Abstract
In high-yielding dairy cows the liver undergoes extensive physiological and biochemical changes during the early postpartum period in an effort to re-establish metabolic homeostasis and to counteract the adverse effects of negative energy balance (NEB). These adaptations are likely to be mediated by significant alterations in hepatic gene expression. To gain new insights into these events an energy balance model was created using differential feeding and milking regimes to produce two groups of cows with either a mild (MNEB) or severe NEB (SNEB) status. Cows were slaughtered and liver tissues collected on days 6–7 of the first follicular wave postpartum. Using an Affymetrix 23k oligonucleotide bovine array to determine global gene expression in hepatic tissue of these cows, we found a total of 416 genes (189 up- and 227 downregulated) to be altered by SNEB. Network analysis using Ingenuity Pathway Analysis revealed that SNEB was associated with widespread changes in gene expression classified into 36 gene networks including those associated with lipid metabolism, connective tissue development and function, cell signaling, cell cycle, and metabolic diseases, the three most significant of which are discussed in detail. SNEB cows displayed reduced expression of transcription activators and signal transducers that regulate the expression of genes and gene networks associated with cell signaling and tissue repair. These alterations are linked with increased expression of abnormal cell cycle and cellular proliferation associated pathways. This study provides new information and insights on the effect of SNEB on gene expression in high-yielding Holstein Friesian dairy cows in the early postpartum period.
Keywords: microarray, pathway analysis, lipid metabolism
fertility problems in the modern high-yielding dairy cow are of considerable economic cost to the dairy industry (44, 58), with both nutritional and metabolic stress contributing to poor reproductive performance. During the early postpartum period high-yielding dairy cows experience negative energy balance (NEB), as the energy demands for lactogenesis exceed energy intake. These increased metabolic demands lead to increased mobilization of body reserves such as fat and protein (86). A prominent feature of the energy-metabolic response to the NEB state involves reliance on fatty acids and ketones as a source of energy and an increased capacity for mitochondrial fatty acid oxidation in tissues with high oxidative energy demands such as the liver (17, 27).
Oxidation of nonesterified fatty acids (NEFA) in the liver results in the increased production of reactive oxygen species (ROS) (90), and NEB can result in the onset of oxidative stress, which can lead to disruption of normal metabolism and physiology (66). The liver undergoes extensive changes during the early postpartum period concomitant with increases in its rate of metabolism and weight (76). In severe cases this may result in the development of production diseases such as hepatic lipidosis and ketosis (25, 41). Increased lipolysis and subsequent triglyceride accumulation is a potential cause of liver damage (77) and is therefore likely to affect liver function.
The liver has been estimated to supply up to 90% of glucose requirements in ruminants through hepatic gluconeogenesis (68) critical to the progression of growth and lactation. Consequently liver is a logical tissue for transcript profiling to identify key regulatory pathways affecting energy generation, carbohydrate, lipid, and amino acid metabolism (88). Indeed recent oligonucleotide array-based studies in cattle have uncovered a host of genes putatively involved in metabolic processes such as energetic efficiency (32), ketosis (56), and adaptation to the onset of lactation (55) in liver tissue. The recent availability of the bovine genome sequence and of bovine-specific microarrays provides an excellent opportunity to study global gene expression in tissues of interest using a well-controlled and validated approach (11).
The experiment described here was designed to test the hypothesis that severe NEB (SNEB) in the early postpartum period has profound effects on the global expression of genes regulating liver metabolic processes and its concomitant ability to function normally.
MATERIALS AND METHODS
Animal model.
The animal model employed in this study has been described previously (19, 66), and all procedures were carried out under license in accordance with the European Community Directive 86-609-EC. The nutritional and lactational management regime employed were designed to create significant divergence in the energy balance (EB) profiles of cows in early lactation.
In brief, multiparous Holstein-Friesian cows (n = 24) were blocked 2 wk prior to expected calving date according to parity, body condition score, and previous lactation yield (average lactation 6,477 ± 354 kg) and randomly allocated to mild (MNEB, n = 12) or severe (SNEB, n = 12) NEB groups. MNEB cows were fed ad libitum grass silage and 8 kg/day concentrates and milked once daily; SNEB cows were fed 25 kg/day silage and 4 kg/day concentrate and milked three times daily. Measurements of body condition score and EB were used to select cows that showed extremes in EB from each group (MNEB, n = 5; SNEB, n = 6). Cows were slaughtered on days 6–7 of the first follicular wave after calving (mean number of days postpartum: MNEB mean 13.6 ± 0.75, range 11–15; SNEB mean 14.3 ± 0.56, range 13–16), based on daily transrectal ultrasonography.
Liver tissue collection for RNA and TAG analysis.
The entire liver was removed within 15–30 min after slaughter and weighed. Samples weighing ∼1 g were dissected, rinsed in RNase-free phosphate buffer, snap-frozen in liquid nitrogen, and stored at −80°C. For triacylglyceride (TAG) analysis, total lipids were extracted from 50 mg samples of liver as previously described (21).
Blood sampling and metabolite assays.
Stabilized (EDTA-treated) whole blood samples were collected on the day of slaughter by jugular venipuncture for hematological analysis. Blood samples were analyzed for glucose, NEFA, β-hydroxybutyrates (BHB), and urea using appropriate kits and an ABX Mira autoanalyzer (ABX Mira, Cedex, France). All metabolite assay coefficients of variation were low and typically <5%.
RNA extraction and quality analysis.
Total RNA was prepared from 100–200 mg of fragmented frozen liver tissue using the TRIzol reagent (Sigma-Aldrich, Dorset, UK). Tissue samples were homogenized in 3 ml of TRIzol reagent and chloroform and subsequently precipitated using isopropanol (Sigma Chemical). RNA samples were stored at −80°C. We treated 20 μg of total RNA from each sample for genomic DNA contamination with the RNase-free DNase set (QIAGEN, Crawley, West Sussex, UK) and purified it using the RNeasy mini kit in accordance with guidelines supplied (QIAGEN). RNA quality and quantity were assessed using automated capillary gel electrophoresis on a Bioanalyzer 2100 with RNA 6000 Nano Labchips according to manufacturer's instructions (Agilent Technologies Ireland, Dublin, Ireland). Samples of RNA had 28S/18S ratios ranging from 1.8 to 2.0 and RNA integrity number values of between 8.0 and 10.0, which indicates that RNA samples are of high quality and suitable for microarray analysis.
cDNA synthesis.
From this reaction, 1 μg of DNase-treated RNA was reverse transcribed using AMV reverse transcriptase and 500 ng random hexamer primers in a 20 μl reaction (reverse transcription system kit; Promega, Madison, WI). A quantity (0.39 ng) of kanamycin mRNA (Promega) was spiked into each sample as an exogenous control (Promega). A master mix of reagents was prepared for the above reaction to minimize potential variation from pipetting. Negative control samples were also prepared by including all reagents as above for the cDNA synthesis, minus the reverse transcriptase enzyme to ensure there was no genomic DNA contamination.
Primer design.
Gene-specific primers (Table 1) were designed online using the Primer3 web-based software program (http://frodo.wi.mit.edu/primer3). Primer specificity was determined using the basic local alignment search tool from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). All oligonucleotides were commercially synthesized as highly purified, salt-free products (Sigma Genosys). For each gene, PCR conditions were optimized by conventional PCR amplification using Go Taq Flexi DNA Polymerase (Promega), with the addition of 1 μl cDNA (1 μg/μl) and 1 μl of 20 μM forward and reverse primer mix. Standards for absolute real time RT-PCR assays were prepared from PCR products generated from cDNA, which was subsequently purified using QIquick PCR purification columns (QIAGEN, Crawley, West Sussex, UK). Exact concentrations of purified PCR product was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and the presence of a single product confirmed by electrophoresis on a 1.5% (wt/vol) ethidium bromide agarose gel.
Table 1.
Oligonucleotide forward and reverse primer sequences (5′-3′), PCR product length, linearity, efficiency, and % intra-assay variation of real-time RT-PCR reactions
| Gene | Primer Sequence | Product Size, bp | Accession No. | r2 | E | % Variation |
|---|---|---|---|---|---|---|
| SAA1 | F CAACTACAGGGGTGCAGACA | 241 | XM_867996 | 0.998 | 1.03 | ± 3.9 |
| R AGCAGGTCTGAAGTGGTTGG | ||||||
| GPX3 | F ACACGAGTACAGGGCCACAT | 201 | NM_174077 | 0.997 | 1.04 | ± 5.5 |
| R GGTAGGGTGAGGTCCTGGTT | ||||||
| FOXA3 | F GCCATGTCATCCCTGTCTCT | 176 | NM_001033119 | 0.996 | 1.08 | ± 0.1 |
| R CCGGCAAGAACCAGACTTTA | ||||||
| ANGPT4 | F GAATTTCCAACGGAACTGGA | 113 | EU599227 | 0.98 | 1.28 | ± 21.0 |
| R GAGAGTAGGTTGCCCTGCTG | ||||||
| ADIPOR2 | F CCAGAGGAGCCCCACAGT | 117 | BT025411 | 1.0 | 0.89 | ± 6.0 |
| R GCCATCTCGCGTCTTCTC | ||||||
| CD36 | F CAGCATCAGCAAGGCATTA | 115 | BC103112 | 0.99 | 1.23 | ± 1.5 |
| R CAGAAGAGCTGTAGAACGTGGA | ||||||
| CLDN1 | F CCATTGCTAAAAATGACTTAGCC | 120 | BT021861 | 0.999 | 1.03 | ± 0.1 |
| R AAGGAATGCTATCTCCCCTCA | ||||||
| CPT1A | F TCCTGGTGGGCTACCAATTA | 181 | FJ415874 | 0.999 | 0.99 | ± 2.6 |
| R TGCGTCTGTAAAGCAGGATG | ||||||
| CPT1B | F GCGCTTTGGGAACCAGAT | 128 | NM_001034349 | 0.998 | 1.08 | ±16.5 |
| R CCCCCTCCTCCACTCTGT | ||||||
| HP | F TCGTCGTTCACGACAAGG | 129 | BC109668 | 0.995 | 1.13 | ± 0.1 |
| R TTTTCCGAACCCAGTCCA | ||||||
| IL8 | F AAAAGACACAAACAGAAAGACCTC | 140 | AF232704 | 0.99 | 1.06 | ± 0.0 |
| R GCATCTCAAAAGTAGGACTTCCA | ||||||
| TNFα | F CATCCTGTCTGCCATCAAGA | 147 | EU276079 | 0.995 | 0.93 | ± 19.4 |
| R TAGTCCGGCAGGTTGATCTC | ||||||
| ACADVL | F ATCGTCCACCAGGAACTGAG | 130 | BT030546 | 0.999 | 1.05 | ± 5.2 |
| R GCTGCAGCAGAAACTGTTCA | ||||||
| PPARA | F TTGTGGCTGCTATCATTTGC | 135 | NM_001034036 | 0.998 | 1.0 | ± 2.9 |
| R AGAGGAAGACGTCGTCAGGA | ||||||
| PPARG | F GTGAAGCCCATTGAGGACAT | 149 | NM_181024 | 0.999 | 0.95 | ± 2.4 |
| R AGCTGCACGTGTTCTGTCAC | ||||||
| PPARD | F CACTCTCACTGCTGGACCAA | 194 | NM_001083636 | 0.99 | 0.98 | ± 2.9 |
| R GCAGATCCGCTCACATTTCT | ||||||
| RxRG | F ATGAAGATATGCCCGTGGAG | 126 | NM_001075408 | 0.991 | 1.01 | ± 11.8 |
| R CAGCGTGGCATATATTGGTG | ||||||
| HSP90 | F GGCGCTGATATCTCCATGAT | 119 | BC151818 | 0.99 | 1.07 | ± 0.9 |
| R GACTCCCAGGCATACTGCTC | ||||||
| FABP1 | F GGAGTTCATGACTGGGGAGA | 135 | NM_175817 | 0.99 | 1.22 | ± 12.7 |
| R CCCTTCGTCATGGTACTGGT | ||||||
| FADS2 | F CTGACTGGTGATGGACCTGA | 127 | NM_001083444 | 0.99 | 1.09 | ± 5.6 |
| R TCCCTATGGATCCAGTCTGC | ||||||
| GSTA4 | F TTCATGCCATAGGACAGCAG | 111 | BT020960 | 0.9997 | 1.02 | ± 4.3 |
| R ACGACAGAGCTGGATCACG |
r2, Linearity; E, efficiency; F, forward; R, reverse.
Real-time RT-PCR.
Real-time RT-PCR analysis was performed using the ABI 7500 Fast real-time PCR System (Applied Biosystems, Warrington, UK). Genes were chosen to be representative of those in the top three Ingenuity Pathways Analysis (IPA) networks and the more biologically interesting of the most up- and downregulated genes. The stability of the expression of several control or housekeeping genes, including GAPDH, β-actin, RPL-19, and 18S rRNA, across all samples in this study was investigated as part of a preliminary study. Resulting expression data were analyzed using GeNorm software package, and as statistically significant differences in expression of these genes existed between samples, it was concluded that none of these genes were suitable as a housekeeping genes and consequently relative real-time PCR could not be conducted. Thus, an absolute real-time RT-PCR methodology employing an exogenous reference control (i.e., a kanamycin resistance gene) to normalize real-time PCR results was performed to measure the expression of genes. The application of this method has previously been described (19, 23, 61, 83). Real-time RT-PCR assays were performed to detect expression of the exogenous kanamycin control in synthesized cDNA. External standards were run on the same plate in triplicate. Nontemplate controls were included on every plate for each gene product. To minimize variation, all samples included in each analysis were derived from the same cDNA batch and prepared under the same conditions, and samples were run in duplicate. All samples for a particular gene were assayed on the same plate, thereby eliminating intra-assay variation, and specificity of the reaction products was also confirmed by dissociation curve analysis and gel electrophoresis.
We amplified 1 μl of first-strand cDNA from standards and samples in a 20 μl volume using 10 μl Power SYBR Master Mix and 1 μl of 20 μM forward and reverse primer mix and 8 μl nuclease-free water (Promega). The real-time PCR temperature profile consisted of 15 min incubation at 95°C for SYBR Green activation. The cycling conditions consisted of 35 cycles of 30 s at 95°C, the optimal annealing temperature for each individual gene for 30 s, 30 s at 72°C for primer extension followed by an amplicon-specific melt curve analysis. A melting curve analysis was performed for each amplicon between 50 and 95°C to ensure the absence of nonspecific products such as primer dimers. Nonreverse transcribed total RNA was included as a control for the presence of genomic DNA contaminants. All samples were assayed on the same run, thereby eliminating interassay variation, and specificity of the reaction products was confirmed by melt curve analysis and gel electrophoresis.
Statistical analysis.
Blood metabolite and real-time RT-PCR data were analyzed using MIXED procedure of SAS (81). The type of variance-covariance structure used was chosen depending on the magnitude of the Akaike criterion (AIC) for models ran under compound symmetry, unstructured, autoregressive, or Toeplitz variance-covariance structures. The model with the lowest AIC was chosen. The PDIFF (predicted difference) and CONTRAST (for orthogonal contrasts) statements in SAS were used, and Tukey-Kramer test was used to examine pair-wise comparisons of treatment means. Real-time RT-PCR data were log-transformed prior to analysis to stabilize variances. Data are presented as back-transformed least square means and their confidence intervals.
The r2 and amplification efficiency (E) values for real-time RT-PCR were calculated from linear regression analysis of log (input cDNA) versus threshold cycle number (Ct) plot (Table 1). The slope for each set of standards was used to determine E = 10(−1/slope) − 1. Intra-assay variation was determined from the average standard deviation across the quantification range and presented as percentage using the formula: ± % variation = [(E + 1)SD − 1] × 100 (78).
Microarray hybridization.
Gene expression was determined using a 24,027 probe set bovine oligonucleotide array (Affymetrix), representing ∼23,000 bovine transcripts based on the original mapping using Unigene build 57 (March 24, 2004). RNA from each cow was hybridized to a separate array. All 11 RNA samples were hybridized and scanned by the German Resource Centre for Genomics Research, Germany (RZPD), according to the manufacturer's instructions.
Microarray analysis.
All microarray analysis including preprocessing, normalization, and statistical analysis was carried out using R (R, 2007) version 2.6 and Bioconductor (22) version 2.1, as described by Morris et al. (66). Data were quality assessed before and after normalization using a number of in-built quality control methods implemented in the Bioconductor affycoretools and associated packages to identify problems if they existed with array hybridization, RNA degradation, and data normalization. Microarray data were preprocessed using the mmgMOS normalization method (38, 72) using the default settings and differential expression (DE) was analyzed using the puma DE method both implemented in the Bioconductor package “puma” (53, 71, 72, 80). The puma method uses a Bayesian hierarchical model to calculate the probability of positive likelihood ratio (PPLR). The PPLR associates probability values of being differentially expressed, which is a measure of the false positive detection of differential expression to each ratio and generates lists of genes ranked by the probability of DE. This PPLR statistic was converted into “P-like values” using the recommended formula in the puma method prior to subsequent analysis. Differentially expressed genes (DEG) detected by the puma method were also compared with those detected by the Limma (84) and rank product (6) methods.
As many of the original annotations for the Affymetrix bovine chip are erroneous (12, 21), Affyprobeminer (52) redefines the chip definition files (CDFs) for Affymetrix chips, taking into account the most recent genomic sequence information. The remapped annotations were determined using the “bovineccdscdf” cdf annotation file downloaded from the Affyprobeminer website that returned Entrez Gene gene name identifiers (18).
This remapped annotation includes mappings to all RefSeq (mature RNA protein coding) transcripts, and validated complete coding sequences in GenBank Annotations were also supplemented by interrogating the Ensembl bos-taurus database version 46 using the biomaRt package in Bioconductor and manual annotation where possible with recent entries in Entrez Gene.
The Entrez Gene IDs of DEG were then submitted to DAVID (14, 34) to determine gene function and to cluster genes into functionally significant gene clusters and also to determine significant biological pathways through KEGG (39). The official gene names corresponding to the Entrez Gene of the remapped annotations was used as the “population” background gene list in DAVID as opposed to the default DAVID bos-taurus gene list. This was to ensure that the calculated EASE scores were not overly conservative, resulting in a failure to detected significant differences.
Pathway analysis.
To examine the molecular functions and genetic networks, the microarray data were further analyzed using IPA (v. 7.5, Ingenuity Systems, Mountain View, CA; http://www.ingenuity.com), a web-based software application that enables identification of biological mechanisms, pathways, and functions most relevant to experimental datasets or genes of interest (5, 56).
A dataset containing gene identifiers and corresponding expression and P-like values was uploaded into the application as described by Morris et al. (66). Briefly, each identifier was mapped to its corresponding gene object in the Ingenuity knowledge base. A P-like value of P < 0.05 from the puma analysis was set to identify genes whose expression was significantly differentially up- or downregulated. These genes, called “focus” genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity knowledge base. Networks of these focus genes were then algorithmically generated based on their connectivity. Network analysis returns a score that ranks networks according to their degree of relevance to the network eligible molecules in the dataset. The score takes into account the number of network eligible molecules in the network and its size, as well as the total number of network eligible molecules analyzed and the total number of molecules in the knowledge base that could potentially be included in networks.
RESULTS
EB model.
Average EB from day 2 postcalving to slaughter for MNEB and SNEB groups was = −1.7 ± 1.17 unité fourragère lait (UFL)/day and −6.1 ± 1.03 UFL/day respectively (P < 0.05) as previously described (93). Systemic concentrations of glucose (4.08 ± 0.15 vs. 2.66 ± 0.15 mmol/l) were higher (P < 0.001) in the MNEB cows, while BHB (3.70 ± 0.70 vs. 0.53 ± 0.09 mmol/l) (P < 0.001), NEFA (1.40 ± 0.14 vs. 0.34 ± 0.05 mmol/l) (P < 0.001), and hepatic TAG concentrations (116.0 ± 7.8 vs. 30.0 ± 11.8 mmol/g) (P < 0.05) were higher in the SNEB cows as reported by Fenwick et al. (19). Urea was not different (P = 0.17) between MNEB and SNEB (4.2 ± 0.46 vs. 5.1 ± 0.31 mmol/l) groups (21).
Gene expression.
From a total of 5,739 Affyprobeminer annotated genes, 5,229 genes were mapped to the IPA database. However, a cut-off P-like value of P < 0.05 resulted in a total of 416 differentially expressed genes (189 up- and 227 downregulated) using the puma method. IPA was used to place the DEG into different function and disease categories. Of the 416 DEG, 341 genes were network eligible with 298 genes being function/pathway or list eligible. Among the network-eligible genes, 154 were upregulated (Supplementary Table S1), while 187 genes were downregulated (Supplementary Table S2).1 A comparison between the puma method and the limma and rank product methods revealed that the puma method included 89% of DEG detected by the Limma method and 60% of the DEG detected by the rank product method with a total of 41 DEG common to all three methods. Overall, the puma method detected the greatest number of DEG.
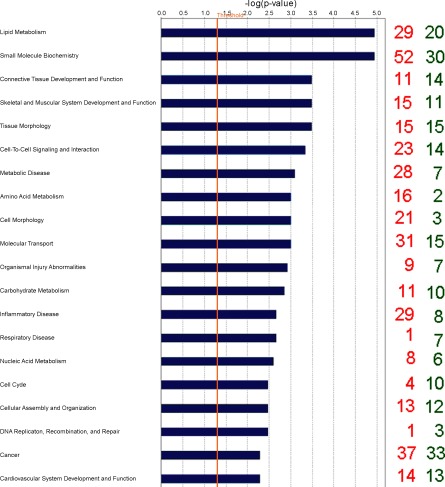
Biological categories with the largest number of downregulated genes were cell signaling, cardiovascular development and function, organ morphology, inflammatory disease, and respiratory disease, while genes associated with cell morphology, amino acid metabolism, small molecule biochemistry, lipid metabolism, and cell cycle were the categories containing the greatest number of genes upregulated by SNEB according to IPA terminology. Gene classification according to canonical signaling pathways revealed that some of the nuclear hormone receptor superfamily including liver X receptors (LXRs) and the retinoid X receptor (RXR) as well as amino acid metabolism were the pathways most affected by EB status (Table 2). The top 20 molecular and cellular functions most significantly affected by EB are shown in Fig. 1.
Table 2.
Gene classification according to canonical signaling pathways using IPA
| Pathway | P Value | %DEG | Genes |
|---|---|---|---|
| Pyruvate metabolism | 0.00018 | 6.2 | ACACA, AKR1A1, ALDH1A1, PKLR, HADHA, HADHB, LDHA, LDHB, PC |
| LXR/RXR activation | 0.00020 | 11.7 | ABCG8, ACACA, CD14, APOA1, ARG2, CCL2, CD36, IL1A, IL1RN, RXRG |
| Fatty acid metabolism | 0.00092 | 5.3 | CPT1B, ACADSB, HADHA, ACADVL, SLC27A4, CPT2, HADHB, ALDH1A1, CYP2E1, HSD17B4 |
| FXR/RXR activation | 0.00098 | 9.0 | ABCG8, APOA1, FOXA3, IL1A, IL1RN, LIPC, OSTALPHA, PKLR, SCARB1 |
| Propanoate metabolism | 0.0019 | 6.4 | ACACA, ACADSB, ACADVL, ALDH1A1, HADHA, HADHB, LDHA, LDHB, |
| Arginine and proline metabolism | 0.0036 | 5.0 | ALDH1A1, ARG1, ARG2, GAMT, OAT, ODC1, OTC, P4HA2, SAT1, |
| Urea cycle and metabolism of amino groups | 0.0037 | 7.5 | OTC, GAMT, OAT, ARG2, ODC1, ARG1 |
| Valine, leucine and isoleucine degradation | 0.0049 | 7.4 | HADHB, ALDH1A1, ACADVL, HIBADH, ACADSB, AOX1, HSD17B4, HADHA |
| β-Alanine metabolism | 0.0074 | 6.1 | HADHB, ALDH1A1, DPYD, ACADVL, ACADSB, HADHA |
| Aryl hydrocarbon receptor signaling | 0.0078 | 6.4 | TGM2, NR2F1, RXRG, FOS, RB1, IL1A, ALDH1A1, NQO2, NFIA, GSTM4 |
| LPS/IL-1-mediated inhibition of RXR function | 0.0107 | 5.5 | ABCG8, ALDH1A1, LIPC, SCARB1, CPT1B, CPT2, GSTM4, FABP1, CD14, CES2 (includes EG:8824), SLC27A4 |
| Tryptophan metabolism | 0.0178 | 3.4 | HADHB, AADAT, ALDH1A1, CYP2E1, DDC, AOX1, HSD17B4, HADHA |
| Fatty acid biosynthesis | 0.0282 | 4.0 | OXSM, ACACA |
| IL-10 signaling | 0.0282 | 8.4 | FOS, IL1A, IL1RN, CD14, ARG2, CHUK |
| Glutamate receptor signaling | 0.0302 | 3.0 | SLC1A4, GNG11, SLC17A2 |
| Glutathione metabolism | 0.0355 | 5.1 | GPX3, GSTM4, GCLM, IDH3A, IDH1 |
| Glycolysis/gluconeogenesis | 0.0372 | 4.2 | AKR1A1, ALDH1A1, PKLR, GALM, LDHB, LDHA |
| Fatty acid elongation in mitochondria | 0.0398 | 4.6 | HADHB, HSD17B4, HADHA |
| Hepatic fibrosis/hepatic stellate cell activation | 0.0498 | 6.6 | SMAD2, VCAM1, IL1A, CTGF, CCL2, CYP2E1, IGFBP3, CD14, AGTR1 |
The %DEG is the proportion of differentially expressed genes (DEG) relative to the total no. of genes in the specific canonical pathway. Downregulated genes are highlighted in boldface, upregulated genes are in lightface. IPA, Ingenuity Pathways Analysis.
Fig. 1.
Classification of differentially expressed genes (DEG) according to top 20 molecular and cellular functions, most significantly affected by energy balance (EB) using Ingenuity Pathways Analysis. The blue bars indicate the likelihood [−log (P-value)] that the specific molecular and cellular function category was affected by negative energy balance compared with others represented in the list of DEG. The number of up- and downregulated genes in each group is represented on the righthand side by red and green numbers, respectively. The cut-off (yellow line) is shown at P < 0.05 (1.301 log scale).
The most dramatically upregulated gene in SNEB animals was d-aspartate oxidase (DDO) with a fold-change of 6.1 (P < 0.05); carnitine palmitoyltransferase 1B (CPT1B) also appeared in the top five with a 5.5-fold higher abundance in SNEB cows, as did angiopoietin-like 4 (ANGPTL4) (5.5-fold upregulated) and retinoid X receptor gamma (RXRG) (4.9-fold upregulated) (P < 0.001). There was also increased expression of other genes involved in lipid transport, apolipoprotein A1 (ApoA1) (P < 0.05), and lipid oxidation, very long chain acyl-Coenzyme A dehydrogenase (ACADVL) (P < 0.001). The gene that displayed the most dramatic downregulation was fatty acid desaturase 2 (FADS2) (19.6-fold decrease) (P < 0.001), and the insulin-like growth factor binding protein acid labile subunit (IGFBPALS) was also in the top five most downregulated genes (6.4-fold decrease) (P < 0.001). Twenty genes were validated by real-time PCR, and a high level of consistency was observed between microarray and real-time PCR in terms of both direction of fold-change and magnitude (Table 3). Of the 41 genes found to be differentially expressed by all three methods of statistical analysis, almost 27% (11 genes) have been validated by real-time RT-PCR. Of these 11 genes, all except one gene (ANGPTL4, nonsignificant) were in agreement with array data in terms of fold direction and significance.
Table 3.
Microarray validation with real-time RT-PCR on selected genes
| Real-time Data |
||||||
|---|---|---|---|---|---|---|
| Gene | MNEB | SNEB | Real-time Fold-change | P Value | Array Fold change | P Value |
| SAA1 | 253 (70–908) | 787 (245–2527) | +3.1 | 0.17 | +1.7 | 0.2 |
| GPX3 | 40.9 (16–102) | 437 (191–1000) | +10.7 | <0.01 | +3.7 | <0.001 |
| FOXA3 | 4.53 (2.07–9.9) | 1.91 (0.93–3.9) | −2.4 | 0.10 | −2.0 | <0.001 |
| ANGPTL4 | 0.176 (0.151–0.205) | 0.199 (0.174–0.228) | +1.13 | 0.25 | +5.4 | <0.001 |
| ADIPOR2 | 0.833 (0.594–1.167) | 1.092 (0.803–1.485) | +1.3 | 0.21 | +1.4 | <0.05 |
| CD36 | 0.05 (0.04–0.07) | 0.12 (0.08–0.16) | +2.4 | <0.01 | +1.8 | <0.001 |
| CLDN1 | 1.53 (1.12–2.09) | 1.33 (1.0–1.77) | −1.2 | 0.48 | −1.3 | 0.32 |
| CPT1B | 0.017 (0.01–0.028) | 0.084 (0.052–0.134) | +4.9 | <0.001 | +5.5 | <0.001 |
| CPT1A | 0.203 (0.034–0.373) | 0.694 (0.539–0.849) | +3.4 | <0.01 | nd | nd |
| HP | 1.37 (0.26–7.35) | 5.32 (1.15–2.46) | +3.9 | 0.21 | +1.0 | 0.82 |
| IL8 | 0.001 (0.001–0.002) | 0.001(0.001–0.002) | 0.79 | +1.1 | 0.43 | |
| TNFα | 0.025 (0.017–0.032) | 0.015 (0.008–0.022) | −1.7 | 0.09 | −1.1 | 0.59 |
| ACADVL | 2.2 (1.774–2.777) | 3.3 (2.715–4.084) | +1.5 | <0.05 | +1.4 | <0.001 |
| PPARA | 0.128 (0.102–0.161) | 0.116 (0.092–0.146) | +1.1 | 0.52 | +1.0 | 0.9 |
| PPARG | 0.006 (0.004–.009) | 0.005 (0.003–.007) | −1.2 | 0.2 | +1.0 | 0.96 |
| PPARD | 0.028 (0.022–0.037) | 0.031 (0.024–.041) | +1.1 | 0.6 | +1.0 | 0.93 |
| RXRγ | 0.073 (0.0–0.292) | 0.514 (0.318–0.71) | +7.04 | <0.05 | +4.9 | <0.001 |
| HSP90 | 13.1 (9.8–17.5) | 10.8 (8.3–14.1) | −1.2 | 0.32 | −1.04 | 0.69 |
| FABP1 | 11.19 (7.94–15.76) | 6.31 (4.64–8.59) | −1.8 | <0.05 | −1.3 | <0.01 |
| FADS2 | 0.236 (0.139–0.398) | 0.027 (0.016–0.043) | −8.74 | <0.001 | −19.6 | <0.001 |
| GSTA4 | 0.319 (0.181–0.559) | 0.2 (0.12–0.334) | −1.6 | 0.2 | −1.2 | 0.27 |
Values are back-transformed least square means followed by the 95% confidence limits and are expressed as pg per μg of reversed transcribed RNA. nd, Not determined; MNEB, mild negative energy balance; SNEB, severe negative energy balance.
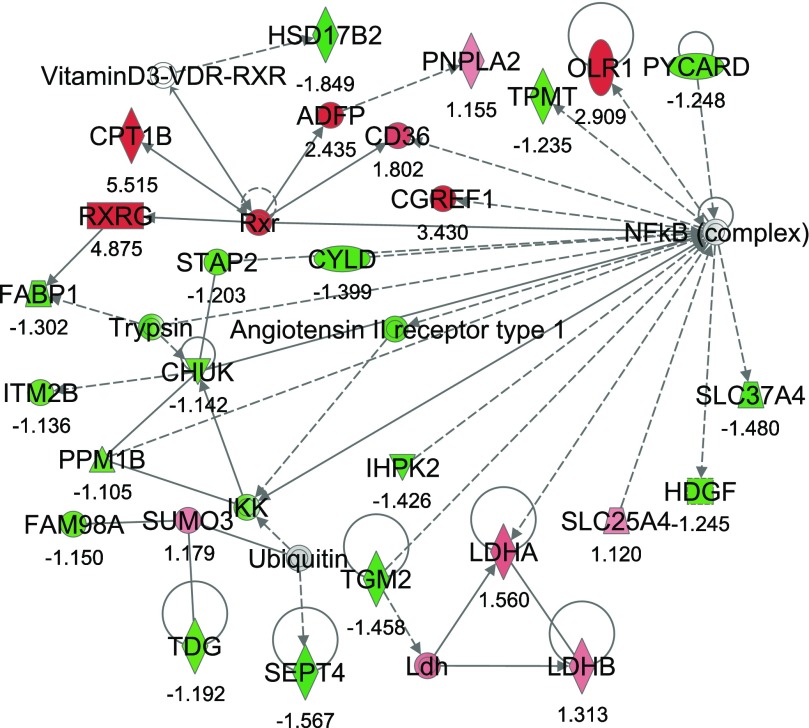
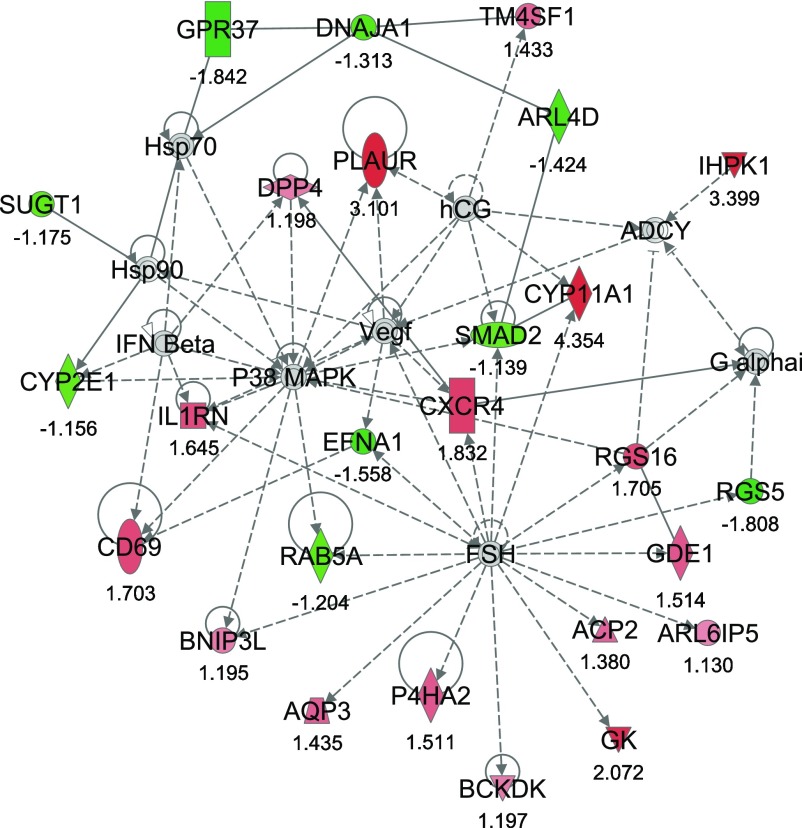
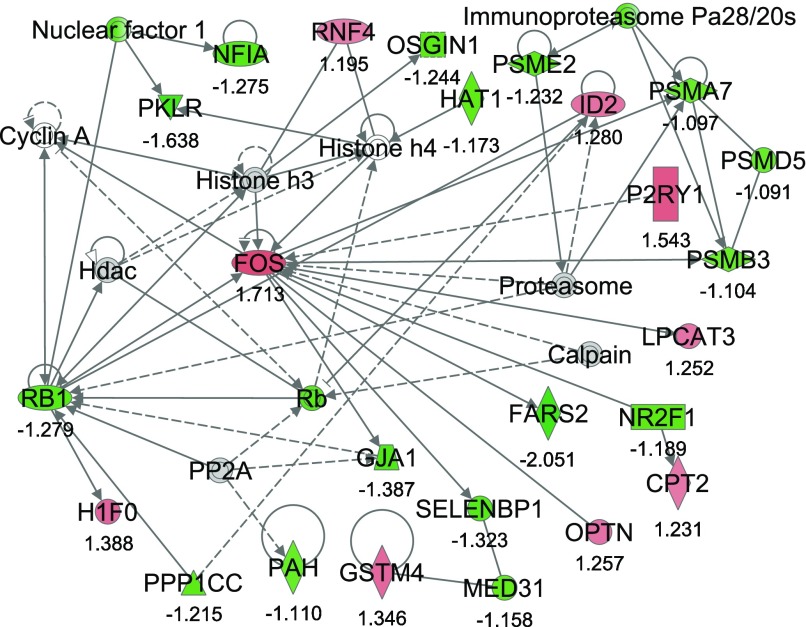
From IPA, 36 networks (P < 0.05) were identified with 20 networks each having 10 or more focus genes among the DEG. The three top networks identified were lipid metabolism, molecular transport small molecule biochemistry (network #1, score 43, 27 focus molecules) (Fig. 2); lipid metabolism, small molecule biochemistry, cell cycle (network #2, score 40, 26 focus molecules) (Fig. 3); and gene expression, cellular compromise, DNA replication, recombination, and repair (network #3, score 38, 25 focus molecules) (Fig. 4).
Fig. 2.
Network #1: lipid metabolism, molecular transport, small molecule biochemistry. The network is displayed graphically as nodes (genes). The node color intensity indicates the expression of genes, with red representing upregulation and green downregulation in severe negative energy balance (SNEB) versus mild negative energy balance (MNEB) liver. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product, and the lines indicate the type of interaction (Supplementary Fig. S1).
Fig. 3.
Network #2: lipid metabolism, small molecule biochemistry, cell cycle. The network is displayed graphically as nodes (genes). The node color intensity indicates the expression of genes, with red representing upregulation and green downregulation in SNEB versus MNEB liver. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction (Supplementary Fig. S1).
Fig. 4.
Network #3: gene expression, cellular compromise, DNA replication, recombination, and repair. The network is displayed graphically as nodes (genes). The node color intensity indicates the expression of genes, with red representing upregulation and green downregulation in SNEB versus MNEB liver. The fold value is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction (Supplementary Fig. S1).
Network #1 included 11 upregulated and 16 downregulated genes in SNEB cows. The upregulated genes included CPT1B, CD36 antigen (CD36), and RXRG. Also upregulated were the patatin-like phospholipase domain containing 2 (PNPLA) gene, the cell growth regulator with EF-hand domain 1 (CGREF1), and lactate dehydrogenase A and B (LDHA/B). Downregulated genes in SNEB cows include fatty acid binding protein 1, liver (FABP1), septin 4 (SEPT4), transglutaminase 2 (TGM2), hydroxysteroid (17-beta) dehydrogenase 2 (HSD17B2), thymine-DNA glycosylase (TDG), thiopurine S-methyltransferase (TPMP), conserved helix-loop-helix ubiquitous kinase (CHUK), and inositol hexakisphosphate kinase 2 (IHPK2).
The second most significant network, network #2, encompassed 17 upregulated and 9 downregulated genes. The upregulated genes in this network include glycerol kinase (GK), branched chain ketoacid dehydrogenase kinase (BCKDK), CD69 molecule (CD69), plasminogen activator, urokinase receptor (PLAUR), and interleukin 1 receptor antagonist (IL1RN). Chemokine (C-X-C motif) receptor 4 (CXCR4) was also observed to have increased expression in SNEB cows as was the cell surface antigen transmembrane 4 L six family member 1 (TM4SF1) and regulator of G-protein signaling 16 (RGS16). Genes downregulated by SNEB in this network include ADP-ribosylation factor-like 4D (ARL4D), RAB5A member RAS oncogene family, SMAD family member 2 (SMAD2), and SGT1 suppressor of G2 allele of SKP1 (SUGT1).
Network #3 included 9 upregulated and 16 downregulated genes. Genes upregulated include carnitine palmitoyl transferase (CPT2); v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS); glutathione S-transferase mu 4 (GSTM4); H1 histone family, member 0 (H1F0); inhibitor of DNA binding 2 (ID2); lysophosphatidylcholine acyltransferase 3 (LPCAT3); optineurin (OPTN); and ring finger protein 4, (RNF4). Some genes downregulated include phenylalanyl-tRNA synthetase 2, mitochondrial (FARS2), junction protein, alpha 1 (GJA1), nuclear factor I/A (NFIA), histone acetyltransferase 1 (HAT1), selenium binding protein 1 (SELENBP1), and retinoblastoma 1 (RB1).
DISCUSSION
As previously discussed the animal model used in this study generated two groups of high-yielding dairy cows divergent in EB status (19, 54). Given the well-documented disparity between feed intake capacity and lactational energy output in the early postpartum period, dairy cows typically experience some degree of NEB. Our aim, therefore, in generating our control group of cows was to establish an EB status as close to parity as was practical within an early postparturient context. Additionally, all animals selected exhibited no detectable signs of clinical conditions such as lameness, hypocalcaemia, ketosis, endometritis, or mastitis, typical of the postparturient period and that might confound the interpretation of the hepatic gene expression profiles generated. Furthermore, expression of genes encoding IL8, Hp, TNFa were not different among treatment lending support to these observations.
Previous studies conducted on liver tissues harvested from this animal model focused on specific adaptations and effects of SNEB on expression of selected genes critical to metabolism (19). However, it is known that biological change is more likely due to coordinated small changes in a large set of genes, and recent studies by Morris et al. (66) and Wathes et al. (93) have examined global affects of NEB on spleen and uterine tissues from this animal model, respectively, using the Affymetrix 23k bovine microarray. In this study we chose to examine the influence of an SNEB condition on gene expression profile in the liver, due to its critical importance to animal metabolism and wellbeing. In contrast to the study conducted by Loor et al. (56), which examined the effects of nutrition-induced ketosis on gene networks in dairy cows, the study reported here uses differential feed intake in conjunction with different milking frequencies to induce NEB in high-yielding dairy cows. This study, for the first time, also discusses SNEB and the consequent alterations in the expression of genes and associated pathways controlling cell proliferation, cell cycle, cell signaling, and tissue repair.
The nature of the DEG indicates that SNEB affects expression of genes encoding proteins and enzymes involved in a broad range of biological functions. Cows in SNEB displayed increased expression of genes involved in lipid transport and catabolism. It has been previously documented that SNEB specifically attenuates expression of IGF-1, IGFBP-3 to IGFBP-6, and IGFALS mRNA, while increasing IGFBP-2 mRNA synthesis (19). In the current study, IGFALS was listed among the top five most downregulated genes by SNEB. In agreement with this finding, expression of this gene was decreased by >14-fold when examined by real-time RT-PCR (19). Although IGF-1 expression remained unaffected on the array, in agreement with the observations of Loor et al. (56), genes encoding the binding proteins IGFBP3 and IGFALS were downregulated, while IGFBP2 was increased in SNEB cows. These results are also consistent with real-time PCR analysis of hepatic tissue from these animals (19).
DDO was the most upregulated gene in SNEB animals. The protein it encodes has been previously detected in the peroxisomes of bovine kidney and liver (96) and catalyses the oxidation of d-aspartate to yield oxaloacetate, ammonia, and hydrogen peroxide. As oxaloacetate deficiency is likely to be limiting the citric acid cycle in SNEB animals, the activation of this gene may provide an alternative mechanism of oxaloacetate production for energy generation during this time period. The FADS2 gene was found to be most downregulated on the microarray. This gene encodes the enzyme responsible for controlling the synthesis of long chain polyunsaturated fatty acids (PUFA) (70); however, a link between SNEB and depressed PUFA synthesis has not been reported thus far.
Also amongst the five most upregulated genes were CPT1B and RXRG. On examination of gene networks, it was observed that network #1 centered on a well-established response to NEB, lipolysis. Expression of CPT1B and RXRG genes was validated here by real-time RT-PCR and is critical to the lipid metabolic process and fatty acid beta-oxidation (7, 30). As CPT1B is involved in transferring long-chain fatty acids from the cytosol to the mitochondrial matrix to undergo beta-oxidation (7, 63) this finding was not surprising. Although the expression of this gene has previously been reported as being muscle specific (3), real-time RT-PCR employed in the current study validated the detection of transcript expression of CPT1B in the liver and confirmed that it is upregulated by SNEB. Expression of the liver-specific CTP1A, which is not represented on the microarray, was also measured using real-time PCR and found to also have increased expression in SNEB animals. This would suggest, for the first time, that at least two forms of CPT are expressed in the liver. However, absolute measurement of the mRNA transcript of the CPT1A gene indicated that its expression was at least 10-fold higher than that of CPT1B, establishing it as the predominant variant. The validation of RXRG was a particularly good indicator of the accuracy of both platforms of expression measurement, as it was found that other isoforms of this transcriptional regulator were not differentially expressed, when examined with both methods.
The PNPLA gene encodes a protein that possesses TAG lipase activity (37). Expression of this gene was upregulated by SNEB, and increased mRNA levels have similarly been observed in fasting mice (92). In addition, LDHA/B genes are involved in the carbohydrate metabolic process, and it has been documented that liver damage results in elevated expression of LDH (40, 48), possibly indicating a clinical sign of SNEB in cows. Microarray results indicated that FABP1 gene expression was reduced in SNEB cows. Similarly, the FABP1 gene was decreased in SNEB cows when analyzed by real time RT-PCR. Bionaz et al. (4) reported reductions in bovine hepatic expression of FABP1 in dairy cows during the early postpartum period. While more recently, Kuhla et al. (45) reported a dramatic downregulation of FABP1 in dairy cows that underwent feed restriction and suggested that this may be an attempt to limit fatty acid oxidation and hepatic TAG accumulation associated with NEB. Expression of IHPK2, which may act as an energy reserve protein (79), was also downregulated in SNEB cows.
Furthermore, within IPA network #1 there appeared to be a general downregulation of genes involved in cell cycle and tissue growth and repair. The increased expression of the cell growth regulator CGREF1 may be in response to the stress that SNEB cows are likely to endure. Coincidentally, it is known that the EF-hand domain of the CGR11 protein is essential for growth inhibitory activity by promoting cell cycle arrest and negative regulation of cellular proliferation (16). The septins are polymerizing scaffold proteins involved in cytoskeletal organization in mitosis, exocytosis, and other cellular processes (42). SEPT4 knockout studies have recently reported that the gene supports suppressive modulation of processes associated with liver diseases (36). The observed reduction in SEPT4 expression in SNEB cows may be a contributing factor in what appears to be a trend toward compromised liver function. Given that Piacentini et al. (74) reported the proliferative phase of hepatocytes to be accompanied by a 10-fold increase in TGM2 mRNA levels, we suggest that the reduction in expression observed here is indicative of reduced capability of liver to recover from SNEB at this time. However, increased levels of expression of this gene are also associated with apoptotic function (60), and reduced expression may signify deregulated cell growth. Likewise, the HSD17B2 gene is involved in estrogen metabolism. However, reduced levels are associated with unregulated cell cycle (28, 29), suggesting that the reduction observed here may have a negative impact on normal liver processes.
Severe NEB also resulted in reduced expression of TDG, which codes for a protein involved in DNA repair (89). TPMT, which catalyses the S-methylation of aromatic and heterocyclic sulfhydryl compounds including the thiopurine drugs (94), also displayed reduced expression in SNEB cows. Considering that in human studies, patients expressing low levels of this gene require dose adjustments when receiving thiopurine drugs to avoid severe toxicity (75), a reduction in the expression of this gene in periparturient cows might suggest suboptimal health status. The CHUK gene is responsible for activation of the transcriptional regulator NF-κB and is therefore critical to this signaling pathway (64). Also, it has been documented that NF-κB is an important transcription factor complex involved in almost every aspect of cell regulation including initiation of immune responses and cellular proliferation (87). NF-κB deregulation is associated with a deviation from normal cell growth (91). Therefore, perhaps SNEB cows are subject to impaired hepatic cell growth and repair during the early postpartum period when CHUK expression is depressed.
In the second most significant network the GK gene is upregulated by SNEB. GK converts glycerol and ATP to glycerol-3-phosphate and ADP (13). This intermediate step is more than likely directed toward glycolysis rather than gluconeogenesis in SNEB cows attempting to deal with a drastic energy deficit. The CD69 gene is usually induced on activation of T lymphocytes and functions as a signal-transmitting receptor in lymphocytes, natural killer cells, and platelets (9). Therefore, increased expression of the CD69 gene may suggest an increased immune response in SNEB cows; however, upregulation of this gene alone is not sufficient to signify alterations in the immune system particularly given that a companion study examining the influence of SNEB on splenic gene expression by Morris et al. (66) reported evidence of a depressed immune system in these cows. The cell surface receptor PLAUR, which is involved in proteolytic degradation by converting plasminogen to the active form plasmin, was also increased in the more metabolically challenged cows, and the recent studies by Morris et al. (66) and Wathes et al. (93) reported similar alterations in expression of this gene in the spleen and uterine tissues, respectively, of SNEB cows. Again, the link between increased abnormal or unexpected gene expression and SNEB is apparent, as Foca et al. (20) previously reported that PLAUR mRNA levels were positively correlated with the invasive potential of endometrial carcinomas. Similarly, the IL1RN gene that inhibits the activities of interleukin 1 alpha (IL1A) and interleukin 1 beta (IL1B) was increased. As a consequence, these cytokines are neutralized in physiological immune and inflammatory responses (1, 10), an event that is consistent with both the reduced expression of CHUK and NF-κB genes associated with IL regulation (59) and the findings of a depressed immune response in SNEB cows reported by Morris et al. (66). Muller et al. (67) found the CXCR4 gene to be highly expressed in cells with abnormal cell cycle but is undetectable in normal tissue. Upregulation of CXCR4, in this study, is indicative of poor animal health or reduced hepatic performance during periods of SNEB. Similarly, expression of the cell surface antigen TM4SF1 was increased in SNEB cows and is known to be highly expressed in different carcinomas (82).
The RGS16 gene, which negatively influences G protein signaling (62), was increased in SNEB cows, thus interrupting normal signal transduction in these animals. Furthermore, SNEB downregulation of ARL4D, thought to be involved in small GTPase-mediated signal transduction and protein secretion (33, 50), and RAB5A member, which is involved in signal transduction, endocytosis, and mitogenesis (65), is concomitant with the large-scale decrease in signaling events. Likewise SMAD proteins mediate TGF-β signaling to regulate cell growth and differentiation (31), however, SMAD2 being downregulated in the SNEB cows may not fulfill its role in the intracellular signaling cascade and positive regulation of transcription (35) to the same extent as in MNEB cows. Notably, another gene downregulated was SUGT1, which is required for the G1/S and G2/M transitions of the cell cycle (43). Hence, this expression pattern is likely to have a negative impact on tissue growth and repair mechanisms. All of these downregulated genes provide evidence that cell growth and proliferation are compromised in SNEB cows.
From analysis of IPA network #3, the common theme in this study is again evident. SNEB cows appear to experience decreased cell signaling, tissue growth, and repair processes, while they contend with an apparent increase in expression of genes associated with abnormal cell cycle or cellular proliferation. The FOS gene encoding an oncoprotein that is involved in promoting the transcription of genes containing AP-1 regulatory elements (2) displayed increased expression in this network. Interestingly FOS is also associated with liver regeneration as expression is induced after partial hepatectomy in mice (85) and may be involved in the cellular response to ROS. Similarly upregulated was GSTM4, a gene important for detoxification of physiological products of oxidative stress (15), which are likely to be present in the SNEB cows (66).
The ID2 gene has proliferative effects and is increased in SNEB cows; however, the gene is also known to affect the cell cycle as it disrupts regulator proteins (26, 46, 47). The upregulation of this gene provides further evidence that any increased expression of growth-promoting genes in SNEB cows appears to be toward those with possible deleterious effects on cow health. The RNF4 gene, which was upregulated in SNEB cows, possesses both growth-inhibiting (73) and growth-promoting properties (69); therefore, it is difficult to assess what its role may be in terms of differential EB status in the cow. FARS2, involved in the processes of translation and tRNA processing (8), was downregulated in SNEB cows, as was GJA1, which is involved in cell-to-cell adhesion and direct intercellular communication and usually increased by the cell cycle (49). Likewise, NFIA, involved in DNA replication and transcriptional regulation (51, 57), and SELENBP1, which acts as a transporter protein, were also downregulated. In mouse models it has been reported that expression of the SELENBP1 gene is reduced in response to PPAR agonists that are known to be in circulation in SNEB cows and are therefore more than likely responsible for SELENBP1 downregulation in these cows. Such downregulation has been reported to enhance cellular proliferation, but this is again associated with some level of carcinogenicity (24). Concomitant with this we also observed reduced expression of RB1, which encodes a tumor suppressor protein (95).
In conclusion, from a global examination of hepatic gene expression in cows with differing EB status, we see that SNEB increases expression of genes associated with lipid catabolism and appears to have a negative or inhibitory impact on cell growth and repair. In addition, there is an apparent decrease in DNA replication, with a tendency toward abnormal or unregulated cell cycle progression in SNEB cows. Taken together there appears to be strong links between the increased circulation of metabolic by-products (PPAR agonists), decreased normal functional activity, and increased susceptibility to abnormal gene expression in hepatic tissue of dairy cows experiencing SNEB.
GRANTS
This work was funded by the Wellcome Trust and the Irish National Development Plan.
DISCLOSURES
The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank staff at Teagasc Moorepark for the management and care of the cows used in this study.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Arend WP. Interleukin 1 receptor antagonist. A new member of the interleukin 1 family. J Clin Invest 88: 1445–1451, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 283: 387–390, 1999. [DOI] [PubMed] [Google Scholar]
- 3. Bernard C, Cassar-Malek I, Le Cunff M, Dubroeucq H, Renand G, Hocquette JF. New indicators of beef sensory quality revealed by expression of specific genes. J Agric Food Chem 55: 5229–5237, 2007. [DOI] [PubMed] [Google Scholar]
- 4. Bionaz M, Drackley JK, Dann HM, Loor JJ. Liver fatty acid binding protein (FABP) and acyl-CoA synthase (ACSL) isoform gene expression due to plane of dietary energy prepartum in dairy cows. J Dairy Sci 90, Suppl 1: 972, 2007. [Google Scholar]
- 5. Bionaz M, Loor JJ. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol Genomics 29: 312–319, 2007. [DOI] [PubMed] [Google Scholar]
- 6. Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92, 2004. [DOI] [PubMed] [Google Scholar]
- 7. Britton CH, Schultz RA, Zhang B, Esser V, Foster DW, McGarry JD. Human liver mitochondrial carnitine palmitoyltransferase I: characterization of its cDNA and chromosomal localization and partial analysis of the gene. Proc Natl Acad Sci USA 92: 1984–1988, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullard JM, Cai YC, Demeler B, Spremulli LL. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol 288: 567–577, 1999. [DOI] [PubMed] [Google Scholar]
- 9. Cambiaggi C, Scupoli MT, Cestari T, Gerosa F, Carra G, Tridente G, Accolla RS. Constitutive expression of CD69 in interspecies T-cell hybrids and locus assignment to human chromosome 12. Immunogenetics 36: 117–120, 1992. [DOI] [PubMed] [Google Scholar]
- 10. Carter DB, Deibel MR, Jr, Dunn CJ, Tomich CS, Laborde AL, Slightom JL, Berger AE, Bienkowski MJ, Sun FF, Harris PKW, Yem AW, Waszak GA, Chosay JG, Sieu LC, Hardee MM, Zurcher-Neely HA, Reardon IM, Heinrikson RL, Truesdell SE, Shelly JA, Eessalu TE, Taylor BM, Tracey DE. Purification, cloning, expression and biological characterization of an interleukin-1 receptor antagonist protein. Nature 344: 633–638, 1990. [DOI] [PubMed] [Google Scholar]
- 11. Connor EE, Kahl S, Elsasser TH, Parker JS, Li RW, Van Tassell CP, Baldwin RLt, Barao SM. Enhanced mitochondrial complex gene function and reduced liver size may mediate improved feed efficiency of beef cattle during compensatory growth. Funct Integr Genomics 10: 39–51, 2010. [DOI] [PubMed] [Google Scholar]
- 12. Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeFrain JM, Hippen AR, Kalscheur KF, Jardon PW. Feeding glycerol to transition dairy cows: effects on blood metabolites and lactation performance. J Dairy Sci 87: 4195–4206, 2004. [DOI] [PubMed] [Google Scholar]
- 14. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3, 2003. [PubMed] [Google Scholar]
- 15. Denson J, Xi Z, Wu Y, Yang W, Neale G, Zhang J. Screening for inter-individual splicing differences in human GSTM4 and the discovery of a single nucleotide substitution related to the tandem skipping of two exons. Gene 379: 148–155, 2006. [DOI] [PubMed] [Google Scholar]
- 16. Devnath S, Kataoka T, Miura K, Kusuda M, Kitamura K, Kumada Y, Mochiduki A, Kaneko K, Adachi A, Inoue K. Cgr11 encodes a secretory protein involved in cell adhesion. Eur J Cell Biol 88: 521–529, 2009. [DOI] [PubMed] [Google Scholar]
- 17. Drackley JK. ADSA Foundation Scholar Award. Biology of dairy cows during the transition period: the final frontier? J Dairy Sci 82: 2259–2273, 1999. [DOI] [PubMed] [Google Scholar]
- 18. Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res 34: D319–D321, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fenwick MA, Fitzpatrick R, Kenny DA, Diskin MG, Patton J, Murphy JJ, Wathes DC. Interrelationships between negative energy balance (NEB) and IGF regulation in liver of lactating dairy cows. Domest Anim Endocrinol 34: 31–44, 2008. [DOI] [PubMed] [Google Scholar]
- 20. Foca C, Moses EK, Quinn MA, Rice GE. Differential mRNA expression of urokinase-type plasminogen activator, plasminogen activator receptor and plasminogen activator inhibitor type-2 in normal human endometria and endometrial carcinomas. Gynecol Oncol 79: 244–250, 2000. [DOI] [PubMed] [Google Scholar]
- 21. Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315, 2004. [DOI] [PubMed] [Google Scholar]
- 22. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilsbach R, Kouta M, Bonisch H, Bruss M. Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. Biotechniques 40: 173–177, 2006. [DOI] [PubMed] [Google Scholar]
- 24. Giometti CS, Liang X, Tollaksen SL, Wall DB, Lubman DM, Subbarao V, Rao MS. Mouse liver selenium-binding protein decreased in abundance by peroxisome proliferators. Electrophoresis 21: 2162–2169, 2000. [DOI] [PubMed] [Google Scholar]
- 25. Goff JP, Horst RL. Physiological changes at parturition and their relationship to metabolic disorders. J Dairy Sci 80: 1260–1268, 1997. [DOI] [PubMed] [Google Scholar]
- 26. Gray MJ, Dallas NA, Van Buren G, Xia L, Yang AD, Somcio RJ, Gaur P, Mangala LS, Vivas-Mejia PE, Fan F, Sanguino AM, Gallick GE, Lopez-Berestein G, Sood AK, Ellis LM. Therapeutic targeting of Id2 reduces growth of human colorectal carcinoma in the murine liver. Oncogene 27: 7192–7200, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grummer RR. Etiology of lipid-related metabolic disorders in periparturient dairy cows. J Dairy Sci 76: 3882–3896, 1993. [DOI] [PubMed] [Google Scholar]
- 28. Gunnarsson C, Hellqvist E, Stal O. 17beta-Hydroxysteroid dehydrogenases involved in local oestrogen synthesis have prognostic significance in breast cancer. Br J Cancer 92: 547–552, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gunnarsson C, Olsson BM, Stal O. Abnormal expression of 17beta-hydroxysteroid dehydrogenases in breast cancer predicts late recurrence. Cancer Res 61: 8448–8451, 2001. [PubMed] [Google Scholar]
- 30. Haugen BR, Jensen DR, Sharma V, Pulawa LK, Hays WR, Krezel W, Chambon P, Eckel RH. Retinoid X receptor gamma-deficient mice have increased skeletal muscle lipoprotein lipase activity and less weight gain when fed a high-fat diet. Endocrinology 145: 3679–3685, 2004. [DOI] [PubMed] [Google Scholar]
- 31. Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390: 465–471, 1997. [DOI] [PubMed] [Google Scholar]
- 32. Herd RM, Arthur PF. Physiological basis for residual feed intake. J Anim Sci 87: E64–71, 2009. [DOI] [PubMed] [Google Scholar]
- 33. Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr Biol 17: 711–716, 2007. [DOI] [PubMed] [Google Scholar]
- 34. Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell 10: 283–294, 2002. [DOI] [PubMed] [Google Scholar]
- 36. Iwaisako K, Hatano E, Taura K, Nakajima A, Tada M, Seo S, Tamaki N, Sato F, Ikai I, Uemoto S, Kinoshita M. Loss of Sept4 exacerbates liver fibrosis through the dysregulation of hepatic stellate cells. J Hepatol 49: 768–778, 2008. [DOI] [PubMed] [Google Scholar]
- 37. Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 279: 48968–48975, 2004. [DOI] [PubMed] [Google Scholar]
- 38. Kadota K, Nakai Y, Shimizu K. Ranking differentially expressed genes from Affymetrix gene expression data: methods with reproducibility, sensitivity, and specificity. Algorithms Mol Biol 4: 7, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanno T, Sudo K, Takeuchi I, Kanda S, Honda N, Nishimura Y, Oyama K. Hereditary deficiency of lactate dehydrogenase M-subunit. Clin Chim Acta 108: 267–276, 1980. [DOI] [PubMed] [Google Scholar]
- 41. Kelly JM, Summers M, Park HS, Milligan LP, McBride BW. Cellular energy metabolism and regulation. J Dairy Sci 74: 678–694, 1991. [DOI] [PubMed] [Google Scholar]
- 42. Kinoshita M. Diversity of septin scaffolds. Curr Opin Cell Biol 18: 54–60, 2006. [DOI] [PubMed] [Google Scholar]
- 43. Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33, 1999. [DOI] [PubMed] [Google Scholar]
- 44. Kossaibati MA, Esslemont RJ. The costs of production diseases in dairy herds in England. Vet J 154: 41–51, 1997. [DOI] [PubMed] [Google Scholar]
- 45. Kuhla B, Albrecht D, Kuhla S, Metges CC. Proteome analysis of fatty liver in feed-deprived dairy cows reveals interaction of fuel sensing, calcium, fatty acid, and glycogen metabolism. Physiol Genomics 37: 88–98, 2009. [DOI] [PubMed] [Google Scholar]
- 46. Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol 16: 2570–2578, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407: 592–598, 2000. [DOI] [PubMed] [Google Scholar]
- 48. Lee MY, Hwang ES, Lee SK. Novel CRE-binding proteins of 11–16 kDa bind to the LDH A-gene CRE in a sequence specific and hepatocyte-growth dependent manner in partially hepatectomized rat liver. Biochem Biophys Res Commun 246: 50–54, 1998. [DOI] [PubMed] [Google Scholar]
- 49. Lee SW, Tomasetto C, Paul D, Keyomarsi K, Sager R. Transcriptional downregulation of gap-junction proteins blocks junctional communication in human mammary tumor cell lines. J Cell Biol 118: 1213–1221, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li CC, Chiang TC, Wu TS, Pacheco-Rodriguez G, Moss J, Lee FJ. ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol Biol Cell 18: 4420–4437, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ling G, Hauer CR, Gronostajski RM, Pentecost BT, Ding X. Transcriptional regulation of rat CYP2A3 by nuclear factor 1: identification of a novel NFI-A isoform, and evidence for tissue-selective interaction of NFI with the CYP2A3 promoter in vivo. J Biol Chem 279: 27888–27895, 2004. [DOI] [PubMed] [Google Scholar]
- 52. Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, Kahn A, Ryan MC, Nuhanovic A, Munson PJ, Reinhold WC, Kane DW, Weinstein JN. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics 23: 2385–2390, 2007. [DOI] [PubMed] [Google Scholar]
- 53. Liu X, Milo M, Lawrence ND, Rattray M. Probe-level measurement error improves accuracy in detecting differential gene expression. Bioinformatics 22: 2107–2113, 2006. [DOI] [PubMed] [Google Scholar]
- 54. Llewellyn S, Fitzpatrick R, Kenny DA, Murphy JJ, Scaramuzzi RJ, Wathes DC. Effect of negative energy balance on the insulin-like growth factor system in pre-recruitment ovarian follicles of post partum dairy cows. Reproduction 133: 627–639, 2007. [DOI] [PubMed] [Google Scholar]
- 55. Loor JJ, Dann HM, Everts RE, Oliveira R, Green CA, Guretzky NA, Rodriguez-Zas SL, Lewin HA, Drackley JK. Temporal gene expression profiling of liver from periparturient dairy cows reveals complex adaptive mechanisms in hepatic function. Physiol Genomics 23: 217–226, 2005. [DOI] [PubMed] [Google Scholar]
- 56. Loor JJ, Everts RE, Bionaz M, Dann HM, Morin DE, Oliveira R, Rodriguez-Zas SL, Drackley JK, Lewin HA. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics 32: 105–116, 2007. [DOI] [PubMed] [Google Scholar]
- 57. Lu W, Quintero-Rivera F, Fan Y, Alkuraya FS, Donovan DJ, Xi Q, Turbe-Doan A, Li QG, Campbell CG, Shanske AL, Sherr EH, Ahmad A, Peters R, Rilliet B, Parvex P, Bassuk AG, Harris DJ, Ferguson H, Kelly C, Walsh CA, Gronostajski RM, Devriendt K, Higgins A, Ligon AH, Quade BJ, Morton CC, Gusella JF, Maas RL. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet 3: e80, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lucy MC. Reproductive loss in high-producing dairy cattle: where will it end? J Dairy Sci 84: 1277–1293, 2001. [DOI] [PubMed] [Google Scholar]
- 59. Makarov SS. NF-kappaB as a therapeutic target in chronic inflammation: recent advances. Mol Med Today 6: 441–448, 2000. [DOI] [PubMed] [Google Scholar]
- 60. Mastroberardino PG, Iannicola C, Nardacci R, Bernassola F, De Laurenzi V, Melino G, Moreno S, Pavone F, Oliverio S, Fesus L, Piacentini M. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington's disease. Cell Death Differ 9: 873–880, 2002. [DOI] [PubMed] [Google Scholar]
- 61. McCarthy SD, Butler ST, Patton J, Daly M, Morris DG, Kenny DA, Waters SM. Differences in the expression of genes involved in the somatotropic axis in divergent strains of Holstein-Friesian dairy cows during early and mid lactation. J Dairy Sci 92: 5229–5238, 2009. [DOI] [PubMed] [Google Scholar]
- 62. McEntaffer RL, Natochin M, Artemyev NO. Modulation of transducin GTPase activity by chimeric RGS16 and RGS9 regulators of G protein signaling and the effector molecule. Biochemistry 38: 4931–4937, 1999. [DOI] [PubMed] [Google Scholar]
- 63. McGarry JD, Woeltje KF, Kuwajima M, Foster DW. Regulation of ketogenesis and the renaissance of carnitine palmitoyltransferase. Diabetes Metab Rev 5: 271–284, 1989. [DOI] [PubMed] [Google Scholar]
- 64. Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278: 860–866, 1997. [DOI] [PubMed] [Google Scholar]
- 65. Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116: 445–456, 2004. [DOI] [PubMed] [Google Scholar]
- 66. Morris DG, Waters SM, McCarthy SD, Patton J, Earley B, Fitzpatrick R, Murphy JJ, Diskin MG, Kenny DA, Brass A, Wathes DC. Pleiotropic effects of negative energy balance in the postpartum dairy cow on splenic gene expression: repercussions for innate and adaptive immunity. Physiol Genomics 39: 28–37, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature 410: 50–56, 2001. [DOI] [PubMed] [Google Scholar]
- 68. Nafikov RA, Beitz DC. Carbohydrate and lipid metabolism in farm animals. J Nutr 137: 702–705, 2007. [DOI] [PubMed] [Google Scholar]
- 69. Niu BY, Ye LZ, Li FE, Deng CY, Jiang SW, Lei MG, Xiong YZ. Identification of polymorphism and association analysis with reproductive traits in the porcine RNF4 gene. Anim Reprod Sci 110: 283–292, 2009. [DOI] [PubMed] [Google Scholar]
- 70. Nwankwo JO, Spector AA, Domann FE. A nucleotide insertion in the transcriptional regulatory region of FADS2 gives rise to human fatty acid delta-6-desaturase deficiency. J Lipid Res 44: 2311–2319, 2003. [DOI] [PubMed] [Google Scholar]
- 71. Pearson RD. A comprehensive re-analysis of the Golden Spike data: towards a benchmark for differential expression methods. BMC Bioinformatics 9: 164, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pearson RD, Liu X, Sanguinetti G, Milo M, Lawrence ND, Rattray M. puma: a Bioconductor package for propagating uncertainty in microarray analysis. BMC Bioinformatics 10: 211, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pero R, Lembo F, Di Vizio D, Boccia A, Chieffi P, Fedele M, Pierantoni GM, Rossi P, Iuliano R, Santoro M, Viglietto G, Bruni CB, Fusco A, Chiariotti L. RNF4 is a growth inhibitor expressed in germ cells but not in human testicular tumors. Am J Pathol 159: 1225–1230, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piacentini M, Autuori F, Dini L, Farrace MG, Ghibelli L, Piredda L, Fesus L. “Tissue” transglutaminase is specifically expressed in neonatal rat liver cells undergoing apoptosis upon epidermal growth factor-stimulation. Cell Tissue Res 263: 227–235, 1991. [DOI] [PubMed] [Google Scholar]
- 75. Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, Pui CH, Evans WE. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst 91: 2001–2008, 1999. [DOI] [PubMed] [Google Scholar]
- 76. Reynolds CK, Durst B, Lupoli B, Humphries DJ, Beever DE. Visceral tissue mass and rumen volume in dairy cows during the transition from late gestation to early lactation. J Dairy Sci 87: 961–971, 2004. [DOI] [PubMed] [Google Scholar]
- 77. Rigazio S, Lehto HR, Tuunanen H, Nagren K, Kankaanpaa M, Simi C, Borra R, Naum AG, Parkkola R, Knuuti J, Nuutila P, Iozzo P. The lowering of hepatic fatty acid uptake improves liver function and insulin sensitivity without affecting hepatic fat content in humans. Am J Physiol Endocrinol Metab 295: E413–E419, 2008. [DOI] [PubMed] [Google Scholar]
- 78. Rutledge RG, Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res 31: e93, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol 9: 1323–1326, 1999. [DOI] [PubMed] [Google Scholar]
- 80. Sanchez-Calderon H, Rodriguez-de la Rosa L, Milo M, Pichel JG, Holley M, Varela-Nieto I. RNA microarray analysis in prenatal mouse cochlea reveals novel IGF-I target genes: implication of MEF2 and FOXM1 transcription factors. PLoS One 5: e8699, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. SAS The SAS System for Windows, Release 9.1. Cary, NC: SAS Institute Inc, 2003. [Google Scholar]
- 82. Shih SC, Zukauskas A, Li D, Liu G, Ang LH, Nagy JA, Brown LF, Dvorak HF. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res 69: 3272–3277, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Smith RD, Brown B, Ikonomi P, Schechter AN. Exogenous reference RNA for normalization of real-time quantitative PCR. Biotechniques 34: 88–91, 2003. [DOI] [PubMed] [Google Scholar]
- 84. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. [DOI] [PubMed] [Google Scholar]
- 85. Sobczak J, Mechti N, Tournier MF, Blanchard JM, Duguet M. c-myc and c-fos gene regulation during mouse liver regeneration. Oncogene 4: 1503–1508, 1989. [PubMed] [Google Scholar]
- 86. Tamminga S, Luteijn PA, Meijer RJM. Changes in composition and energy content of liveweight loss in dairy cows with time after parturition. Livestock Prod Sci 52: 129–155, 1997. [Google Scholar]
- 87. Tergaonkar V. NFkappaB pathway: a good signaling paradigm and therapeutic target. Int J Biochem Cell Biol 38: 1647–1653, 2006. [DOI] [PubMed] [Google Scholar]
- 88. Timperio AM, D'Alessandro A, Pariset L, D'Amici GM, Valentini A, Zolla L. Comparative proteomics and transcriptomics analyses of livers from two different Bos taurus breeds: “Chianina and Holstein Friesian”. J Proteomics 73: 309–322, 2009. [DOI] [PubMed] [Google Scholar]
- 89. Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell 9: 265–277, 2002. [DOI] [PubMed] [Google Scholar]
- 90. Turk R, Juretic D, Geres D, Svetina A, Turk N, Flegar-Mestric Z. Influence of oxidative stress and metabolic adaptation on PON1 activity and MDA level in transition dairy cows. Anim Reprod Sci 108: 98–106, 2008. [DOI] [PubMed] [Google Scholar]
- 91. Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev 9: 2723–2735, 1995. [DOI] [PubMed] [Google Scholar]
- 92. Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem 279: 47066–47075, 2004. [DOI] [PubMed] [Google Scholar]
- 93. Wathes DC, Cheng Z, Chowdhury W, Fenwick MA, Fitzpatrick R, Morris DG, Patton J, Murphy JJ. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiol Genomics 39: 1–13, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Watters JW, Zhang W, Meucci MA, Hou W, Ma MK, McLeod HL. Analysis of variation in mouse TPMT genotype, expression and activity. Pharmacogenetics 14: 247–254, 2004. [DOI] [PubMed] [Google Scholar]
- 95. Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 81: 323–330, 1995. [DOI] [PubMed] [Google Scholar]
- 96. Zarr K. Light and electron microscopic localisation of d-aspartate oxidase in peroxisomes of bovine kidney and liver: an immunocytochemical study. J Histochem Cytochem 44: 1013–1019, 1996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.