-terminus of the HIVHXBc2 gp120 core were deleted for the crystallization studies and were also omitted from the computational model of the HIVSF2 gp120 (9, 12). However, all additional residues characterized in this study are exposed on the protein surface.
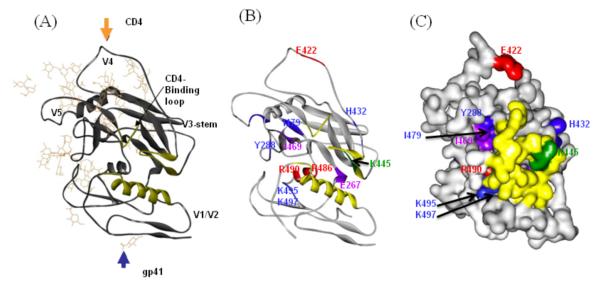
The amino acids characterized in this study were also mapped on the structure of the SIVgp120 core, which was crystallized in the absence of interacting proteins such as CD4 or Abs (27, 44). Residues I469, A486, and R490 corresponding to the significantly affected residues R456, R476 and R480 in the HIVHXBc2 env-protein (27) are located within or in the vicinity of the CD4-interaction site (Figure 5A, same orientation as for Figure 4A), and the distances between these residues are similar to the distances determined for HIVHXBc2 (I469 – A486: 18.16Å; I469 – R490: 18.67Å; A486 – R490: 5.55Å between Cα-atoms). However, in the crystal structure these residues are partly covered by the residues N-terminal of the helix α1 (Figure 5C). Stricher, et al., recently studied the structural basis of immune evasion at the CD4-BS on HIV-1 gp120 and found that several antibodies recognize the CD4-BS, but are not broadly neutralizing, and these less broadly neutralizing antibodies assumed a conformation that was poorly compatible with the functional trimeric viral spike(24).
FIGURE 5.
(A) The structure of the glycosylated unliganded SIV gp120-core (32) is shown with the protein core as gray ribbon and the carbohydrate moieties as tan stick models. The orientation is the same as for the HIV gp120 core in Figure 4A. (B) Amino acids at the same position in the linear sequence corresponding to the residues characterized in this study were mapped to the ribbon model of the SIV gp120 core. (C) Amino acids corresponding to the residues characterized in this study were mapped to the surface model of the SIV gp120 core. The color code of the relative reactivity of residues affected by the MAb 559/64-D is as in Figure 4.
Although neutralizing antibodies such as 2F5 can recognize a linear protein-based epitope on the env-protein (45-47) (though the linear peptide corresponding to the epitope cannot generate neutralizing antibodies), the majority of broadly neutralizing antibodies recognize conformational (e.g., 2F5) and/or discontinuous epitopes ((1, 14, 20) and references therein). According to the distribution of the amino acids in the linear sequence of gp120, the residues with decreased relative reactivity for derivatizing agents upon binding of the MAb 559/64-D, the epitope recognized by this neutralizing Ab is also discontinuous. Based on the crystal structure of the gp120 core from the HIV strain HXBc2 (9, 45) and the assignment of the immunogenic areas “neutralizing face,” “non-neutralizing face,” and “silent face” (17, 20), the MAb 559/64-D binds to the CD4-BS within the neutralizing face on gp120 as inferred from the significantly reduced reactivity of K409, R456, R476 and R480. The structure of the Fab fragment of MAb F105, another broadly neutralizing Ab that does not neutralize primary isolates, has been published and its interaction with the gp120 CD4-BS has been modeled (36). The MAb was predicted to interact with residues 255-7, 368, 370, 375, 384, 421, 470 and 473-477. Our results indicate that 559/64-D binds, but not identical, manner in that R476 and 480 are significantly affected by the Ab, but R469 is not. In contrast, the broadly neutralizing MAb b12 does not interact with residues 473-476 (the N-terminal end of helix α5), consistent with the hypothesis that interactions with residues 473-477 may be a major difference between antibodies recognizing primary isolates and those recognizing gp120 epitopes on TCLA strain (36).
As discussed previously, differences in the environment of specific residues, e.g., salt bridges, can lead to differences in reactivity not directly associated with surface reactivity (see, for example, (40, 48). Conformational changes have been similarly invoked to explain changes in differential reactivity of specific residues (49). Experimental evidence from a number of researchers indicates that gp120 undergoes a conformational change upon CD4 binding (24, 50-53) and upon binding of certain antibodies (25). Our results also implicate conformational effects on our differential surface modification data. As residues distant from the CD4-BS were also affected, this approach to epitope mapping is also sensitive to factors that affect surface reactivity in addition to direct interaction between the antigen and the antibody. The complexity of the glycoprotein gp120 may complicate the differential reactivity approach to epitope mapping in various ways. Due to the extensive glycosylation of gp120, steric hindrance for the derivatizing agent might lead to reduced reactivity of amino acids which do not interact with the Ab. In addition, Ab binding to the CD4-BS might induce conformational changes of the gp120 core, which results in newly formed intermolecular interaction sites. Although the reduced accessibilities of these residues would be specific for the Ab-bound conformation they would not be indicative of the intermolecular interaction site between Ab and antigen. Indeed, using a similar approach, we have observed significant changes between chemical reactivity in HCV E2 glycoprotein in the presence and absence of an Ab that can only be explained by significant Ab-induced conformational changes (49). However, comparison of the results obtained for the epitope of the MAb 559/64-D and reports for epitopes of other Abs in the literature show the validity of our approach for epitope mapping using glycosylated full-length gp120 (44, 46).
In summary, we conclude from our experiments of chemical modification of lysine and arginine residues followed by mass spectrometric analyses that the epitope recognized by the MAb 559/64-D overlaps with the CD4-binding site of gp120. Moreover, binding of the MAb to its antigen seems to have an effect on the conformation of gp120.
As mentioned above, modifications of residues that lead to change of the formal charge on the residue side chain may also influence the solubility of the modified protein, leading to reduced information about the reactivity of some residues. We are currently investigating alternative derivatizing reagents to circumvent this problem.
Supplementary Material
Table 2.
Relative Reactivities of Arginine Residues
| Arginine Residuea | Ratio of modified to unmodified In Antibody-Bound vs. Antibody Free HIV-1 gp120 |
Changeb |
|---|---|---|
| 59 47EATTTLFCASDARAY61 |
1.11 | m↑ |
| 166 160NITTSIR166 |
0.92 | — |
| 177 172ENALFRNL179 |
0.87 | — |
| 252 c237/241GP-TNVST VQCTHGIRPIVSTQLLL261 |
0.81 | m↓ |
| 273 262NGSLAEEEVVIRS274 |
1.01 | — |
| 298 276NFTNNAKTI IVQLNE--SV AINCTRPN300 |
1.02 | — |
| 335 332NISRAQW338 |
1.18 | — |
| 395a 392NNTWRL396ins |
1.05 | — |
| 419 410GNDTIILPCRIK421 |
0.95 | — |
| 456 d448NITGLLLT RD--GGT456/462 ins 459a=T |
0.78 | m↓ |
| 469 463NDTEVFRPGGG473 |
1.11 | — |
| 476 463NDTEVFRPGGGDMR476 |
0.40 | s↓ |
| 480 477DNWRSE LYK485 |
0.25 | s↓ |
Italicized N indicates that this residue was originally glycosylated.
m=moderate (between 1 and 2 standard deviations; s= strong (greater than two standard deviations); - = no significant change
Two peptides were found that included R252, one starting with AA 237 and the second starting with AA241.
Two peptides were found that included R456, one ending with AA 456 and the second ending with AA459a ins
Acknowledgements
This research was supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences/National Institutes of Health (z050150) and The NIH/NHLBI (grant HL59725 to Susan Zolla-Pazner).
Abbreviations
- HIV
human immunodeficiency virus
- Ab/Abs
antibody/antibodies
- CD4-BS
CD4-binding site
- TCLA
T-cell line adapted
- MAb
monoclonal antibody
- R
arginine residue
- K
lysine residue
- HPG
hydroxyphenylglyoxal
- RT
room temperature
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- Ag
antigen
- DTT
dithiothreitol
- LC
liquid chromatography
- ESI
electrospray ionization
- N
aspartic acid
References
- 1.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nature Reviews Immunology. 2004;4(3):199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poignard P, Klasse PJ, Sattentau QJ. Antibody neutralization of HIV-1. Immunology Today. 1996;17(5):239–246. doi: 10.1016/0167-5699(96)10007-4. [DOI] [PubMed] [Google Scholar]
- 3.Center RJ, Leapman RD, Lebowitz J, Arthur LO, Earl PL, Moss B. Oligomeric structure of the human immunodeficiency virus type I envelope protein on the virion surface. Journal of Virology. 2002;76(15):7863–7867. doi: 10.1128/JVI.76.15.7863-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalgleish AG, Beverley PCL, Clapham PR, Crawford DH, Greaves MF, Weiss RA. the CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 5.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. lymphocyte-T T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312(5996):767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 6.Gagne N. Demonstration of a thermoreversible gel for the prevention of HIV and herpes transmission. Universite Laval,QC,Can. 2000:111. [Google Scholar]
- 7.Zaitseva M, Peden K, Golding H. HIV coreceptors: role of structure, posttranslational modifications, and internalization in viral-cell fusion and as targets for entry inhibitors. Biochimica Et Biophysica Acta-Biomembranes. 2003;1614(1):51–61. doi: 10.1016/s0005-2736(03)00162-7. [DOI] [PubMed] [Google Scholar]
- 8.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC, Wongstaal F. identification and characterization of conserved and variable regions in the envelope gene of HTLV-III LAV, the retrovirus of AIDS. Cell. 1986;45(5):637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 9.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type-1 recombinant human-immunodeficiency-virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. Journal of Biological Chemistry. 1990;265(18):10373–10382. [PubMed] [Google Scholar]
- 11.Cutalo JM, Deterding LJ, Tomer KB. Characterization of glycopeptides from HIV-I-SF2 gp120 by liquid chromatography mass spectrometry. Journal of the American Society for Mass Spectrometry. 2004;15(11):1545–1555. doi: 10.1016/j.jasms.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu XG, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39(37):11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 13.Bouma P, Leavitt M, Zhang PF, Sidorov IA, Dimitrov DS, Quinnan GV. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. Journal of Virology. 2003;77(14):8061–8071. doi: 10.1128/JVI.77.14.8061-8071.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. Journal of Virology. 2002;76(18):9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. Journal of Virology. 1996;70(3):1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyambi PN, Mbah HA, Burda S, Williams C, Gorny MK, Nadas A, Zolla-Pazner S. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. Journal of Virology. 2000;74(15):7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 18.Zwick MB, Kelleher R, Jensen R, Labrijn AF, Wang M, Quinnan GV, Parren P, Burton DR. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. Journal of Virology. 2003;77(12):6965–6978. doi: 10.1128/JVI.77.12.6965-6978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwick MB, Parren P, Saphire EO, Church S, Wang M, Scott JK, Dawson PE, Wilson IA, Burton DR. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. Journal of Virology. 2003;77(10):5863–5876. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 21.Nunberg JH, Follis KE, Trahey M, LaCasse RA. Turning a corner on HIV neutralization? Microbes and Infection. 2000;2(2):213–221. doi: 10.1016/s1286-4579(00)00267-7. [DOI] [PubMed] [Google Scholar]
- 22.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(16):9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang LP, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. Journal of Virology. 2002;76(19):9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stricher F, Huang CC, Descours A, Duquesnoy S, Combes O, Decker JM, Do Kwon Y, Lusso P, Shaw GM, Vita C, Kwong PD, Martin L. Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein. Journal of Molecular Biology. 2008;382(2):510–524. doi: 10.1016/j.jmb.2008.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Kwon YD, Zhou TQ, Wu XL, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural Basis of Immune Evasion at the Site of CD4 Attachment on HIV-1 gp120. Science. 2009;326(5956):1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan YP, Ma BY, Keskin O, Nussinov R. Characterization of the conformational state and flexibility of HIV-1 glycoprotein gp120 core domain. Journal of Biological Chemistry. 2004;279(29):30523–30530. doi: 10.1074/jbc.M404364200. [DOI] [PubMed] [Google Scholar]
- 27.Chen B, Vogan EM, Gong HY, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 28.McKeating JA, Thali M, Furman C, Karwowska S, Gorny MK, Cordell J, Zolla-Pazner S, Sodroski J, Weiss RA. Amino acid residues of the human immunodeficiency virus type I gp120 critical for the binding of rat and human neutralizing antibodies that block the gp120-sCD4 interaction. Virology. 1992;190(1):134–142. doi: 10.1016/0042-6822(92)91199-5. [DOI] [PubMed] [Google Scholar]
- 29.Karwowska S, Gorny MK, Buchbinder A, Gianakakos V, Williams C, Fuerst T, Zollapazner S. production of human monoclonal-antibodies specific for conformational and linear non-V3 epitopes of gp120. Aids Research and Human Retroviruses. 1992;8(6):1099–1106. doi: 10.1089/aid.1992.8.1099. [DOI] [PubMed] [Google Scholar]
- 30.Chien PC, Cohen S, Tuen M, Arthos J, Chen PD, Patel S, Hioe CE. Human immunodeficiency virus type 1 evades T-helper responses by exploiting antibodies that suppress antigen processing. Journal of Virology. 2004;78(14):7645–7652. doi: 10.1128/JVI.78.14.7645-7652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HL, Chien PC, Tuen M, Visciano ML, Cohen S, Blais S, Xu CF, Zhang HT, Hioe CE. Identification of an N-linked glycosylation in the C4 region of HIV-1 envelope gp120 that is critical for recognition of neighboring CD4 T cell epitopes. Journal of Immunology. 2008;180(6):4011–4021. doi: 10.4049/jimmunol.180.6.4011. [DOI] [PubMed] [Google Scholar]
- 32.Li HL, Xu CF, Blais S, Wan Q, Zhang HT, Landry SJ, Hioe CE. Proximal Glycans Outside of the Epitopes Regulate the Presentation of HIV-1 Envelope gp120 Helper Epitopes. Journal of Immunology. 2009;182(10):6369–6378. doi: 10.4049/jimmunol.0804287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visciano ML, Tuen M, Gorny MK, Hioe CE. In vivo alteration of humoral responses to HIV-1 envelope glycoprotein gp120 by antibodies to the CD4-binding site of gp120. Virology. 2008;372(2):409–420. doi: 10.1016/j.virol.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nandi A, Lavine CL, Wang PC, Lipchina I, Goepfert PA, Shaw GM, Tomaras GD, Montefiori DC, Haynes BF, Easterbrook P, Robinson JE, Sodroski JG, Yang XZ, Immunol NCHAV. Epitopes for broad and potent neutralizing antibody responses during chronic infection with human immunodeficiency virus type 1. Virology. 396(2):339–348. doi: 10.1016/j.virol.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatatos L, Chengmayer C. structural modulations of the envelope gp120 glycoprotein of human-immunodeficiency-virus type-1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor-binding. Journal of Virology. 1995;69(10):6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson RA, Piscitelli C, Teintze M, Cavacini LA, Posner MR, Lawrence CM. Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120. Journal of Virology. 2005;79(20):13060–13069. doi: 10.1128/JVI.79.20.13060-13069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glocker MO, Borchers C, Fiedler W, Suckau D, Przybylski m. molecular characterization of surface-topology in protein tertiary structures by amino-acylation and mass-spectrometric peptide-mapping. Bioconjugate Chemistry. 1994;5(6):583–590. doi: 10.1021/bc00030a014. [DOI] [PubMed] [Google Scholar]
- 38.Hochleitner EO, Borchers C, Parker C, Bienstock RJ, Tomer KB. Characterization of a discontinuous epitope of the human immunodeficiency virus (HIV) core protein p24 by epitope excision and differential chemical modification followed by mass spectrometric peptide mapping analysis. Protein Science. 2000;9(3):487–496. doi: 10.1110/ps.9.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner RF, Albaugh S, Fenselau C, Murphy C, Vestling M. a mass-spectrometry method for mapping the interface topography of interacting proteins, illustrated by the melittin-calmodulin system. Analytical Biochemistry. 1991;196(1):120–125. doi: 10.1016/0003-2697(91)90127-f. [DOI] [PubMed] [Google Scholar]
- 40.Suckau D, Mak M, Przybylski M. protein surface topology-probing by selective chemical modification and mass-spectrometric peptide-mapping. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(12):5630–5634. doi: 10.1073/pnas.89.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hager-Braun C, Tomer KB. Characterization of the tertiary structure of soluble CD4 bound to glycosylated full-length HIVgp120 by chemical modification of arginine residues and mass spectrometric analysis. Biochemistry. 2002;41(6):1759–1766. doi: 10.1021/bi011626k. [DOI] [PubMed] [Google Scholar]
- 42.Wood TD, Guan ZQ, Borders CL, Chen LH, Kenyon GL, McLafferty FW. Creatine kinase: Essential arginine residues at the nucleotide binding site identified by chemical modification and high-resolution tandem mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3362–3365. doi: 10.1073/pnas.95.7.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korber BT, Foley BT, Kuiken CL, Pillai SK, Sodroski JG. Numbering Positions in HIV Relative to HXB2CG. 1998 cited 1998; Available from: http://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/1998/III/HXB2.pdf.
- 44.Hager-Braun C, Katinger H, Tomer KB. The HIV-neutralizing monoclonal antibody 4E10 recognizes N-terminal sequences on the native antigen. Journal of Immunology. 2006;176(12):7471–7481. doi: 10.4049/jimmunol.176.12.7471. [DOI] [PubMed] [Google Scholar]
- 45.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8(12):1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 46.Parker CE, Deterding LJ, Hager-Braun C, Binley JM, Schulke N, Katinger H, Moore JP, Tomer KB. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. Journal of Virology. 2001;75(22):10906–10911. doi: 10.1128/JVI.75.22.10906-10911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren P. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. Journal of Virology. 2001;75(22):10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hobbs CA, Deterding LJ, Perera L, Bobay BG, Thompson RJ, Darden TA, Cavanagh J, Tomer KB. Structural Characterization of the Conformational Change in Calbindin-D-28k upon Calcium Binding Using Differential Surface Modification Analyzed by Mass Spectrometry. Biochemistry. 2009;48(36):8603–8614. doi: 10.1021/bi900350q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iacob RE, Keck Z, Olson O, Foung SKH, Tomer KB. Structural elucidation of critical residues involved in binding of human monoclonal antibodies to hepatitis C virus E2 envelope glycoprotein. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2008;1784(3):530–542. doi: 10.1016/j.bbapap.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 51.Lusso P. HIV and the chemokine system: 10 years later. Embo Journal. 2006;25(3):447–456. doi: 10.1038/sj.emboj.7600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, ChengMayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 53.Wu LJ, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384(6605):179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 54.Sanchezpescador R, Power MD, Barr PJ, Steimer KS, Stempien MM, Brownshimer SL, Gee WW, Renard A, Randolph A, Levy JA, Dina D, Luciw PA. nucleotide-sequence and expression of an AIDS-associated retrovirus (ARV-2) Science. 1985;227(4686):484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.