Abstract
To characterize gene expression networks linked to AT1 angiotensin receptors in the kidney, we carried out genome-wide transcriptional analysis of RNA from kidneys of wild-type (WT) and AT1A receptor-deficient mice (KOs) at baseline and after 2 days of angiotensin II infusion (1,000 ng·kg−1·min−1). At baseline, 405 genes were differentially expressed (>1.5×) between WT and KO kidneys. Of these, >80% were upregulated in the KO group including genes involved in inflammation, oxidative stress, and cell proliferation. After 2 days of angiotensin II infusion in WT mice, expression of ≈805 genes was altered (18% upregulated, 82% repressed). Genes in metabolism and ion transport pathways were upregulated while there was attenuated expression of genes protective against oxidative stress including glutathione synthetase and mitochondrial superoxide dismutase 2. Angiotensin II infusion had little effect on blood pressure in KOs. Nonetheless, expression of >250 genes was altered in kidneys from KO mice during angiotensin II infusion; 14% were upregulated, while 86% were repressed including genes involved in immune responses, angiogenesis, and glutathione metabolism. Between WT and KO kidneys during angiotensin II infusion, 728 genes were differentially expressed; 10% were increased and 90% were decreased in the WT group. Differentially regulated pathways included those involved in ion transport, immune responses, metabolism, apoptosis, cell proliferation, and oxidative stress. This genome-wide assessment should facilitate identification of critical distal pathways linked to blood pressure regulation.
Keywords: transgenic mice, hypertension, oxidative stress, microarray analysis
the renin-angiotensin system (RAS) modulates many physiological processes and plays a central role in mechanisms of chronic renal disease (20). The major peptide mediator of the RAS, angiotensin II (Ang II), affects function of cells through specific, seven-transmembrane spanning receptors that can be divided into two pharmacological classes: AT1 and AT2. Most of the classically recognized functions of the RAS are mediated by AT1 receptors. The murine AT1 receptors, AT1A and AT1B, are products of separate genes, sharing 94% homology at amino acid level and are pharmacologically indistinguishable (36). AT1A receptors predominate in most organs and the AT1A receptor is considered the closest murine homolog to the single human AT1 receptor. Intracellular signaling pathways linked to AT1 receptor activation have been well characterized and include signaling through Gq proteins, PLC, and increases in intracellular calcium (7). In addition, AT1 receptors also activate JAK/STAT pathways through actions that may be independent of G proteins (9) and have been shown to transactivate EGF receptor signaling (27). These signaling pathways trigger programs of gene expression that determine physiological and pathological responses.
Hypertension is a highly prevalent disorder affecting >50 million Americans, and its complications, including heart disease, stroke, and kidney disease, cause significant morbidity and mortality. Despite decades of scrutiny, the pathogenesis of essential hypertension has been difficult to delineate. The majority of patients with essential hypertension experience blood pressure lowering following treatment with specific AT1 receptor blockers (12), suggesting that activation of AT1 receptors plays a key role in human hypertension. Nonetheless, the precise pathways that are responsible for hypertension in humans have not been clearly characterized.
In previous studies, we developed an experimental model to separate AT1 receptor pools in the kidney from those in other tissues (5, 6). Using this model, we found that AT1 receptors within the kidney play a key role in normal blood pressure control (6) and are absolutely required for the development of Ang II-dependent hypertension (5). Furthermore, our findings suggested that AT1 receptors cause cardiac hypertrophy by increasing blood pressure, not by direct effects in cardiac myocytes but through direct actions of AT1 receptors in the renal parenchyma. Yet the networks of downstream genes triggered by AT1 receptor activation in the kidney that are critical for modulating renal excretory function and generating hypertension are not known.
To explore this issue, we performed a comprehensive analysis of gene expression networks linked to renal AT1 receptors. Specifically, we carried out genome-wide transcriptional profiling of RNA from kidneys of wild-type (WT) and AT1A receptor-deficient mice (KOs) at baseline and after 2 days of Ang II infusion during the initiation phase of hypertension. We find evidence for activation of a series of downstream gene expression pathways linked to the AT receptor that are likely to promote hypertension pathogenesis.
MATERIALS AND METHODS
Mice.
The generation and phenotypical characterization of mice with targeted disruption of the Agtr1a gene locus encoding the AT1A receptor were described previously (17). F1 (C57BL6 × 129/SvEv) Agtr1a+/+ and −/− mice were used in this study. All of the experiments were conducted with 2 to 3 mo old male mice and included three mice in each experimental group. Mice were fed a 0.4% NaCl diet ad libitum. To reveal genes triggered by AT1 receptor activation with Ang II, osmotic minipumps were implanted subcutaneously into mice to infuse Ang II (1,000 ng·kg−1·min−1) for 2 days. Mice were maintained according to the National Institutes of Health Guide for the Care and the Use of Laboratory Animals. Procedures used in this study were approved by the Institutional Animal Care and Use Committees of Duke University and the Durham Veterans Affairs Medical Center.
RNA isolation.
Kidneys were harvested from Agtr1a+/+ and −/− mice and from separate groups that had been infused with Ang II as described above. The kidney capsule was removed and a ≈4 mm central slice containing cortex and medulla was snap-frozen in liquid nitrogen. Total RNA was extracted with RNeasy Mini Kit (Qiagen) according to manufacturer's protocol. Concentration and purity of all RNA samples was determined by the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE), and RNA integrity was verified by using standard gel electrophoresis and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Microarray expression analysis.
To define gene expression patterns in kidney triggered by of activation AT1 receptors, we used GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix, Santa Clara, CA). These chips contain >45,000 probe sets that analyze the expression >39,000 transcripts and variants representing >34,000 well-characterized mouse genes. Microarray expression analysis of kidney was conducted in the Duke University Microarray Core Facility. Briefly, 5 μg of total RNA was amplified with the One-Cycle cDNA synthesis and target labeling kit according to manufacturer's instructions (Affymetrix). Biotin-labeled target complementary RNA (20 μg) was fragmented and hybridized to each array according to the manufacturer's instructions. The arrays were scanned using Affymetrix equipment and protocols. Microarray data are available via the Gene Expression Omnibus, accession #GSE18430.
The expression data were processed with robust multiarray analysis algorithms using GeneSpring GX version 7.3.1 (Agilent Technologies, Santa Clara, CA), and per-gene normalization to the median was applied using the cross-error model to enhance the statistical power of the analysis. Transcripts that were “present” in at least two out of 12 samples were used for further statistical analysis. Accordingly, a probe set of 27,805 was selected from a total of >45,000. These probe sets were filtered using Volcano plot with a parametric test, assuming equal variances, and with a specified minimum 1.5-fold change. In general, the P values were controlled for multiple testing [false discovery rate (FDR)] with P < 0.05. However, for the baseline comparison between AT1A-deficient and WT mice, a specified threshold P value of <0.005 was used to increase the number of genes available for analysis. Genes with expression differences that were statistically significant genes were then subjected to GO (Gene Ontology) analysis (P < 0.05). To identify biological pathways triggered by Ang II, the Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Redwood City, CA) was used. To determine effects of genotype, Ang II treatment and the interaction between genotype and treatment, a two-way ANOVA analysis (FDR < 0.005) was used.
Real-time PCR analysis.
Differences in expression of selected genes were confirmed using quantitative real-time PCR analysis. Briefly, total RNA was prepared from kidneys as described above using RNeasy minicolumns (Qiagen). Extracted RNA was reverse-transcribed with random hexamers using the Invitrogen SuperScript II kit (Invitrogen, Carlsbad, CA). Real-time quantitative PCR amplifications were performed in duplicate in a 96-well plate (Bio-Rad iCycler iQ Real Time PCR System). Primers and probes for genes were purchased from Applied Biosystems (Foster City, CA). For normalization, Taqman 18 S rRNA Control Reagent was used as an internal control. The levels of gene expression were calculated using a standard curve.
Analysis of protein levels by immunoblotting.
To determine serum/glucocorticoid-regulated kinase (Sgk)1 protein levels in kidney, samples (40 μg total protein) were denatured, separated on NuPage 4–12% Bis-Tris gel (Invitrogen), and transferred to Invitrolon PVDF membrane (Invitrogen). The membranes were incubated with rabbit monoclonal anti-Sgk1 antibody [Millipore (Chemicon/Upstate/Linco); clone Y238, 1:250 dilution] overnight at 4°C, followed by incubation with a peroxidase-labeled anti-rabbit antibody for 1 h. Protein was detected using ECL Plus Western blotting analysis system (Amersham Pharmacia). α-Tubulin (Santa Cruz Biotechnology) was used as a loading control.
RESULTS
Comparison of gene expression profiles between kidneys from KO and WT control mice.
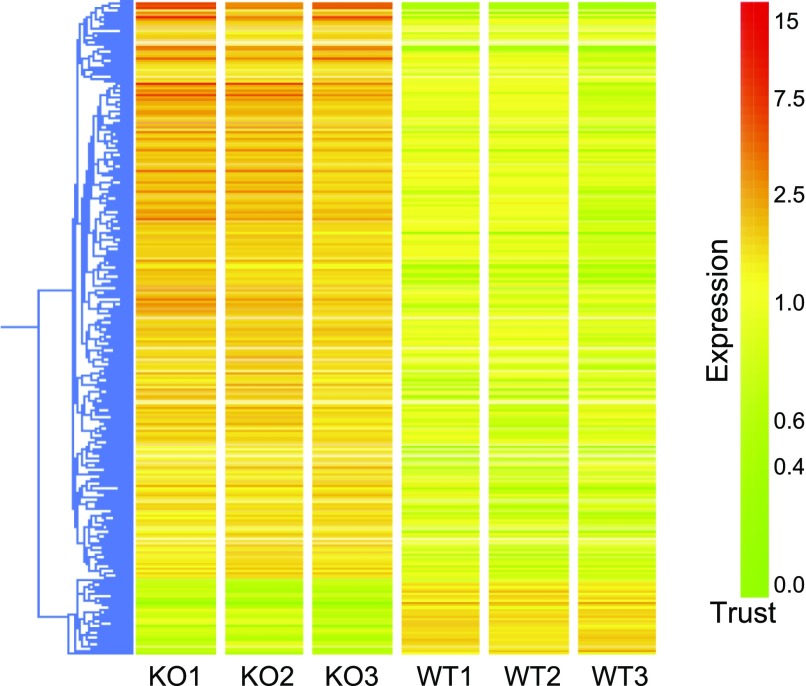
Our analysis showed that there were 406 genes with significantly altered expression (by 1.5-fold or more) in kidneys from Agtr1a−/− mice compared with WT controls at baseline. Of these, 359 genes (≈88%) were upregulated in the KOs, while 47 genes (≈12%) were repressed (Fig. 1). Highly represented GO categories among these genes included cell communication, response to stimulus, physiological processes, developmental response to biotic stimulus, defense response, immune response, response to stress, response to external stimulus, organ development, and cell differentiation. Specific examples of genes that were upregulated in the AT1A receptor-deficient mice are shown in Supplemental Table S11 and included genes associated with chemokine pathways, inflammatory mediators such as interferon-γ, leukotrienes, and various adhesion molecules. We also observed upregulation of other genes involved in the immune response including genes in the histocompatibility complex, the complement system, lysosomes, and the proteosome. Expression of several growth factors was also upregulated in KO mice including early growth response 1 (Egr1); early growth response 2 (Egr2); transforming growth factor, β1 (Tgfb1); transforming growth factor, β-induced (Tgfbi); transforming growth factor, β-receptor III (Tgfbr3), connective tissue growth factor (Ctgf), cysteine rich protein 61 (Cyr61). Notably, TGF-β (Tgfb1) is a prominent mediator in the cell-to-cell signaling and interaction and cellular movement pathway with the highest score (38) identified by the IPA among differentially expressed genes (Fig. 2).
Fig. 1.
Hierarchical clustering of differentially expressed genes between kidneys from AT1A receptor-deficient [knockout (KO)] and wild-type (WT) control mice. More than 80% of genes were upregulated in the kidneys of KO compared with WT mice including genes involved in inflammation, oxidative stress, and cell proliferation. Yellow depicts no change, whereas red represents upregulation, and green represents downregulation.
Fig. 2.
Network 1 created by Ingenuity Pathway Analysis from differentially expressed genes between kidneys from AT1A receptor-deficient and WT control mice. Blue indicates downregulation and red indicates upregulation. Square, cytokine; vertical oval, transmembrane receptor; rectangle, nuclear receptor; diamond, enzyme; rhomboid, transporter; hexagon, translation factor; horizontal oval, transcription factor; circle, others. A line indicates gene products that bind one another. A line with arrow indicates that a gene products is acting on another.
Finally, our analysis revealed that kidneys from AT1A receptor-deficient mice manifested a marked increase in expression of genes implicated in oxidative stress (Supplemental Table S1). These included an inhibitor of cellular antioxidant of thioredoxin-thioredoxin interacting protein (Txnip, 1.76-fold), oxidative stress-induced genes such as xanthine dehydrogenase (Xdh, 1.61-fold), ceruloplasmin (Cp, 2.93-fold), and the dual-specificity phosphatase 1 (Dusp1, 1.67-fold). Expression of oxygen transporters, such as cytoglobin (Cygb), hemoglobin β-chain complex (Hbb, Hbb-b1, Hbb-b2), and the adult chain of hemoglobin-α (Hba-a1), were also increased in the Agtr1a−/− mice compared with controls.
Altered gene expression profiles caused by infusion of Ang II in WT mice.
To examine the effects of chronic stimulation of the kidney by Ang II at concentrations known to cause hypertension, we compared gene expression profiles from kidneys of WT mice under baseline conditions and after a 2-day infusion of Ang II. This analysis showed that expression of 805 genes was altered by 1.5-fold or more by the Ang II infusion. Of these genes, 145 (≈18%) were upregulated, whereas the vast majority (660 genes, ≈82%) were repressed. To define common pathways among genes with altered expression, we carried out GO analysis, which demonstrated changes in a number of biological pathways with Ang II infusion including physiological and cellular physiological processes, metabolism, localization, transport, biosynthesis, lipid and carbohydrate metabolism, and ion transport (Supplemental Table S2).
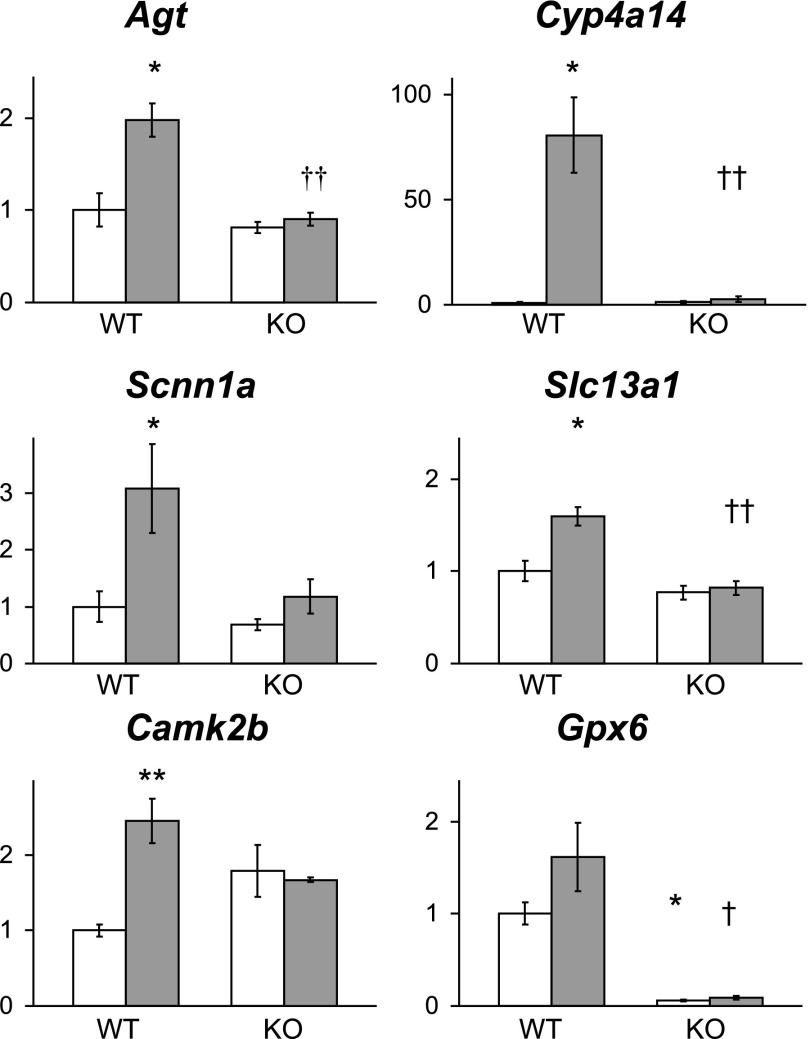
Previous studies by our group and others have suggested that Ang II controls blood pressure primarily by activation of AT1 receptors in the kidney and thereby affecting fluid and solute reabsorption (4). In this regard, infusion of Ang II was associated with upregulation of several sodium transporters such as the sodium/sulphate symporter (Slc13a1, 2.81-fold), sodium/bile acid cotransporter (Slc10a6, 1.75-fold), and the α-subunit of the type I nonvoltage-gated sodium channel (Scnn1a, known as α-ENaC, 1.59-fold). In parallel with enhanced α-ENaC expression, we found that expression of the upstream kinase Sgk1 was similarly altered. To confirm these changes in Sgk1 mRNA expression and to determine whether they resulted in increased levels of Sgk1 protein, we carried out RT-PCR and Western blotting. As shown in Fig. 3A, levels of Sgk1 mRNA were significantly higher in WT mice during infusion of Ang II than WT controls (P < 0.01) or KO mice during Ang II infusion (P < 0.01). As depicted in Fig. 3B, a similar pattern was observed with Sgk1 protein levels. SGK1 protein levels were also higher in the WT + Ang II group than in WT controls (P < 0.001) or KO mice during Ang II infusion (P < 0.01). Expression of Sgk1 in the kidneys of the KO mice during Ang II was higher compared with WT control mice (P = 0.05) (Fig. 3B).
Fig. 3.
Expression of serum/glucocorticoid-regulated kinase (Sgk1) in kidneys of KO and WT mice at baseline and after angiotensin (Ang) II infusion. A: Sgk1 mRNA levels (relative to WT control mice as 100%). Data were normalized to 18S RNA from the same samples (n = 3 for each group). Open column, control mice; gray column, Ang II infusion. B: comparison of the levels of Sgk1 protein using Western immunoblotting analysis. The values above the bands are the means of relative changes in WT and KO mice after Ang II infusion compared with WT control mice. ***P < 0.01 compared with WT control mice; ***P < 0.001 compared with WT control mice; ††P < 0.01 compared with WT mice infused with Ang II.
On the other hand, expression of several sodium transporters was downregulated, including the sodium/glucose cotransporter (Slc5a2, 0.61-fold), type IIc Na+/Pi cotransporter (Slc34a3, 0.52-fold), and a sodium/hydrogen exchanger (Slc9a8, 0.26-fold). With the exception of Scnn1a (ENaC-α), these transporters make minimal contributions to overall handling of sodium by the kidney.
Expression of other major renal sodium transporters tended to be reduced or unchanged including: Na+/H+ exchanger 3 (Slc9a3, NHE3: 0.79-fold), type II sodium Pi cotransporters (Slc34a1, NaPi2: 0.82-fold), β-subunit of the type I nonvoltage-gated sodium channel (Scnn1b; ENaC-β: 1.00-fold), and γ-subunit of the type I nonvoltage-gated sodium channel (Scnn1g, known as ENaC-γ: 1.02-fold). On the other hand, expression of the thiazide-sensitive Na-Cl cotransporter (Slc12a3, NCC) was modestly increased (1.23-fold).
There is evidence supporting the involvement of cytochrome P450 (CYP) enzymes in regulation of sodium homeostasis and blood pressure (2, 33). We found that infusion of Ang II in WT mice was associated with strong upregulation of CYP of Cyp4A subfamily (Cyp4a14, 44.66-fold; and Cyp4a10, 2.73-fold). These enzymes catalyze synthesis of 20-hydroxyeicosatetranoic acids (HETEs), potent vasoconstrictors and modulators of renal sodium reabsorption (33). On the other hand, expression of CYP enzymes of the Cyp2J subfamily (Cyp2j11) is downregulated (0.62-fold). These enzymes produce epoxyeicosatrienoic acids (EETs), which may have vasodilator and natriuretic effects (25).
A critical role for oxidative stress in Ang II-dependent hypertension is well documented (15). With Ang II infusion, we found reduced expression of genes with protective effects against oxidative stress including: glutathione synthetase (Gss, 0.65-fold), mitochondrial superoxide dismutase 2 (Sod2, 0.57-fold), and soluble isocitrate dehydrogenase (Idh1, 0.63-fold), which is critical in the production of NADPH for regeneration of reduced glutathione (37). Furthermore, we observed strong upregulation of downstream genes induced by oxidative stress such as hemoxygenase-1 (Hmox1, 6.24-fold) and Cp (3.78-fold).
Activation of AT1 receptors by Ang II triggers cell growth and proliferation in a number of settings (41). We noticed that a number of genes influencing cell growth and proliferation were differentially expressed in kidneys of WT mice during Ang II infusion including: myelocytomatosis oncogene (Myc, also known as c-Myc, 1.61-fold), citron (Cit, rho-interacting, serine/threonine kinase 21, 1.60-fold), N-myc downstream regulated gene 1 or N-myc downstream regulated-like (Ndrg1, Ndrl; 1.58-fold), and purine-rich element binding protein A (Pura, 1.55-fold). We also found downregulation of genes with a dampening or repressive effect on cell proliferation including: large tumor suppressor 2 (Lats2, 0.60 fold), growth arrest-specific 2 (Gas2, 0.58-fold), retinoblastoma-like 1 (Rbl1, 0.56-fold), and cell growth regulator with EF hand domain 1 (Cgref1, 0.59-fold).
A network IPA analysis identified 13 significant networks with scores of >11. The five top-scoring networks are shown in Supplemental Table S3 and were categorized as gene expression, nervous system development and function, cell cycle, inflammatory response, posttranslational modification, and others.
Ang II affects gene expression in kidneys of mice lacking AT1A receptors.
Infusion of Ang II altered expression profiles of 251 genes in kidneys of KO mice. Of these, 36 (≈14%) were upregulated, while 215 (≈86%) were repressed. The largest groups of GO terms with significant associations were: metabolism, development, biopolymer modification, organ development, negative regulation of cellular process, and response to stress (Supplemental Table S4).
As we had seen in the WT group, expression of cellular antioxidant genes was altered by Ang II infusion in KO mice. For example, genes involved in glutathione metabolism such as members of glutathione S-transferase family [Gstp1 (0.66-fold), Gsta2 (0.44-fold), Gsta1 and/or Gsta2 (0.39-fold), Gstm1 (0.60-fold), Gstm6 (0.64-fold)] were downregulated along with microsomal glutathione S-transferase (Mgst1, 0.62-fold) and thioredoxin interacting protein (Txnip, 0.52-fold). In contrast to WT mice, expression of several genes typically induced by oxidative stress were downregulated including: Hmox1 (0.71-fold), Cp (0.47-fold), and Dusp1 (0.50-fold).
IPA network analysis identified 10 genetic networks with significant scores (>11). The five top-scoring networks are shown in Supplemental Table S5 and were associated with functions of organ development, drug metabolism, cell cycle, mediated immune response, amino acid metabolism, and others.
Differential gene expression between kidneys of WT and AT1A receptor-deficient mice during infusion of Ang II.
We and others have shown that physiological responses to chronic Ang II infusion are quite different between WT and KO mice (6, 34). In WT mice, Ang II infusion causes robust hypertension and renal sodium retention (5). By contrast, Ang II infusion has minimal effects on blood pressure in mice lacking AT1A receptors and they maintain natriuresis. Accordingly, we were interested in defining differences in gene expression patterns between these groups to understand the mechanisms underlying these different responses.
Venn diagram analysis showed that only five genes were upregulated by Ang II infusion in both KO and WT mice, while 46 genes were downregulated in both groups (data not shown). Downregulated genes in both KO and WT mice included genes involved in metabolism, RNA processing, RNA splicing, and protein ubiquitination. A total of 728 transcripts were differentially expressed between KO and WT kidneys during Ang II infusion. Of these, ≈10% were upregulated and ≈90% were downregulated in the WT group. Differentially expressed pathways included genes involved in ion transport, immune responses, metabolism, apoptosis, cell proliferation, and oxidative stress (Supplemental Table S6).
As another approach for analyzing genes with differing responses to Ang II between WT and KO mice, genotype-by-treatment interaction was calculated using two-way ANOVA. This analysis revealed that expression of 1,455 gene sets was significantly changed (P < 0.005). The differentially expressed gene sets were predominantly affected by Ang II treatment (1,026 gene sets) followed by genotype (405 gene sets). The 316 transcripts with significant genotype and treatment interaction are candidates for significant pathways mediating functional actions of AT1 receptors in the kidney. Notably, most genes with genotype and treatment interaction (Fig. 4) were changed in opposite direction between the groups after Ang II treatment. IPA network analysis identified several networks with highly significant associations including: free radical scavenging, cell death and cancer (score 40) and cardiovascular system development and function, and cellular development (score 39; Supplemental Fig. S1).
Fig. 4.
Cluster analysis of genes that showed interaction of genotype and Ang II treatment. The mean appears yellow, whereas red represents upregulation, and green represents downregulation.
Validation of array results by real-time quantitative PCR.
To validate our microarray data, we carried out additional studies using quantitative RT-PCR. Using this approach, we confirmed the results for several selected genes with the same RNA samples that were used for microarrays. As shown in Fig. 5, the patterns of changes in gene expression identified by qRT-PCR were very similar to those detected in our analysis of the microarray data.
Fig. 5.
Differential gene expression confirmed by qRT-PCR. Data were normalized to 18S RNA from the same samples (n = 3 for each group). Open column, control mice; gray column, Ang II infusion. Mean of expression levels in WT control mice were assigned a value of unity. *P < 0.05, **P < 0.01 compared with WT control mice; †P < 0.05, ††P < 0.01 compared with WT mice infused with Ang II. Agt, angiotensinogen; Cyp4a14, cytochrome P450, family 4, subfamily a, polypeptide 14; Scnn1a, sodium channel, nonvoltage-gated 1α (ENaC-α); Slc13a1, solute carrier family 13 (sodium/sulfate symporters), member 1; Camk2b, calcium/calmodulin-dependent protein kinase IIβ; Gpx6, glutathione peroxidase 6 (olfactory).
DISCUSSION
Our previous studies demonstrated a critical role for AT1 receptors within the kidney in hypertension pathogenesis (5). To define genetic networks associated with activation of renal AT1 receptors, we carried out a genome-wide analysis of gene expression profiles from kidneys of AT1A receptor-deficient and WT mice at baseline and after 2 days of Ang II infusion. Using this approach, we identified differentially expressed genes linked to deficiency of AT1A receptors and to the development of Ang II-dependent hypertension.
Our microarray analysis of gene expression patterns identified >400 genes that were differentially expressed at baseline in kidneys of KO mice compared with controls; >80% of these were upregulated. In these kidneys from mice lacking the major murine AT1 receptor isoform, there was broad upregulation of pathways previously associated with AT1 receptor signaling including inflammation, oxidative stress, growth, and proliferation (26). Nonetheless, our data are consistent with results from a study by Ouyang and associates (30), who used real-time PCR analyses to document increased expression of a panel of profibrotic, proinflammatory, and oxidative stress mediators in kidneys of mice lacking both AT1A and AT1B receptor isoforms. Baseline blood pressures are dramatically reduced in mice lacking AT1A receptors (17). Thus, enhanced expression of some of these genes in KO mice may reflect compensatory mechanisms triggered by low blood pressure, perhaps mediated by activation of the sympathetic nervous system or other homeostatic elements. Our previous studies have shown that kidneys from AT1A receptor-deficient mice on the F1 (129 × C57BL/6) background appear normal, lacking significant infiltration by inflammatory cells or fibrosis (21). Thus, upregulation of genes associated with inflammatory pathways, growth factors, and oxidative stress do not appear to be sufficient to promote kidney injury in this setting.
The large number of upregulated genes in AT1A receptor-deficient mice may also reflect stimulation of residual components of the RAS. We and others have previously shown that renin production and Ang II levels are significantly increased in mice lacking AT1A receptors (6, 24, 29), likely due to loss of short-loop feedback inhibition of renin by AT1A receptors in the juxta-glomerular apparatus. Our current microarray analysis confirmed that renin expression was increased more than sevenfold in the kidneys of KO mice compared with controls. This activation of the RAS could produce stimulation of AT2, AT1B, and/or other receptors for Ang peptides that may still be present in kidneys of AT1A receptor-deficient mice.
Chronic infusion of Ang II causes robust and sustained hypertension in normal mice (42). Substantial increases in blood pressure are seen within the first 48 h after the infusion is begun, reaching maximal levels within 5–7 days (5, 14). We therefore carried out our analysis of gene expression during the early phase of the development of hypertension. Our assumption was that changes in gene expression during the initial 48 h of the infusion would likely reflect physiological alterations associated with blood pressure elevation, rather than structural or pathological consequences of elevated blood pressure seen at later time points in this model. Changes in gene expression at this early time point could thus potentially be involved in the pathogenesis of hypertension rather than a result of kidney injury induced by chronic blood pressure elevation. In WT mice after 2 days of Ang II infusion, expression of 805 genes was altered compared with baseline. Activation of AT1 receptors in cells is linked to transcriptional activation of a number of distal effector pathways linked to physiological functions (3), yet our study showed that the majority (≈82%) of differentially expressed genes were repressed. We speculate that suppression of gene expression in kidneys from WT mice may represent protective and compensatory mechanisms in response to high blood pressure.
We have previously documented the importance of AT1 receptors in the kidney to the development of hypertension, promoting reduced excretion of sodium in urine and consequent increases in blood pressure (5). Accordingly, we were interested in the effects of Ang II infusion on expression of genes encoding various sodium transporters. While we found changes in expression of several sodium transporters in WT mice after ANG II infusion, the α-subunit of ENaC was the only major renal sodium transporter component with significantly increased expression compared with baseline. This was accompanied by upregulation of the Sgk1 gene, known to mediate many effects of aldosterone including its actions to modulate expression of ENaC (22). On the other hand, while expression of other major renal sodium transporters such as NHE3 and NaPi2 was modestly reduced, and expression of NCC was increased with Ang II infusion, these differences did not achieve the 1.5× threshold specified by our analysis. Moreover, regulation of these key transporters is complex and includes substantial posttranslational modifications (11, 35). Thus, the actions of AT1 receptors to control the activity and levels of these transporter proteins may also occur through proximal effects on genes that regulate posttranslational processing such as phosphorylation and/or trafficking (10, 13, 19, 32).
A number of previous studies have documented the importance of oxidative stress in AT1 receptor signaling and in the pathogenesis of Ang II-induced hypertension (26, 31). In kidneys of WT mice infused with Ang II, we found strong upregulation of genes previously known to be induced by cellular oxidative stress (1). At the same time, there was attenuated expression of genes involved in the protection against oxidative stress, including mitochondrial SOD2 and Idh1, which is critical for producing NADPH for regeneration of reduced glutathione and Gss. In this regard, previous studies have shown that infusion of Ang II reduces levels of total glutathione by ≈30% in WT mice (40), and glutathione depletion induced by direct inhibition of glutathione synthase causes oxidative stress and hypertension in rats (39).
The differences in gene expression in the cytochrome P450 enzymes of the Cyp4a subfamily were among the most robust that we observed: Ang II infusion in the WT mice was associated with a 45-fold upregulation of Cyp4a14 and a threefold upregulation of Cyp4a10, both of which were statistically significant. The role of Cyp4a enzymes in blood pressure regulation is likely to be complex. These enzymes catalyze the conversion of arachidonic acid to biologically active compounds such as 20-HETE and EETs, which have well-documented actions to modulate vascular tone and sodium excretion (25, 33). Moreover, Cyp4a14 gene may affect plasma androgen levels, also impacting blood pressure (16). Our studies clearly show that Ang II has potent actions to modulate these enzymes and these effects depend on the presence of AT1A receptors. As such, they represent important candidate pathways that may shape the hypertensive response to Ang II.
AT1A receptor-deficient mice are highly resistant to Ang II-dependent hypertension (34). Despite the negligible change in blood pressure typically induced by chronic Ang II infusion in these animals, expression of >250 genes was altered in kidneys of KO mice compared with baseline. The majority of these were repressed (≈86%), with only a minority (≈14%) being upregulated. It seems likely that changes in gene expression caused by Ang II in the KO mice were mediated by activation of residual Ang receptors including AT1B, AT2 or perhaps others. For example, we have previously shown that AT1B receptors mediate modest vascular responses to acute Ang II infusion in KO mice (28). Likewise, although incompletely characterized, there are distinct signaling pathways linked to AT2 receptors (38). Nonetheless, our assessment of the overall patterns of expression through Ingenuity Network Analysis, compared with the WT group, was consistent with a generalized attenuation of signaling through AT1 pathway-associated genes in kidneys of KO mice infused with Ang II.
To begin to identify genetic pathways linked to AT1A receptor activation in the kidney and by extension to the development of hypertension, we compared gene expression patterns in kidneys of WT and KO mice during infusion of Ang II. Thus our microarray analysis showed that elevation of blood pressure in WT during Ang II infusion was associated with differential expression of genes involved in metabolism, transport, growth, cell death, glutathione metabolism, immune response, steroid biosynthesis. In this comparison, ≈70 genes were upregulated with Ang II in the kidneys of WT compared with KO mice. This group of genes is likely to reflect pathways that are specifically activated by AT1A receptors and are candidates for a causative role in hypertension. One of genes of particular interest in this group was Sgk. The increase in Sgk1 mRNA expression was confirmed by PCR and increased protein levels were shown by Western blot (Fig. 3). Through its actions as an intermediary of aldosterone and glucocorticoids, SGK promotes reabsorbtion of sodium and simultaneous secretion of potassium by activating proteins such as ENaC, TRPV5, ROMK, Kv1.3, KCNE1/KCNQ1, NHE3, and the Na+-K+-ATPase (23). Genes involved in cell growth and proliferation such as fibroblast growth factor binding protein 1 (Fgfbp1), insulin-like growth factor binding protein 1 (Igfbp-1), inhibin β-B (Inhbb), growth arrest-specific 5 (Gas5), and a serine/threonine kinase and rho effector-citron (Cit) essential for cytokinesis (8) were also upregulated.
If the stringency of our analysis is relaxed so that we query genes that are upregulated 1.3-fold or more, the number of genes with increased expression in kidneys of WT compared with KO mice during Ang II infusion is larger, up to 179. Among this group are several transporters including aquaporin 1 (Aqp1, 1.45-fold), chloride channel 5 (Clcn5, 1.34-fold), as well as angiotensinogen (Agt, 1.42-fold) (Fig. 5). Kobori and associates (18) have previously demonstrated increased angiotensinogen expression, especially in proximal tubules, during Ang II infusion and have suggested that this may promote increased intrarenal generation of Ang II, promoting hypertension.
In summary, our microarray analysis demonstrated a paradoxical increase in expression of a number of genes in kidneys of AT1A receptor-deficient mice at baseline. Some of these have previously been linked to AT1 receptor signaling and may represent a compensatory response to low blood pressure. The increase in blood pressure induced by Ang II infusion in WT mice was associated with upregulation of genes involved transport, proliferation, and oxidative stress, with attenuated expression of genes with protective effects against oxidative stress. Despite its minimal hemodynamic effects, infusion of Ang II in KO mice altered expression of a number of genes involved in various biological pathways including immune responses, migration, angiogenesis, and glutathione metabolism, likely through activation of other response elements of the RAS. Further pathway analysis of differentially regulated genes between WT and KO kidneys with Ang II infusion should be useful in identifying distal pathways in the kidney linked to AT1 receptor stimulation promoting blood pressure elevation. As such, they should provide insights into the pathogenesis of hypertension and may suggest new targets for therapy.
GRANTS
This study was supported by National Heart, Lung, and Blood Institute Grant HL-56122, by funding from the Medical Research Service of the Veterans Administration, and by the Edna and Fred L. Mandel Center for Hypertension and Atherosclerosis Research.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Abraham NG, Asija A, Drummond G, Peterson S. Heme oxygenase-1 gene therapy: recent advances and therapeutic applications. Curr Gene Ther 7: 89–108, 2007. [DOI] [PubMed] [Google Scholar]
- 2. Capdevila JH, Falck JR, Harris RC. Cytochrome P450 and arachidonic acid bioactivation: molecular and functional properties of the arachidonate monooxygenase. J Lipid Res 41: 163–181, 2000. [PubMed] [Google Scholar]
- 3. Chung O, Kühl H, Stoll M, Unger T. Physiological and pharmacological implications of AT1 versus AT2 receptors. Kidney Int 54: S95–S99, 1998. [DOI] [PubMed] [Google Scholar]
- 4. Coffman TM, Crowley SD. Kidney in hypertension: guyton redux. Hypertension 51: 811–816, 2008. [DOI] [PubMed] [Google Scholar]
- 5. Crowley SD, Gurley SB, Herrera MJ, Ruiz P, Griffiths R, Kumar AP, Kim HS, Smithies O, Le TH, Coffman TM. Angiotensin II causes hypertension and cardiac hypertrophy through its receptors in the kidney. Proc Natl Acad Sci USA 103: 17985–17990, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crowley SD, Gurley SB, Oliverio MI, Pazmino AK, Griffiths R, Flannery PJ, Spurney RF, Kim HS, Smithies O, Le TH, Coffman TM. Distinct roles for the kidney and systemic tissues in blood pressure regulation by the renin-angiotensin system. J Clin Invest 115: 1092–1099, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Gasparo M, Catt KJ, Inagami T, Wright JW, Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol Rev 52: 415–472, 2000. [PubMed] [Google Scholar]
- 8. Di Cunto F, Calautti E, Hsiao J, Ong L, Topley G, Turco E, Dotto GP. Citron rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J Biol Chem 273: 29706–29711, 1998. [DOI] [PubMed] [Google Scholar]
- 9. Doan TN, Ali MS, Bernstein KE. Tyrosine kinase activation by the angiotensin II receptor in the absence of calcium signaling. J Biol Chem 276: 20954–20958, 2001. [DOI] [PubMed] [Google Scholar]
- 10. Donowitz M, Li X. Regulatory binding partners and complexes of NHE3. Physiol Rev 87: 825–872, 2007. [DOI] [PubMed] [Google Scholar]
- 11. Du Cheyron D, Chalumeau C, Defontaine N, Klein C, Kellermann O, Paillard M, Poggioli J. Angiotensin II stimulates NHE3 activity by exocytic insertion of the transporter: role of PI 3-kinase. Kidney Int 64: 939–949, 2003. [DOI] [PubMed] [Google Scholar]
- 12. Fabia MJ, Abdilla N, Oltra R, Fernandez C, Redon J. Antihypertensive activity of angiotensin II AT1 receptor antagonists: a systematic review of studies with 24 h ambulatory blood pressure monitoring. J Hypertens 25: 1327–1336, 2007. [DOI] [PubMed] [Google Scholar]
- 13. Féraille E, Doucet A. Sodium-potassium-adenosinetriphosphatase-dependent sodium transport in the kidney: hormonal control. Physiol Rev 81: 345–418, 2001. [DOI] [PubMed] [Google Scholar]
- 14. Gurley SB, Allred A, Le TH, Griffiths R, Mao L, Philip N, Haystead TA, Donoghue M, Breitbart RE, Acton SL, Rockman HA, Coffman TM. Altered blood pressure responses and normal cardiac phenotype in ACE2-null mice. J Clin Invest 116: 2218–2225, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harrison DG, Cai H, Landmesser U, Griendling KK. Interactions of angiotensin II with NAD(P)H oxidase, oxidant stress and cardiovascular disease. J Renin Angiotensin Aldosterone Syst 4: 51–61, 2003. [DOI] [PubMed] [Google Scholar]
- 16. Holla VR, Adas F, Imig JD, Zhao X, Price E, Jr, Olsen N, Kovacs WJ, Magnuson MA, Keeney DS, Breyer MD, Falck JR, Waterman MR, Capdevila JH. Alterations in the regulation of androgen-sensitive Cyp 4a monooxygenases cause hypertension. Proc Natl Acad Sci USA 98: 5211–5216, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ito M, Oliverio MI, Mannon PJ, Best CF, Maeda N, Smithies O, Coffman TM. Regulation of blood pressure by the type 1A angiotensin II receptor gene. Proc Natl Acad Sci USA 92: 3521–3525, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kobori H, Prieto-Carrasquero MC, Ozawa Y, Navar LG. AT1 receptor mediated augmentation of intrarenal angiotensinogen in angiotensin II-dependent hypertension. Hypertension 43: 1126–1132, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kocinsky HS, Girardi AC, Biemesderfer D, Nguyen T, Mentone S, Orlowski J, Aronson PS. Use of phospho-specific antibodies to determine the phosphorylation of endogenous Na+/H+ exchanger NHE3 at PKA consensus sites. Am J Physiol Renal Physiol 289: F249–F258, 2005. [DOI] [PubMed] [Google Scholar]
- 20. Le TH, Crowley SD, Gurley SB, Coffman TM. The renin-angiotensin system. In: Seldin and Giebisch's The Kidney: Physiology & Pathophysiology 1–2 (vol. 1), edited by Alpern RJ, Hebert SC. Burlington, MA: Elsevier, 2007, p. 343–357. [Google Scholar]
- 21. Le TH, Fogo AB, Salzler HR, Vinogradova T, Oliverio MI, Marchuk DA, Coffman TM. Modifier locus on mouse chromosome 3 for renal vascular pathology in AT1A receptor-deficiency. Hypertension 43: 445–451, 2004. [DOI] [PubMed] [Google Scholar]
- 22. Lee IH, Campbell CR, Cook DI, Dinudom A. Regulation of epithelial Na+ channels by aldosterone: role of Sgk1. Clin Exp Pharmacol Physiol 35: 235–241, 2008. [DOI] [PubMed] [Google Scholar]
- 23. Loffing J, Flores SY, Staub O. Sgk kinases and their role in epithelial transport. Annu Rev Physiol 68: 461–490, 2006. [DOI] [PubMed] [Google Scholar]
- 24. Matsusaka T, Nishimura H, Utsunomiya H, Kakuchi J, Niimura F, Inagami T, Fogo A, Ichikawa I. Chimeric mice carrying ‘regional’ targeted deletion of the angiotensin type 1A receptor gene. Evidence against the role for local angiotensin in the in vivo feedback regulation of renin synthesis in juxtaglomerular cells. J Clin Invest 98: 1867–1877, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McGiff JC, Quilley J. 20-hydroxyeicosatetraenoic acid and epoxyeicosatrienoic acids and blood pressure. Curr Opin Nephrol Hypertens 10: 231–237, 2001. [DOI] [PubMed] [Google Scholar]
- 26. Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol 292: C82–C97, 2007. [DOI] [PubMed] [Google Scholar]
- 27. Miura S, Zhang J, Matsuo Y, Saku K, Karnik Activation of extracellular signal-activated kinase by angiotensin II-induced Gq-independent epidermal growth factor receptor transactivation. Hypertens Res 27: 765–770, 2004. [DOI] [PubMed] [Google Scholar]
- 28. Oliverio MI, Best CF, Kim HS, Arendshorst WJ, Smithies O, Coffman TM. Angiotensin II responses in AT1A receptor-deficient mice: a role for AT1B receptors in blood pressure regulation. Am J Physiol Renal Physiol 272: F515–F520, 1997. [DOI] [PubMed] [Google Scholar]
- 29. Oliverio MI, Madsen K, Best CF, Ito M, Maeda N, Smithies O, Coffman TM. Renal growth and development in mice lacking AT1A receptors for angiotensin II. Am J Physiol Renal Physiol 274: F43–F50, 1998. [DOI] [PubMed] [Google Scholar]
- 30. Ouyang X, Le TH, Roncal C, Gersch C, Herrera-Acosta J, Rodriguez-Iturbe B, Coffman TM, Johnson RJ, Mu W. Th1 inflammatory response with altered expression of profibrotic and vasoactive mediators in AT1A and AT1B double-knockout mice. Am J Physiol Renal Physiol 289: F902–F910, 2005. [DOI] [PubMed] [Google Scholar]
- 31. Reckelhoff JF, Romero JC. Role of oxidative stress in angiotensin-induced hypertension. Am J Physiol Regul Integr Comp Physiol 284: R893–R912, 2003. [DOI] [PubMed] [Google Scholar]
- 32. Riquier-Brison AD, Leong PK, Pihakaski-Maunsbach K, McDonough AA. Angiotensin II stimulates trafficking of NHE3, NaPi2, and associated proteins into the proximal tubule microvilli. Am J Physiol Renal Physiol 298: F177–F186, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev 82: 131–185, 2002. [DOI] [PubMed] [Google Scholar]
- 34. Ryan MJ, Didion SP, Mathur S, Faraci FM, Sigmund CD. Angiotensin II-induced vascular dysfunction is mediated by the AT1A receptor in mice. Hypertension 43: 1074–1079, 2004. [DOI] [PubMed] [Google Scholar]
- 35. Sandberg MB, Riquier AD, Pihakaski-Maunsbach K, McDonough AA, Maunsbach AB. ANG II provokes acute trafficking of distal tubule Na+-Cl− cotransporter to apical membrane. Am J Physiol Renal Physiol 293: F662–F669, 2007. [DOI] [PubMed] [Google Scholar]
- 36. Sasamura H, Hein L, Krieger JE, Pratt RE, Kobilka BK, Dzau VJ. Cloning, characterization, and expression of two angiotensin receptor (AT-1) isoforms from the mouse genome. Biochem Biophys Res Commun 185: 253–259, 1992. [DOI] [PubMed] [Google Scholar]
- 37. Schiff D, Purow BW. Neuro-oncology: isocitrate dehydrogenase mutations in low-grade gliomas. Nat Rev Neurol 5: 303–304, 2009. [DOI] [PubMed] [Google Scholar]
- 38. Steckelings UM, Kaschina E, Unger T. The AT2 receptor-a matter of love and hate. Peptides 26: 1401–1409, 2005. [DOI] [PubMed] [Google Scholar]
- 39. Vaziri ND, Wang XQ, Oveisi F, Rad B. Induction of oxidative stress by glutathione depletion causes severe hypertension in normal rats. Hypertension 36: 142–146, 2000. [DOI] [PubMed] [Google Scholar]
- 40. Widder JD, Guzik TJ, Mueller CF, Clempus RE, Schmidt HH, Dikalov SI, Griendling KK, Jones DP, Harrison DG. Role of the multidrug resistance protein-1 in hypertension and vascular dysfunction caused by angiotensin II. Arterioscler Thromb Vasc Biol 27: 762–768, 2007. [DOI] [PubMed] [Google Scholar]
- 41. Wolf G. Role of reactive oxygen species in angiotensin II-mediated renal growth, differentiation, and apoptosis. Antioxid Redox Signal 7: 1337–1345, 2005. [DOI] [PubMed] [Google Scholar]
- 42. Xue B, Pamidimukkala J, Hay M. Sex differences in the development of angiotensin II-induced hypertension in conscious mice. Am J Physiol Heart Circ Physiol 288: H2177–H2184, 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.