Abstract
The oncogenic phenotype is complex, resulting from the accumulation of multiple somatic mutations that lead to the deregulation of growth regulatory and cell fate controlling activities and pathways. The ability to dissect this complexity, so as to reveal discrete aspects of the biology underlying the oncogenic phenotype, is critical to understanding the various mechanisms of disease as well as to reveal opportunities for novel therapeutic strategies. Previous work has characterized the process of anchorage-independent growth of cancer cells in vitro as a key aspect of the tumor phenotype, particularly with respect to metastatic potential. Nevertheless, it remains a major challenge to translate these cell biology findings into the context of human tumors. We previously used DNA microarray assays to develop expression signatures, which have the capacity to identify subtle distinctions in biological states and can be used to connect in vitro and in vivo states. Here we describe the development of a signature of anchorage-independent growth, show that exhibits characteristics of deregulated mitochondrial function, and then demonstrate that the signatures identifies human tumors with the potential for metastasis.
Keywords: Anchorage-independent Cell Growth, Phenotype, Expression Signature, Metastatic Potential, Heterogeneity
Introduction
Many cancers, particularly those originating in the breast, lung, and ovary are exceedingly heterogeneous, representing not one disorder but a large array of diseases with distinct etiologies (Nevins & Potti, 2007). This complexity is reflected in variations in the phenotypes of individual tumors, including hormone dependence, proliferative activity, drug resistance, and the potential for metastasis. The ability to dissect this complexity so as to understand the unique characteristics of the individual patient is key to understanding disease mechanisms and to developing effective therapeutic strategies. Multiple approaches that include in vitro tissue culture system have been used to provide a rational basis for dissecting and understanding the complexity of these diseases (Hanahan & Weinberg, 2000).
Cultured cancer cells also exhibit substantial phenotypic heterogeneity when measured in a variety of ways such as sensitivity to drugs or the capacity to grow under various conditions. Among these, the ability to exhibit anchorage-independent cell growth (colony forming capacity in semisolid media), has been considered to be fundamental in cancer biology, because it has been connected with tumor cell aggressiveness in vivo such as tumorigenic and metastatic potentials, and also utilized as a marker for in vitro transformation. Although multiple genetic factors for anchorage-independence have been identified, the molecular basis for this capacity is still largely unknown (Cifone & Fidler, 1980; Weinberg, 2007).
Many examples highlight the power of gene expression microarray analysis to inform an understanding of biological phenotypes of cancer (Nevins & Potti, 2007). As a measure of 30,000 or more genes, reflecting the activity of the entire genome, it has the capacity to identify otherwise unrecognized biological distinctions. Underlying the concept of an expression signature is the realization that virtually any biological condition is reflected in changes in gene expression. The enormous complexity of the expression data provides the opportunity to identify patterns of expression that connote subtle distinctions in biology. Moreover the expression signature is portable in the sense that it can be assayed in varied contexts (Nevins & Potti, 2007). To investigate the molecular mechanisms underlying anchorage independent cell growth, we have used genome-wide DNA microarray studies to develop an expression signature associated with this phenotype. Using this signature, we identify a program of activated mitochondrial biogenesis associated with the phenotype of anchorage-independent growth and importantly, we demonstrate that this phenotype predicts potential for metastasis in primary breast and lung tumors.
Results
A strategy to connect in vitro and in vivo cancer biology phenotypes
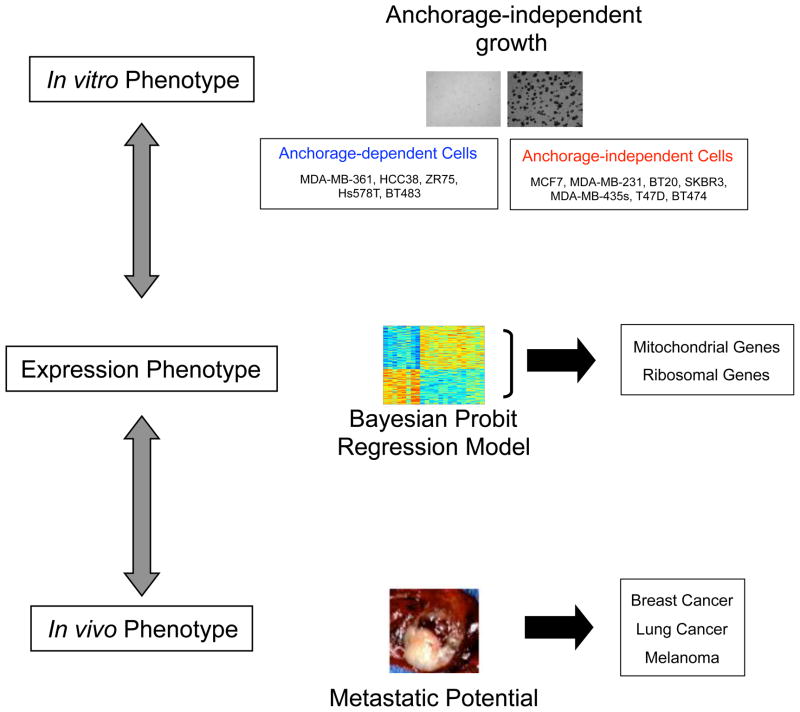
The study of cancer biology, taking advantage of the power of experimental manipulation, has revealed many of the genes and pathways critical for various aspects of the oncogenic phenotype. In parallel, the study of cancer through analysis of patient samples and clinical outcomes has been equally important and powerful, providing the true in vivo context of the disease. The challenge is to bring these two disparate experimental systems together, to facilitate a better understanding of the disease. For instance, the study of anchorage-independent growth as a measure of the behavior of cancer cells has detailed the role for Integrins and RAS as critical for this process (Weinberg, 2007). Although it is believed that these activities are likely important for developing metastatic capacity (Campbell & Der, 2004; Guo & Giancotti, 2004), it has been difficult to extrapolate these observations to the characteristics of human tumors directly. Importantly, a gene expression signature provides an opportunity to bridge this gap as a surrogate phenotype that can be measured in both the in vitro and in vivo states (Nevins & Potti, 2007). To explore the relevance of the anchorage-independent phenotype for human cancer, we made use of a strategy to generate a signature for anchorage-independent growth capability from in vitro assays and then used the signature to assess the phenotype in a collection of human tumor samples (Figure 1).
Figure 1. Strategy to characterize an anchorage-independent growth in vitro phenotype in combination with in vivo phenotypes.
To explore the relevance of the anchorage-independent phenotype for human cancer, we made use of a strategy to generate a signature for anchorage-independent growth capability from in vitro assays by a Bayesian probit regression model and then used the signature to assess the phenotype in a collection of human tumor samples.
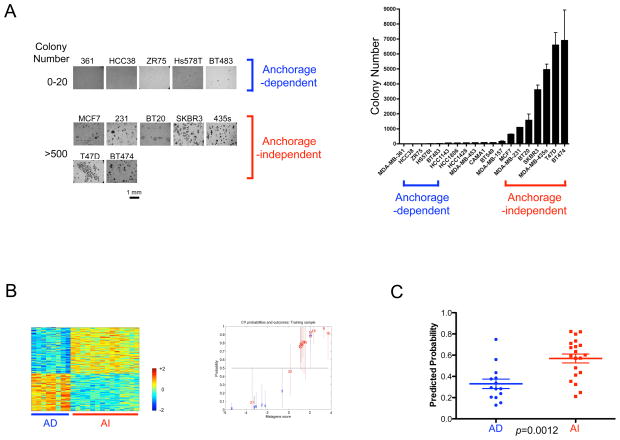
To develop an expression signature reflecting the capacity for anchorage-independent cell growth, we first carried out colony formation assays with 19 breast cancer cell lines in suspension culture dish with methyl-cellulose containing media. Starting with 20,000 plated cells, five cell lines (MDA-MB-361, HCC38, ZR75, Hs578T and BT483) gave rise to less than 20 colonies, while 8 cell lines (MCF7, MDA-MB-231, BT20, SKBR3, MDA-MB-435s, T47D and BT474) showed formation of more than 500 colonies. The rest of the cell lines showed an intermediate phenotype in colony forming ability (20–200 colonies; HCC1143, HCC1806, HCC1428, MDA-MB-453, CAMA1, BT549 and MDA-MB-157) (Figure 2A). We confirmed that this growth ability was not associated with either proliferation rate of cells in anchorage-dependent asynchronous growing culture condition, or breast cancer basal/luminal subtype (data not shown).
Figure 2. Development of a gene expression signature representing anchorage-independent cell growth ability of cultured breast cancer cells.
A. Colony forming ability of cultured breast cancer cells. Left panel: Morphological appearance of human breast cancer cell colonies grown in methylcellulose. Photographs by inverted microscopy are shown with cell line names and number of colonies. We designated cells that form fewer than 20 and more than 500 colonies after plating of 20,000 cells as anchorage-dependent and anchorage-independent, respectively. Right panel: Bar graph of colony numbers for 19 breast cancer cell lines. Error bars indicate standard deviations.
B. A gene signature for anchorage-independent growth of breast cancer cells. Left panel: The expression pattern of genes that distinguish anchorage-independent cells (AI) from anchorage-dependent cells (AD). The expression pattern of a two hundred gene signature is shown as a heatmap (red=high and blue=low expression). Right panel: A leave-one-out cross validation of probabilities for anchorage-independent growth ability (blue=anchorage-dependent cells, red=anchorage-independent cells). The accuracy of this signature was 86.4% using 0.5 as a cutoff probability. Outliers in this leave-one-out cross validation analysis are Hs578T (#6; having AD phenotype but resembling AI cells transcriptionally) and BT474 (#21 and #22 from two different batches; having AI phenotype but resembling AD cells transcriptionally)
C. Prediction of anchorage-independent growth ability of ovarian cells by the gene signature derived from breast cancer cells. Predicted probabilities are plotted for the categories with anchorage-dependent (AD) and anchorage-independent (AI) growth ability, and then are statistically evaluated using Mann-Whitney U test using GraphPad’s Prism. We use the same cut-off (AD<20 and AI>500 colonies) that is used to breast cancer cells. A bar indicates the mean value for each group.
Using expression data generated from this panel of cell lines, we then made use of statistical methodologies to develop a predictive model that maximally distinguishes the cellular ability of anchorage-independence (Bild et al., 2006). Importantly the data was taken from asynchronous growing cells in the anchorage-dependent condition but not from cells in anchorage-independent condition. Therefore the profile reflects the intrinsic capacity of the cell to exhibit anchorage-independent growth and not the state of growth in anchorage-independent condition. To train the model, we selected cell lines with an extreme anchorage-dependence phenotype (less than 20 colonies) or an extreme anchorage-independence (more than 500 colonies). The expression pattern of the selected 200 genes showed a clear distinction between anchorage-dependent and independent cells (Figure 2B). In contrast, sets of randomly selected 200 genes did not retain an ability to differentiate these two states (Supplementary Figure 1A and Supplementary Table 1). The results from leave-one-out cross validation demonstrated the capacity of the selected genes to accurately distinguish the two states, exhibiting an accuracy of 86.4% (Figure 2B). Furthermore, the predicted probability of the cell lines with intermediate abilities to form colonies in methylcellulose was indeed intermediate between that of the cells with anchorage-dependence and -independence, providing another validation of this signature (Supplementary Figure 1B).
To independently validate the signature, we applied it to an expression data set of 42 cultured ovarian cancer cell lines accompanied with an analysis of the colony-forming capacity of the cell lines (Matsumura unpublished data). The predicted probability for growth in an anchorage-independent condition shows statistically significant distinction with the measured colony number of cultured ovarian cell lines in the same condition and category (Figure 2C). Conversely, the gene signature generated from ovarian cancer cells can also predict the anchorage-independent growth ability of breast cancer cells (data not shown). These observations provide an independent validation of the gene signature and also suggest that breast cancer cells and ovarian cancer cells share the expression profile that indicates an ability to form colonies. To further validate the signature, we randomly split the ovarian cell line training samples into two halves and found that either set has the capacity to predict colony forming ability in the other (Supplementary Figure 2).
During the process of in vitro tumorigenesis, various oncogenes with distinct pathways have been shown to transform anchorage-dependent cells to anchorage-independent (Hanahan & Weinberg, 2000). For example, transfer of either c-Myc (a transcription factor), v-Src (a tyrosine kinase) or H-Ras (a small GTPase) into spontaneously immortalized mouse embryonic fibroblasts (MEFs) provides the cells an ability to grow in an anchorage-independent manner (Cifone & Fidler, 1980; Weinberg, 2007). To examine whether in vitro transformed murine fibroblasts exhibit the anchorage-independent cell growth signature generated above, we performed expression microarray analysis of MEFs that were transduced with retroviruses that deliver c-myc or v-src in comparison with that of empty vector control. We first confirmed that immortalized MEFs ectopically expressing c-Myc or v-Src form colonies in methylcellulose whereas control cells did not (Supplementary Figure 3A). It was true that there were several distinct phenotypes induced by these two different oncogenes. We observed that c-Myc and v-Src overexpression rendered MEFs more and less proliferative, respectively. Additionally v-Src expressing MEFs lost an ability to arrest the cell cycle in response to contact inhibition, while c-Myc and control cells retained the ability (data not shown). Importantly, the signature for anchorage-independent cell growth detected the difference between cells with and without colony-forming property but did not distinguish c-Myc expressing cells from MEFs with v-Src expression (Supplementary Figure 3B). This observation supports the conclusion that anchorage-independent cell growth ability in vitro is a common cancer cell phenotype that is induced by various distinct oncogenes (Cifone & Fidler, 1980; Weinberg, 2007). Taken together, the gene expression analysis provides very clear evidence for distinct profiles between anchorage-dependent and -independent cells.
Biological properties that characterize the capacity for anchorage-independent cell growth
An initial inspection of the genes forming the anchorage-independent signature revealed many that were mitochondrial genes, suggesting that the anchorage-independent phenotype could relate to an increased activity of mitochondrial biogenesis. Indeed, one such gene was Tfam, a gene required for mitochondrial biogenesis (Kang & Hamasaki, 2005). A further analysis of the genes encoding proteins predicted to localize to the mitochondrion revealed a statistically significant enrichment for anchorage-independent phenotype (data not shown).
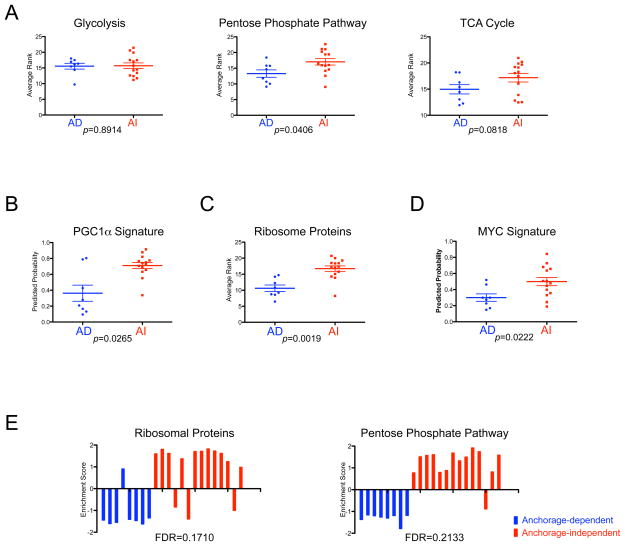
Aerobic glycolysis, or the Warburg effect, has long been considered as a component of the malignant phenotype, in which the tumor is more dependent on glycolysis than the mitochondrial TCA cycle, even in the presence of sufficient level of oxygen (Warburg, 1956). However, this view has been challenged over the years, and it remains contentious whether the increased glycolysis in cancer cells is primarily the result of transformation events or due to the hypoxic tumor microenvironment (Zu & Guppy, 2004). Indeed, several lines of evidence have suggested that more aggressive cancer cells exhibit an increased oxidative phosphorylation due to the TCA cycle in mitochondria to produce energy source as well as up-regulation of the pentose phosphate pathway in cytosol to generate nucleotide pool in the cell (Chen et al., 2007; Funes et al., 2007; Li et al., 2005; Telang et al., 2007; Tong et al., 2009). Our observation, that of increased level of mitochondrial genes in cells with anchorage-independence as mentioned above (Supplementary Table 1), prompted us to investigate the link of the anchorage-independence with metabolic pathways regarding glucose catabolism. We used a list of genes known to play a role in cell metabolism and analyzed those gene expression patterns using the average rank of cells for each pathway (Funes et al., 2007). Figure 3A shows the pattern of the glycolytic and pentose phosphate pathways, and the TCA cycle. Among the three metabolic pathways, the expression profile of glycolytic genes was not significantly correlated, whereas genes in the pentose phosphate pathway were significantly up-regulated, and the overall genes in the TCA cycle showed a trend toward up-regulation (Figure 3A).
Figure 3. Characterization of the anchorage-independent growth expression signatures.
A. Comparison of the patterns of gene sets that reflect glucose metabolism between breast cancer cells with anchorage-dependent (AD<20 colonies) and anchorage-independent (AI>500 colonies) growth ability. Based on the gene list in previous work (Funes et al., 2007), we make the average rank of the gene sets for glucose metabolism pathways to estimate the activity of the pathway. Relative expression in the form of average rank is plotted and compared between breast cancer cells with AD and AI growth ability by Mann-Whitney U test using GraphPad’s Prism. A bar indicates the mean value for each group.
B. Prediction of the peroxisome proliferator-activated receptor-gamma co-activator 1 α (PGC1α) activity in breast cells. The predicted probabilities of PGC1α are plotted and compared between breast cancer cells with anchorage-dependent (AD<20 colonies) and anchorage-independent (AI>500 colonies) growth ability by Mann-Whitney U test using GraphPad’s Prism. A bar indicates the mean value for each group.
C. Comparison of ribosomal protein expression between breast cancer cells with anchorage-dependent (AD<20 colonies) and anchorage-independent (AI>500 colonies) growth ability. Based on gene symbols from Affymetrix, we compute the average rank of the ribosomal genes to estimate a state of ribosomal biogenesis. Relative expression in the form of average rank is plotted and compared between breast cancer cells with AD and AI growth ability by Mann-Whitney U test using GraphPad’s Prism. A bar indicates the mean value for each group.
D. Prediction of the MYC activity in breast cells. The predicted probabilities of MYC are plotted and compared between breast cancer cells with anchorage-dependent (AD<20 colonies) and anchorage-independent (AI>500 colonies) growth ability by Mann-Whitney U test using GraphPad’s Prism. A bar indicates the mean value for each group.
E. Gene Set Enrichment Analysis (GSEA) of anchorage-independent growth ability. GSEA revealed enrichment of ribosomal proteins and pentose phosphate pathway with statistical significance (false discovery rate: FDR<0.25). Enrichment score for each sample is further calculated by ASSESS (analysis of sample set enrichment scores) and plotted as bar graphs.
Peroxisome proliferator-activated receptor-gamma co-activator 1 α (PGC1α) overexpression significantly enhances mitochondria biogenesis in the model of myogenesis, and this model has been used to identify genes whose products localize to mitochondria (Calvo et al., 2006). Making use of publicly available gene expression data, we generated a PGC1α signature and then applied it to the breast cancer cell line data to confirm the observed relation between mitochondria activity and the ability to form colonies in methyl-cellulose by an independent method. PGC1α overexpressing cells are indeed enriched in mitochondria genes as expected (Supplementary Figure 4). Applying this PGC1α signature to cultured breast cancer cells demonstrated strong positive correlation with the ability of anchorage-independent cell growth, further validating our observation by an independent method (Figure 3B). Likewise increased expression of ribosomal protein genes in the anchorage-independence signature was also confirmed by the averaged rank test (Supplementary Table 1, 2 and Figure 3C).
To further explore the possible relation of a specific pathway with a cellular ability to grow in anchorage-independent condition, we applied a series of oncogene pathway signatures that included MYC, RAS, SRC, β-Catenin, PI3-kinase, AKT, STAT3, HER2, TP53, TP63 and TNF-α to evaluate the breast cancer cell line dataset (Bild et al., 2006) (M. Gatza unpublished result). This analysis revealed a significant enrichment of the MYC pathway in cells with anchorage-independent growth among 11 pathways examined (Figure 3D). This observation suggests the possibility that the MYC pathway is directly involved in the cellular process to acquire anchorage-independence through induction both of ribosomal and mitochondrial biogenesis (Dang et al., 2006). The MYC predicted probabilities, however, did not significantly correlate with average ranks of metabolic, mitochondrial, ribosomal gene expression or the predicted probabilities of PGC1α in our 19 breast cells in Spearman correlation analysis (Supplementary Table 3).
Furthermore, consistent with the notions above, a Gene Set Enrichment Analysis using 522 functional test gene sets (C2 set from Broad Institute) (Subramanian et al., 2005) revealed enrichment of only two gene sets for biological properties, ribosomal proteins and pentose phosphate pathway with statistical significance, while anchorage-dependent cells did not show enrichment of any gene sets (Figure 3E).
The anchorage-independent signature predicts the potential for metastasis
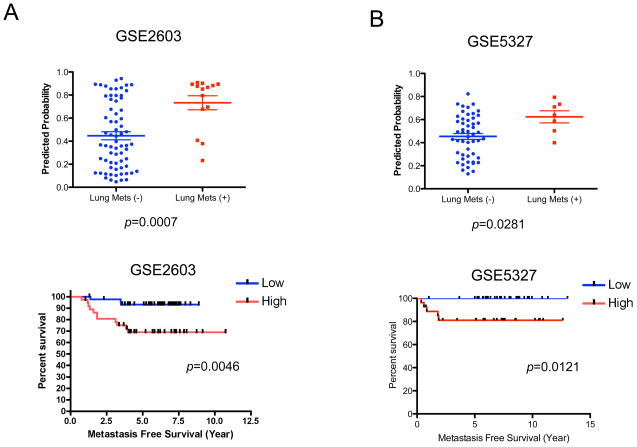
Clearly, the study of in vitro systems to explore oncogenic mechanisms is ultimately for the purpose of better understanding the clinical cancer process. Similarly, the study of clinical cancer, including the potential of a cancer to become aggressive and to metastasize, is to characterize and understand the disease process. We analyzed two breast cancer datasets in which DNA microarray data was available together with clinical outcome information. We then made use of the anchorage-independent signature to predict the probability of the anchorage-independent phenotype within the tumor samples and then relate to clinical outcomes. As shown in Figure 4A, primary tumors with the potential for lung metastasis exhibit the anchorage-independent signature more strongly than those without this metastatic potential. Likewise, the overall outcome for patients with tumors exhibiting the anchorage independent signature is worse than for those without the signature. This same relationship was seen in a second cohort of estrogen receptor negative breast cancer with lung metastasis (Figure 4B). These observations supported the hypothesis that the expression pattern of in vivo tumors with metastatic potential has characteristics of in vitro cultured cancer cells with colony-forming ability. No other property of the tumors including lymph node status, Her2 positivity or tumor size exhibited a significant correlation with the signature, while both estrogen receptor negativity and progesterone receptor negativity correlated with higher probability of anchorage-independence signature in GSE2603 (data not shown).
Figure 4. Metastatic potential of breast cancer is predicted by an anchorage-independent growth signature.
A and B. Prediction of lung metastasis of primary breast tumors by the gene signature for in vitro anchorage-independent growth ability. Upper panels: Mann-Whitney U test based on anchorage-independent growth ability of GSE2603 (includes both estrogen receptor positive and negative breast cancers; left panel) (A) and GSE5327 (only estrogen receptor negative breast cancers; right panel) (B). The predicted probabilities for anchorage-independent cell growth signature are plotted and compared between breast tumors with and without lung metastasis (Lung Mets) by Mann-Whitney U test using GraphPad’s Prism. A bar indicates the mean value for each group. Lower panels: Kaplan-Meier analysis based on anchorage-independent growth ability of GSE2603 (both of estrogen receptor positive and negative breast cancers; left panel) (A) and GSE5327 (only estrogen receptor negative breast cancers; right panel) (B). X-axis indicates lung metastasis free survival. “Low” (blue) and “High” (red) are defined by being below and above 0.5 of predicted probabilities respectively, the optimal cut-off as determined by a receiver-operator curves (ROC) analysis using GraphPad’s Prism (data not shown). A log rank test was used to evaluate the result statistically using GraphPad’s Prism.
We then explored the extent to which the anchorage-independent phenotype could be seen in primary tumors derived from a different tissue type. We evaluated the signature in a lung cancer data set that included data from tumors with or without recurrence and metastasis. Consistent with the results from the breast cancer datasets, the analysis of primary lung tumors revealed that the metastatic tumors exhibited the anchorage-independent signature (Figure 5A). Importantly, this analysis further demonstrated that there was a distinction between those tumors that recurred locally and those that metastasized. As such, the result would suggest that the anchorage-independent phenotype, as measured with the gene expression signature, is not simply a measure of tumor aggressiveness and the capacity to recur but rather truly reflects the metastatic potential of the tumor. Furthermore, detection of similarity in the expression profiling between cultured breast cancer cells with primary lung tumor as well as cultured ovarian cancer cells implies that this phenotype is not tissue-type restricted (Figure 2C and Figure 5A). Indeed, we detected a higher probability of this signature in metastatic melanomas derived from very different cell of origin (Figure 5B).
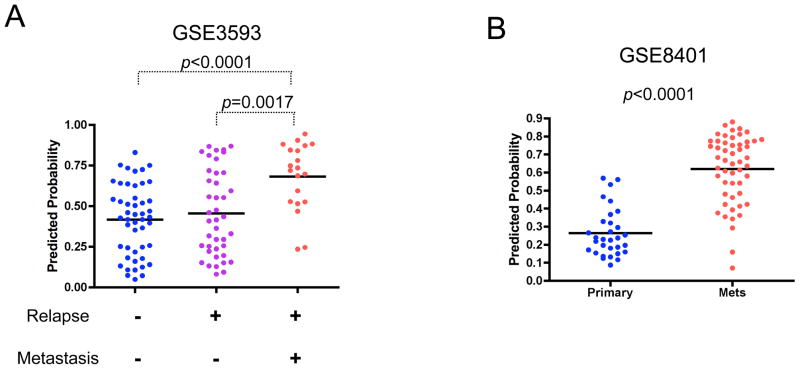
Figure 5. Metastatic potential of lung cancer and melanoma is predicted by the anchorage-independent growth signature.
A. Prediction of metastasis of primary lung tumors by the gene signature for in vitro anchorage-independent growth ability. The predictions based on the anchorage-independent signature are shown for a lung tumor dataset that includes tumors with relapse but not with metastasis, and tumors with relapse and metastasis (GSE3593). Predicted probabilities are plotted for the groups with the defined outcome and statistically evaluated using Mann-Whitney U test. A bar indicates the mean value for each group.
B. Link with metastatic melanomas and anchorage-independent growth ability of breast cells (GSE8401). Predicted probabilities are plotted for the groups for primary and metastatic tumors, and statistically evaluated using Mann-Whitney U test. A bar indicates the mean value for each group.
Discussion
A dominant characteristic of virtually all cancers is heterogeneity and complexity – diseases described as ‘breast cancer’ or ‘lung cancer’ or ‘ovarian cancer’ are actually collections of disease with distinct molecular mechanisms and clinical characteristics (Nevins & Potti, 2007). The challenge is to dissect this complexity to facilitate an understanding of the discrete mechanisms involving multiple mutations and alterations that generate the cancer phenotype and to facilitate an ability to define therapeutic strategies that can match the complexity with equally complex combination regimens (Hanahan & Weinberg, 2000). This complexity of phenotypes has been addressed in a variety of ways, making use of in vitro cultured cancer cells, mouse models of disease, and then also the ability to study the disease process directly in patients. Such studies have given rise to a detailed understanding of the disease process including the various oncogenic and tumor suppressor activities important for the oncogenic phenotype (Vogelstein & Kinzler, 2004). Nevertheless, it is also true that in many instances, these studies are isolated from one another, providing details but not necessarily a coherent view of the role of specific genes and pathways in determining the clinical phenotype. For example, a cell-culture phenotype, such as growth properties, activation of various signaling pathways, or sensitivity to therapeutics, can be difficult to translate to an in vivo tumor setting, although in vitro cultured cancer cells have been extensively used to model human primary cancer. The approach we describe here is one strategy to bring these diverse experimental conditions into a common focus – an ability to translate a cellular phenotype studied in great detail to the context of human cancer.
The concept of gene expression signatures
An expression signature is simply a representation of a biological state in the form of a pattern of gene expression unique to that circumstance. Underlying the concept is the realization that virtually any biological condition, whether a developmental state, a cellular response to extracellular ligands, or an in vitro experimental state such as anchorage-independent growth, is reflected in changes in gene expression. While no one single gene would have the power to define the biological state, the ability to measure and identify patterns of gene expression provides an opportunity to develop these signatures reflecting biological phenotypes. The power of the expression signature is two-fold. First, the enormous complexity of the expression data that can be sampled provides the opportunity to identify patterns of expression that reflect very subtle distinctions in biology. The main limitation is the capacity to define the biological state of interest, whether through the generation of distinct experimental states that can then be used to train an analysis of expression or by taking advantage of existing biological conditions that can be used as the training opportunity. Second, the expression signature is portable in the sense that it can be assayed in varied contexts. That is, an expression signature developed in a cell culture context can be measured in a tumor or a histological section. As such, the expression signature provides the capacity to link otherwise heterologous systems. A cell culture phenotype such as pathway activation is difficult to represent in a diverse sample such as a tumor. In contrast, an expression profile provides a mechanism to link these two states – the profile represents pathway activation in the cell culture and then can be used to interrogate the expression data from a tumor.
Metastasis is a critical event in cancer progression and one of the most life-threatening events for patients with cancer. Substantial research has implicated molecules relevant for the process such as matrix metalloproteinases and their regulators, and for a number of signaling pathways such as RAS and PI3K (Campbell & Der, 2004; Lopez-Otin & Matrisian, 2007; Samuels & Ericson, 2006). The process of metastasis in vivo has been linked with the anchorage-independent cell growth property in vitro, although there have been very limited numbers of experimental validations that are very often controversial. In animal transplantation models, cells with higher colony formation activity show more metastatic potential, and in humans, metastatic tumors are more anchorage-independent in primary culture, while many research experimentally failed to detect the positive link between metastasis and anchorage-independent cell growth ability (Cifone & Fidler, 1980; Johns & Mills, 1983; Mattox & Von Hoff, 1980; Nicolson et al., 1988; Nomura et al., 1989; Price, 1986; Sutherland et al., 1983). In the literature, four of five anchorage-dependent cell lines (MDA-MB-361, Hs578T, ZR75 and BT483) are reported to have metastatic potential to various distant sites such as lung, bone and brain in several different murine experimental models. On the other hand, there is no report of the metastatic potential of BT20 and SKBR3 among the 8 anchorage-independent cell lines (Hackett et al., 1977; Shafie & Liotta, 1980; Thompson et al., 1992; van Slooten et al., 1995; Yin et al., 2003)(Mukhopadhyay et al., 1999; Zhang et al., 1991). Clearly direct experimental link of cellular metastatic potential with anchorage-independent cell growth ability remains to be elucidated for human multiple breast cancer cell lines described in the current study. Relying on an expression signature, however, this study provides a clear link between in vitro colony-forming ability with metastatic potential of tumor. Furthermore, expression similarity of anchorage-independent cells to metastatic but not to relapsed tumors is consistent with the observation, in which anchorage-independent growth ability of human primary breast cancer correlates with its metastatic ability but not with tumor recurrence (Nomura et al., 1989), therefore, may suggest that this in vitro character mimics a narrow aspect of features of in vivo cancer. Considering the fact that metastasis is a still life-threatening event in the life of patient and the difficulty in evaluation of pharmacological effects against metastasis (Eccles & Welch, 2007), we believe that our observation provides the logical basis to use in vitro colony-forming ability as a surrogate phenotype of metastasis, for further study in cancer biology and pharmaceutical field.
Materials and Methods
Cell culture
Methods for culture of 19 breast cancer cell lines are described previously (Bild et al., 2006). For colony formation assay we plated 20,000 cells per well into 6-well plate (Corning, Ultra Low Attachment Surface, #3471) containing 15% methyl-cellulose (Sigma, M-0387) media, and after three weeks, counted the number of colonies at least 100 mm in the diameter.
Transformation of mouse embryonic fibroblasts for expression analysis
Mouse embryonic fibroblasts (MEFs) were cultured and immortalized according to a 3T3 protocol (Giangrande et al., 2004). Three independent cell lines were established and designated as WA, WB and WC cells. pBabe-puro, pBabe-human c-myc (gifts from Martin Eilers) and pBabe-v-src (a gift from Hidesaburo Hanafusa) were used to generate retroviral vectors with Plat-E cells (a gift from Toshio Kitamura) as previously described (Giangrande et al., 2004). For each vector, infection and selection by puromycin were performed in quadriplicate as previously described (Giangrande et al., 2004). Using one of four infections, we confirmed all three (WA, WB and WC) MEFs infected with c-myc or v-src viruses exhibit oncogene-specific morphological alterations and colony forming ability by the same criteria as that of human breast cancer cell lines (Supplementary Figure 3A and data not shown). We extracted RNA from cells of three infections for each vector and performed expression microarray assays using Affymetrix Mouse 430.2 Gene Chips. Finally, we obtained the expression data for 9 empty vector, 9 c-myc and 8 v-src infections. The expression data is available in GEO (http://www.ncbi.nlm.nih.gov/geo; GSE15161).
Statistical analyses of microarray data
Analysis of expression data was described previously (Bild et al., 2006). In summary, we collected training sets consisting of gene expression values of samples where an ability of anchorage-independent cell growth or a pathway activation status was known. We created gene expression signatures by choosing the genes whose expression profiles across the training samples most highly correlated with the phenotype. Then, to predict status of the cells or samples, we fit a Bayesian probit regression model that assigned scores indicating the probability that a cell line exhibited evidence of the phenotype, based on the concordance of its gene expression values with the signature. We evaluated the statistical relationship between sets of genes with significant Gene Ontology terms or transcription factor binding motifs using GATHER (http://gather.genome.duke.edu/) (Chang & Nevins, 2006).
We generated gene expression signatures reflecting the ability of cells to grow in methyl-cellulose using gene expression data for 5 anchorage-dependent and 8 anchorage-independent breast cancer cell lines. We collected gene expression data of the cells growing asynchronously in an anchorage-dependent condition. In total, due to duplicates derived from two different batches, we collected 22 expression samples, eight and fourteen for anchorage-dependent and anchorage-independent cell lines, respectively. We removed the batch effect of Affymetrix expression data using ComBat according to the instruction of http://statistics.byu.edu/johnson/ComBat/Abstract.html (Johnson et al., 2007). Complete datasets are available in GEO with original format data (http://www.ncbi.nlm.nih.gov/geo; GSE15026; a part of this data is also available as GSE3156) (Bild et al., 2006). Data with MAS5 format was used for Binary Regression analysis, while other analyses were performed with RMA normalized data. We also used a microarray data set of 42 cultured ovarian cancer cell lines with the information of colony forming ability for validation (Matsumura not published).
To test whether the anchorage-independent growth signature provides a unique representation of the underlying biological phenotype, we compared the ability of that signature against random sets of genes. We generated 10,000 signatures of randomly selected sets of 200 genes, the same size as the anchorage-independent growth signature. Then, using the 22 arrays consisting of the anchorage-independent and anchorage-dependent cell lines, we applied each signature to assess its ability to distinguish the two sets of cell lines. We calculated receiver operating characteristic curves (ROC) using R to measure the sensitivity and specificity of the ability of the signatures to model the AI phenotype (Supplementary Figure 1A).
The methods to use gene set enrichment analysis (GSEA) and analysis of sample set enrichment scores (ASSESS) were described previously (Edelman et al., 2006).
Datasets of human cancer samples and mitochondrial biogenesis
In this study we used four independent datasets of cancer with information for metastasis (breast; GSE2603 and GSE5327, melanoma; GSE8401 and lung; GSE3593) (Bild et al., 2006; Minn et al., 2007; Minn et al., 2005). We used GSE4330 to generate mitochondrial biogenesis signature (Calvo et al., 2006).
Subcellular localization of the proteins
To predict the subcellular localization of the proteins represented on the Affymetrix chip, we used the pSLT and pTARGET algorithms (Guda & Subramaniam, 2005; Scott et al., 2004). We downloaded the pSLT localization predictions for known human proteins from their website (http://www.mcb.mcgill.ca/~hera/PSLT/) and mapped them to microarray probe set IDs using the annotations provided by Affymetrix. For the pTARGET predictions, we first obtained from SWISS-PROT the protein sequences for each known protein represented on the microarray. To obtain the localization predictions, we submitted the sequences to the pTARGET predictions server available online (http://bioinformatics.albany.edu/~ptarget/), and then mapped the predictions back to the probe sets on the microarray. We collected the genes that putatively localize at an organelle according to the prediction mentioned above, ranked the cancer cell lines according to the expression levels, and then used the averaged ranks as a signature that roughly reflected the cellular organelle state.
Supplementary Material
Acknowledgments
We thank Drs. J.-T. Chi, T. Hallstrom, L. Kong, R. Rempel, J. Freedman, M. Gatza, B. Balakumaran, H. Dressman, Y. Yokota, S. Akiyama, T. Inoue, M. Oshimura, M. Araki, H. Saya, Aburatani, A. Niida, K. Araki, Y. Taya and Y. Ito for helpful discussions; A. Bild, T. Kitamura, M. Eilers and H. Hanafusa for providing materials; L. Jakoi for help with experiments; and T. Henry and K. Culler for assistance with the preparation of the manuscript. S. M. was a research fellow of the Uehara Memorial Foundation and is a visiting scholar of Riken Genome Science Center.
References
- Bild A, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi M-B, Harpole D, Lancaster JM, Berchuck A, Olson JA, Marks JR, Dressman HK, West M, Nevins JR. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Nat Genet. 2006;38:576–82. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- Campbell PM, Der CJ. Semin Cancer Biol. 2004;14:105–14. doi: 10.1016/j.semcancer.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Chang JT, Nevins JR. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, 3rd, Felding-Habermann B. Cancer Res. 2007;67:1472–86. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- Cifone MA, Fidler IJ. Proc Nat’l Acad Sci USA. 1980;77:1039–1043. doi: 10.1073/pnas.77.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. Semin Cancer Biol. 2006;16:253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Eccles SA, Welch DR. Lancet. 2007;369:1742–57. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman E, Porrello A, Guinney J, Balakumaran B, Bild A, Febbo PG, Mukherjee S. Bioinformatics. 2006;22:e108–116. doi: 10.1093/bioinformatics/btl231. [DOI] [PubMed] [Google Scholar]
- Funes JM, Quintero M, Henderson S, Martinez D, Qureshi U, Westwood C, Clements MO, Bourboulia D, Pedley RB, Moncada S, Boshoff C. Proc Natl Acad Sci U S A. 2007;104:6223–8. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande P, Zhu W, Schlisio S, Sun XH, Mori S, Gaubatz S, Nevins JR. Genes & Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C, Subramaniam S. Bioinformatics. 2005;21:3963–9. doi: 10.1093/bioinformatics/bti650. [DOI] [PubMed] [Google Scholar]
- Guo W, Giancotti FG. Nat Rev Mol Cell Biol. 2004;5:816–26. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Hackett AJ, Smith HS, Springer EL, Owens RB, Nelson-Rees WA, Riggs JL, Gardner MB. J Natl Cancer Inst. 1977;58:1795–806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Johns ME, Mills SE. Cancer. 1983;52:1401–4. doi: 10.1002/1097-0142(19831015)52:8<1401::aid-cncr2820520811>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kang D, Hamasaki N. Ann N Y Acad Sci. 2005;1042:101–8. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Mol Cell Biol. 2005;25:6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Matrisian LM. Nat Rev Cancer. 2007;7:800–8. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- Mattox DE, Von Hoff DD. Am J Surg. 1980;140:527–30. doi: 10.1016/0002-9610(80)90205-6. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, Kreike B, Zhang Y, Wang Y, Ishwaran H, Foekens JA, van de Vijver M, Massague J. Proc Natl Acad Sci U S A. 2007;104:6740–5. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Theriault RL, Price JE. Clin Exp Metastasis. 1999;17:325–32. doi: 10.1023/a:1006659230585. [DOI] [PubMed] [Google Scholar]
- Nevins JR, Potti A. Nat Rev Genet. 2007;8:601–9. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- Nicolson GL, Lembo TM, Welch DR. Cancer Res. 1988;48:399–404. [PubMed] [Google Scholar]
- Nomura Y, Tashiro H, Hisamatsu K. Cancer Res. 1989;49:5288–93. [PubMed] [Google Scholar]
- Price JE. J Natl Cancer Inst. 1986;77:529–35. [PubMed] [Google Scholar]
- Samuels Y, Ericson K. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- Scott MS, Thomas DY, Hallett MT. Genome Res. 2004;14:1957–66. doi: 10.1101/gr.2650004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafie SM, Liotta LA. Cancer Lett. 1980;11:81–7. doi: 10.1016/0304-3835(80)90097-x. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gilletee MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland CM, Mather FJ, Carter RD, Cerise EJ, Krementz ET. Surgery. 1983;94:370–5. [PubMed] [Google Scholar]
- Telang S, Lane AN, Nelson KK, Arumugam S, Chesney J. Mol Cancer. 2007;6:77. doi: 10.1186/1476-4598-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME, et al. J Cell Physiol. 1992;150:534–44. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. Curr Opin Genet Dev 2009 [Google Scholar]
- van Slooten HJ, Bonsing BA, Hiller AJ, Colbern GT, van Dierendonck JH, Cornelisse CJ, Smith HS. Br J Cancer. 1995;72:22–30. doi: 10.1038/bjc.1995.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Warburg O. Science. 1956;124:269–70. [PubMed] [Google Scholar]
- Weinberg RA. The biology of cancer. Garland Science; New York: 2007. [Google Scholar]
- Yin JJ, Mohammad KS, Kakonen SM, Harris S, Wu-Wong JR, Wessale JL, Padley RJ, Garrett IR, Chirgwin JM, Guise TA. Proc Natl Acad Sci U S A. 2003;100:10954–9. doi: 10.1073/pnas.1830978100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RD, Fidler IJ, Price JE. Invasion Metastasis. 1991;11:204–15. [PubMed] [Google Scholar]
- Zu XL, Guppy M. Biochem Biophys Res Commun. 2004;313:459–65. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.