Abstract
Stressful environmental factors, such as a high-fat diet, can induce responses in the expression of genes that act to maintain physiological homeostasis. We observed variation in plasma concentrations of high-density lipoprotein (HDL) cholesterol across inbred mouse strains in response to high dietary fat intake. Several strains, including C57BL/6J, have stable levels of plasma HDL independent of diet, whereas other strains, including DBA2/J, show marked changes in plasma HDL. To explore this phenomenon further, we used publicly available data from a C57BL/6J × DBA/2J intercross to identify genetic factors that associate with HDL under high-fat diet conditions. Our analysis identified an epistatic interaction that plays a role in the buffering of HDL levels in C57BL/6J mice, and we have identified Arl4d as a candidate gene that mediates this effect. Structural modeling further elucidates the interaction of genetic factors that contribute to the robustness of HDL in response to high-fat diet in the C57BL/6J strain.
Keywords: structural equation model
the genetic architecture of complex diseases can involve multiple genes, with epistasic and gene-environment interactions. Atherosclerotic cardiovascular disease (CVD) is a major health problem. In the past decades, studies in mice and human have demonstrated that elevated low-density lipoprotein (LDL) cholesterol levels and reduced high-density lipoprotein (HDL) cholesterol levels are important risk factors for CVD (19, 28). Several single gene mutations have been shown to have significant effects on plasma cholesterol levels in humans (4, 20, 27). However, most common forms of atherosclerotic CVD are caused by multiple genetic and environmental factors and their interactions (2, 7, 22, 42).
In a robust biological system, natural selection has maintained a favorable genetic architecture to accommodate genetic and environmental perturbations. Robustness (or buffering) is the insensitivity of phenotypes to genetic or environmental perturbations (40). It can arise from mutation-selection equilibrium in a population or by stabilizing selection that favors polygenic genotypes contributing to robustness (14, 29). There are many ways in which morphological or physiological traits can be buffered from genetic or environmental perturbations (1, 30), including feedback control of cholesterol synthesis in humans (4).
Genetic mechanisms can also modulate phenotypic variation and epistatic interaction is one of the possible mechanisms that has been proposed (5, 15, 41). For example, genetic interaction involving a receptor-ligand pair has recently been used to predict risk of carotid artery disease in humans (9). Another mechanism of buffering involves balancing of pleiotropic effects that are mediated by multiple pathways with opposing directions of effect.
Genome-wide association (GWA) studies have provided a powerful approach to identify common genetic variants associated with human diseases and complex traits including plasma lipids (16). However, the variants that have been identified explain only a small fraction of the overall genetic contribution to disease risk (10). This may be explained in part by the undetected effects of gene-gene and gene-environment interactions. Environmental effects, such as dietary fat intake, are difficult to control in human GWA studies. Therefore, studies in animal models under controlled conditions provide an essential complement to GWA studies.
Mice have been established as a model system for human diseases including CVD (25). Inbred mouse strains show a broad range of variation in plasma lipids (24). However, there is limited information on the genetic mechanisms that contribute to this variation or to the response of plasma lipids to dietary fat intake. We use the publicly available C57BL/6J × DBA/2J (B6 × D2) intercross data to investigate HDL response to dietary fat intake. Our results indicate that epistatic interactions and balancing of allelic effects across multiple pathways can play a role in shaping the response of HDL to dietary fat.
MATERIAL AND METHODS
Plasma Lipid Profiling of Inbred Mouse Strains
Shockley et al. (35) used genome-wide expression profiling and measures of plasma lipids to study the effect of diet on 10 inbred mouse strains including C57BL/6J (B6) and DBA/2J (D2). Male and female mice (five animals for each sex) were raised on a high-fat diet (21) (30% Kcal from dairy fat containing by weight 1% cholesterol and 0.5% cholic acid) or on a low-fat diet (product 5K52 from LabDiets, St. Louis, MO) for 4 wk starting at 8 wk of age. At 12 wk of age blood samples were drawn from each mouse after fasting for 4 h. Plasma concentrations of total cholesterol and HDL cholesterol were assayed using a CX5 Delta Chemistry Analyzer (Bechman Coulter, Fullerton, CA). Gene expression profiling was performed on the liver tissue at 12 wk of age using Affymetrix Mouse 430 v2 arrays, with three animals per treatment group. These data are publicly available (http://cgd.jax.org/datasets/expression/10strain.shtml).
C57BL/6J × DBA/2J Intercross
We used publicly available data from a C57BL/6J × DBA/2J (B6 × D2) intercross consisting of 111 female mice (8, 32). These mice and the two parental strains were fed a low-fat diet until 12 mo of age, and were then switched to a high-fat diet for 4 mo. Blood samples were collected from mice at 16 mo following an overnight fast. Gene expression profiling on liver samples from this cross has been previously described. These data are available from (http://www.diabetesgenome.org/thirdpartydata/).
Quantitative Trait Locus Analysis
We performed linkage analyses for HDL cholesterol levels using single-quantitative trait locus (QTL) and two-QTL models to identify main effect and interacting QTL. Significance thresholds for the genome scans were estimated based on 1,000 permutations (6). We fit multiple regression models using all significant and suggestive QTL. All QTL analyses were carried out using R/qtl software (3). The HDL data were transformed in a logarithm base 10 scale.
Filtering Transcript Data
We carried out a series of filtering steps to eliminate genes from consideration as QTL candidates based on the genome-wide transcription data.
Step 1. Correlation analysis.
We identified transcripts that had significant correlation with HDL in the B6 × D2 cross population. With false discovery rate (FDR) control at the q<0.05 level (36), the significance threshold of the absolute correlation coefficient (r) is 0.31.
Step 2. expression QTL analysis.
We performed genome-wide scans for all of the expression traits based on a single-locus model. We selected transcripts that had significant expression QTL (eQTL) peaks falling within the 95% confidence intervals of any HDL QTL. Significance of individual transcripts was established using permutation analysis to obtain a genome-wide adjusted P value. We then applied an FDR adjustment to these P values to account for multiple testing across all transcripts (36). eQTL exceeding the q<0.05 level of significance were retained for further analysis.
Step 3. Genome scans with covariates.
In addition to the single-locus genome scan for HDL, we computed a second set of genome scans using each transcript as a covariate and computed change in logarithm of the odds (LOD) scores (ΔLOD) along the genome. This amounts to a comparison of two regression models
| (1) |
| (2) |
where Y denotes HDL cholesterol phenotype; Q indicates a QTL; T denotes a transcript; β0, β1, and β2 are coefficients; and ε is residual error. Significant ΔLOD values identify transcripts that are candidates for causal interactions with HDL. This is a generalization of methods previously proposed (31). A drop in the LOD score provides evidence to support a causal relationship (Q→T→Y). We also consider cases with an increase in LOD score, which provides evidence for more complex causal interaction involving all three variables (18).
We used permutation analysis to determine the ΔLOD threshold. In genome scans contrasting model 1 and model 2, we can compute the maximum ΔLOD for each QTL. We then permute T and repeat the scans 1,000 times, to obtain the null distribute of the genome-wide maximum ΔLOD. We transformed all transcripts using van der Waerden scores (17), and thus permutation analysis of a single transcript provides a null distribution for all of the transcripts. After multiple-test adjustment using FDR at q<0.05, the significant ΔLOD is 1.4.
Step 4. Epistasis affecting transcripts.
We performed genome-wide two-locus scans to identify QTL interactions affecting transcripts. Due to the computational demands of the all-pairs genome scans, we restricted attention to transcripts that have met the criteria of steps 1–3. Significant epistatic interactions were determined based on permutation thresholds with genome-wide adjustment (6).
Structural Equation Modeling
Structural equation models (SEMs) are a generalization of linear regression to include multiple response variables. We use the R/nlme package (26) to fit the component regression models and to compute the Bayesian information criteria (BIC) score for each SEM. BIC provides a model comparison criterion that penalizes models with larger numbers of free parameters and allows for the comparison of nonnested models (33). Changes in BIC >10 are considered highly significant, the 3–10 range provides strong evidence in favor of the model with the lower score, and models falling within 2 BIC units are considered to be more or less equivalent alternatives (12).
In the SEM, genotype effects can be treated as additive (coding the genotypes B/B, B/D, D/D as −1, 0, and 1) or as a three level factor (the free genetics model). Similarly, epistasic interactions can be treated as additive-by-additive, as a free genetic model with nine parameters, or as a mixed linear-by-discrete interaction. We fit SEMs using all three approaches (additive, free, and mixed) and arrived to similar conclusions in each case. For simplicity we describe the additive models below. We used the expected genotype at the peak LOD of each QTL in the regression (13). Path coefficients were estimated using the R/sem package (11).
RESULTS
Differential Response of Inbred Mouse Strains to Dietary Fat Intake
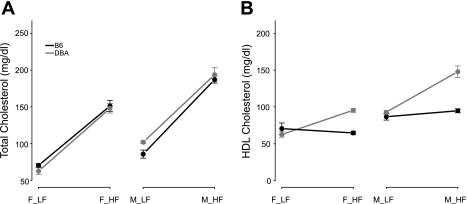
We have previously reported (35) a survey of 10 inbred mouse strains in which we observed that plasma HDL levels in several strains, including C57BL/6J (B6), are insensitive to a dietary perturbation, while other strains including DBA2/J (D2) respond significantly (P < 0.001) to the high-fat diet with increased HDL cholesterol levels (Fig. 1). Stable HDL levels in B6 mice and significant change in HDL in D2 in response to high-fat diet were also seen in the parents of the B6 × D2 cross (8). The B6 × D2 intercross progeny shows an average decrease in HDL of 7 mg/dl, intermediate between the parental values. Interestingly, the changes in HDL in D2 mice occur in different directions between these two studies. This might be explained by differences in the age of the mice, the duration of high-fat diet, or the specific components of diets. For example, cholic acid was incorporated in the high-fat diet (21) in our study (35).
Fig. 1.
Total and high-density lipoprotein (HDL) cholesterol in B6 and D2 mice. Inbred mouse strains respond differentially to fat intake. The mean levels of total (A) and HDL (B) cholesterol in female (F) and male (M) mice of strain C57BL/6J (B6) and DBA/2J (DBA) are shown for low-fat (LF) and high-fat (HF) diet conditions. The error bars in each panel are SE.
QTL Analysis for HDL Cholesterol
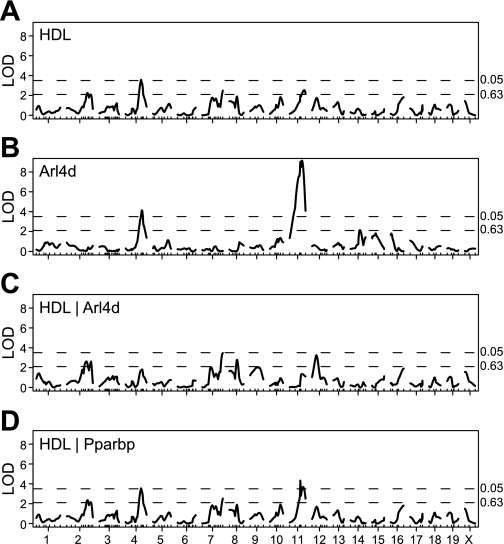
A genome scan for HDL cholesterol in the B6 × D2 cross (Fig. 2A) identified a significant QTL on Chromosome 4 at 126 Mb and three suggestive QTL on chromosome 2 at 164 Mb, chromosome 7 at 118 Mb, and on chromosome 11 at 115 Mb. The QTL on chromosomes 2, 4, and 7 have been reported previously (8, 37).
Fig. 2.
Genome scans. We scanned the genome for quantitative trait loci (QTL) in the B6 × D2 cross at a 2 cM resolution. Four scans are shown: HDL cholesterol with no covariates (A), Arl4d transcript with no covariates (B), HDL with Arl4d transcript as a covariate (C), and HDL with Pparbp transcript as a covariate (D). Dotted lines indicate significant (P = 0.05) and suggestive (P = 0.63) thresholds based on 1,000 permutations. Transcript data were transformed using van der Waerden scores (17). LOD, logarithm of the odds.
A two-locus genome scan of HDL cholesterol identified an epistatic interaction involving the main effect loci on chromosomes 4 and 11 and a second interaction between the chromosome 4 locus and a new QTL on chromosome 12 at 112 Mb. A multiple regression (Table 1) including all of the main effects and interactions, explains 57% of the variance in the HDL cholesterol, although this is almost certainly an overestimate due to model selection. The 4 × 11 interaction alone explains 6% of the total variance, and the 4 × 12 interaction accounts for 10% of the total variance after accounting for other terms in the model.
Table 1.
Multiple regression of all QTL and interactions explaining HDL cholesterol levels
| Term | df | SS (adj) | %Var. | P Value |
|---|---|---|---|---|
| Q2@164 | 2 | 0.827 | 3.8 | 2.1e-02 |
| Q4@126 | 10 | 6.981 | 32.2 | 7.7e-08 |
| Q7@118 | 2 | 1.485 | 6.8 | 1.2e-03 |
| Q11@115 | 6 | 2.133 | 9.8 | 4.0e-3 |
| Q12@112 | 6 | 2.727 | 12.6 | 5.5e-04 |
| Q4@126*Q11@115 | 4 | 1.323 | 6.1 | 1.6e-02 |
| Q4@126*Q12@112 | 4 | 2.233 | 10.3 | 5.6e-04 |
| Error | 90 | 9.2 | ||
| Total | 108 | 21.7 |
QTL, quantitative trait locus; HDL, high-density lipoprotein; df, degree of freedom; SS (adj), adjusted sum of square.
Analysis of Transcript Data
We identified 399 transcripts having significant correlations with HDL cholesterol (| r | > 0.31; FDR = 0.05). Of these, 205 are positively correlated and 194 are negatively correlated with HDL cholesterol. Among these 399 transcripts we found 95 significant eQTL (LOD > 3.5; P < 0.05), of which 89 were local and 6 were distant relative to the chromosome of the corresponding genes. We identified 25 eQTL within the 95% confidence intervals of the HDL QTL (Table 2). To further narrow the list of candidate genes, we used each of these 25 transcripts as a covariate in genome scans for HDL cholesterol. We identified 13 transcripts (Table 2) with ΔLOD >1.4 in these scans (Methods). Additionally, we carried out two-locus genome scans for these 13 transcripts, and we identified a single transcript, Arl4d that shared a genetic interaction with HDL.
Table 2.
Candidate QTL genes identified by using our procedure
| Chr* | Colocalize† | ΔLOD‡ | Gene Symbols |
|---|---|---|---|
| 2 | 8 | 3 | Cst3, Hsbp1, 9030622O22Rik |
| 4 | 11 | 8 | Arl4d, Cox7b, Fkbp5, Eif4 g3, Dsip1, Gnb1, Nudc, 5330417C22Rik |
| 7 | 3 | 1 | Arl6ip1 |
| 11 | 3 | 1 | Arl4d |
Chromosome of the HDL QTL.
Number of significant (q <0.05) expression QTL (eQTL) with logarithm of the odds (LOD) score peaks that fall within the 95% confidence interval.
Number of eQTL that exceed 1.4 ΔLOD when the corresponding transcript is included as a covariate.
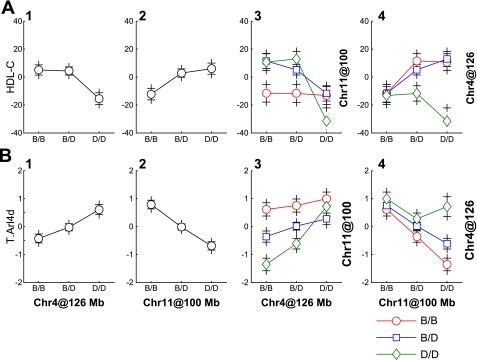
The Arl4d gene is physically located on chromosome 11. It has a large local QTL on chromosome 11 and a distant QTL on chromosome 4 (Fig. 2B), both of which correspond to HDL LOD peaks. Moreover, we identified a significant epistatic interaction between these loci. A visual comparison of the QTL effects on HDL cholesterol and on the Arl4d transcript reveals a similar pattern (Fig. 3). The reversed direction of the effects of the 4 × 11 interaction on HDL cholesterol compared with the Arl4d transcript is consistent with the observed negative correlation between these variables (r = −0.45; P < 0.001). The interaction pattern indicates that in the presence of homozygous B6 alleles (BB) on chromosome 11, the QTL on chromosome 4 no longer has an effect on the Arl4d transcript or on HDL cholesterol levels. The masking of the chromosome 4 QTL effect by BB genotypes on chromosome 11 suggests a genetic mechanism explaining the stability of HDL cholesterol in B6 mice on the high-fat diet.
Fig. 3.
Epistatic interactions. The interaction effects between QTL on chromosomes 4 and 11 for HDL levels (A) and the Arl4d transcript (B) are shown. Columns 1 and 2 show the main effects for QTL on chromosomes 4 and 11, respectively. Column 3 shows their joint effects with chromosome 4 genotypes on the x-axis and chromosome 11 genotypes indicated by grouping of points (BB, red; BD, blue; and DD, green). Column 4 shows the same joint effects with the roles of chromosomes 4 and 11 reversed. The transcript levels were transformed using van der Waerden scores (17). Values are shown as means ± SE. The HDL levels are expressed as deviations from the population mean value of 0.
Arl4d Transcript Mediates Genetic Effects on HDL Cholesterol
Genome scans with covariates can help to clarify the causal relationship among a QTL, a transcript, and a phenotype (18). When the Arl4d transcript is included as a covariate in a genome scan for HDL cholesterol, the LOD scores of both QTL on Chromosomes 4 and 11 decrease significantly (Fig. 2C). We were concerned that this decrease could be due to the large local QTL effect at Arl4d (115 Mb on Chr 11) acting as a surrogate for the genotype. To rule out this possibility we considered another nearby transcript, Pparbp, with a large local eQTL. In genome scans with the Pparbp transcript as a covariate, neither of the QTL peaks (on chromosomes 4 and 11) decreases (Fig. 2D), suggesting that Pparbp is not likely to be a mediator of the effect of the chromosome 11 QTL and alleviating concerns about the Arl4d result being due to a strong local QTL.
Interestingly, we detect a new QTL at 40 Mb on chromosome 12 in the genome scan of HDL with the Arl4d transcript as a covariate (Fig. 2C). The new locus is distinct from the interaction effect on HDL detected at 112 Mb on chromosome 12. An increased LOD score indicates that the chromosome 12 QTL (at 40 Mb) may have opposing effects, with the direct effect on HDL balanced by an indirect effect through the Arl4d transcript (18). As a result the QTL is masked in the standard genome scan (representing the sum of direct and indirect effects). When Arl4d is included as a covariate, the genome scan reflects only the effects of the direct path and the QTL is unmasked. D alleles at the chromosome 12 locus (at 40 Mb) are associated with decreased HDL and decreased Arl4d. The negative correlation (r = −0.45; P < 0.001) of HDL and Arl4d results in the net cancellation of these effects. Another new QTL, on chromosome 8 appears as a suggestive peak in the covariate scan, although it does not meet the ΔLOD > 1.4 criterion. The chromosome 8 locus was only marginally significant in the SEM analysis below.
SEM
We used SEM to gain further insight in the relationships among genetic loci, the Arl4d transcript, and HDL. We used BIC as a criterion to identify the best-fitting regression models for four cases: 1) Arl4d transcript with only genetic predictors, 2) HDL with Arl4d transcript as a predictor, 3) HDL with only genetic predictors, and 4) Arl4d transcript with HDL as a predictor (Table 3). In some cases, more than one model provided a reasonable fit to the data (within 2 BIC units of the best model). These near optimal models included the suggestive QTL on chromosome 8 or the 4 × 12 interaction.
Table 3.
Best-fitting regression models for SE analysis
| df | SS | F Value | P Value | Coef. | ||
|---|---|---|---|---|---|---|
| Model 1: Arld4 | ||||||
| Q4@126 | 1 | 15.54 | 27.54 | 8.169e-07 | 0.45 | |
| Q11@115 | 1 | 26.97 | 47.78 | 3.956e-10 | −0.70 | |
| Q12@40 | 1 | 3.13 | 5.55 | 2.031e-02 | −0.23 | |
| Q4:Q11 | 1 | 3.67 | 6.50 | 1.224e-02 | 0.35 | |
| Residuals | 104 | 58.69 | ||||
| Model 2: HDL | Arl4d | ||||||
| Arl4d | 1 | 22.15 | 36.93 | 2.039e-08 | −0.50 | |
| Q2@164 | 1 | 7.88 | 13.14 | 4.488e-04 | 0.30 | |
| Q7@118 | 1 | 8.99 | 15.00 | 1.881e-04 | −0.42 | |
| Q12@40 | 1 | 6.59 | 10.99 | 1.262e-03 | −0.38 | |
| Residuals | 104 | 62.38 | ||||
| Model 3: HDL | ||||||
| Q2@164 | 1 | 7.15 | 10.02 | 2.0e-03 | 0.35 | |
| Q4@126 | 1 | 10.99 | 15.40 | 1.6e-04 | −0.44 | |
| Q7@118 | 1 | 8.42 | 11.81 | 8.5e-04 | −0.42 | |
| Q11@115 | 1 | 4.37 | 6.13 | 1.5e-02 | 0.24 | |
| Q4:Q11 | 1 | 3.61 | 5.06 | 2.7e-02 | −0.35 | |
| Residuals | 103 | 73.47 | ||||
| Model 4: Arl4d | HDL | ||||||
| HDL | 1 | 22.15 | 44.56 | 1.2e-09 | −0.35 | |
| Q12@40 | 1 | 6.13 | 12.32 | 6.6e-04 | −0.39 | |
| Q4@126 | 1 | 4.97 | 9.99 | 2.1e-03 | 0.27 | |
| Q11@115 | 1 | 23.06 | 46.38 | 6.5e-10 | −0.67 | |
| Residuals | 104 | 51.70 | ||||
SS, sum of square; Coef., regression coefficients.
The key question that we wished to address is whether Arl4d is causally upstream of HDL. The best-fitting casual model (Arl4d → HDL), obtained by combing regression models (1) and (2), has score BIC = 362.08. The best-fitting reactive model (HDL → Arl4d), obtained by combing regressions (3) and (4) has score BIC = 370.84. The difference ΔBIC = 8.76 strongly supports the causal model.
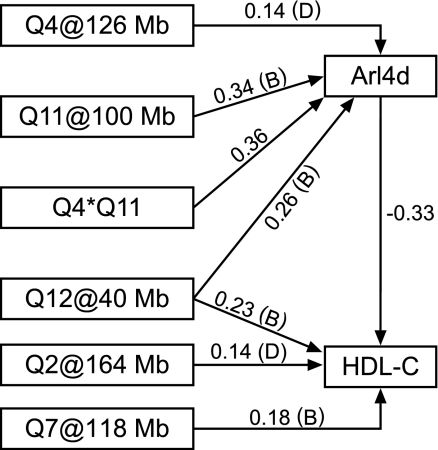
On the basis of model comparisons, we arrived at the best-fitting causal SEM (Fig. 4), the 4 × 11 interaction directly affects Arl4d transcript, which in turn affects HDL. There is no direct path from the 4 × 11 interaction term to HDL. The effect is mediated entirely through Arl4d. The QTL on chromosome 12 at 40 Mb has a pleiotropic effect on the Arl4d transcript and on HDL cholesterol. The overall effect of this QTL on HDL cholesterol is positive (B alleles increase HDL) along the direct path and the negative along the indirect path (B alleles decrease HDL). This balancing of effects explains the unmasking of the QTL when the Arl4d transcript is included as a covariate in the genome scan.
Fig. 4.
Structural equation model. The best-fitting structural equation model relating HDL cholesterol and the Arl4d transcript is shown. Genetic loci, indicated as Q(chromosome)@(position), are treated as additive effects in the regression. The interaction term (Q4*Q11) is also additive-by-additive. Connecting edges in the graph are directed indicating the hypothesized causal structure of the model. Numerical edge labels are standardized path coefficients indicating the relative magnitude of the regression coefficients. Parenthetic labels indicate the high allele for genetic effects.
DISCUSSION
Previous studies have indicated that genetic factors can have a significant influence on plasma concentration of HDL cholesterol in response to dietary fat (23, 35, 38). Our previous study of 10 inbred mouse strains, fed high- and low-fat diets indicated that plasma HDL cholesterol levels in several strains, including C57BL/6J, are insensitive to dietary fat intake, compared with the HDL cholesterol levels in responsive strains such as DBA/2J (35). We have identified two genetic mechanisms that help to explain the robustness of HDL to dietary perturbation in C57BL/6J mice.
The first genetic buffering mechanism involves the 4 × 11 epistatic interaction that was identified directly using the HDL trait but on further analysis appears to be mediated entirely through the Arl4d transcript. The pattern of interaction suggests that in the presence of a homozygous BB genotype at the chromosome 11 QTL, HDL levels are no longer sensitive to QTL genotype of the chromosome 4 locus.
A second genetic mechanism that contributes to genetic buffering of HDL also involves the Arl4d transcript. By including Arl4d as a covariate in the genome scan for HDL, we identified a previously undetected QTL on chromosome 12 (Fig. 2C). Structural modeling confirmed that this QTL has pleiotropic effects on both the Arl4d transcript and HDL cholesterol. The two paths that mediate the QTL effect on HDL act in opposite directions. In the direct path (it does not mean a direct biological pathway), B alleles are associated with increased HDL. In the indirect path, B alleles are associated with increased Arl4d, which in turn is negatively correlated with HDL.
The small size of the B6 × D2 intercross population limits the power for detection of QTL and epistatic interactions. Nonetheless, we were able to detect effects that individually explain 5% or more of total variation. Structural model of multiple transcripts based on limited data runs the risk of overfitting. Therefore, in our analysis of the B6 × D2 cross, we have focused on elucidating the relationship of Arl4d transcript to HDL. Multiple lines of evidence drew attention to the Arl4d transcript as a likely mediator of the genetic effects on HDL. It remains a likely possibility that additional QTL and transcripts could play key roles in the regulation of HDL in this cross (Table 2).
Causal interpretation of structural equation models that are obtained by fitting models to data must be approached with caution (34). The setting of an experimental cross between inbred strains is ideal for causal inference because hidden confounders can be ruled out due to the random assignment of genotypes during meiosis. We propose the hypothesis that intervention in Arl4d expression will alter dietary response in HDL cholesterol levels. However, establishing the validity of Arl4d as a potential therapeutic target for cholesterol-related CVD will require further experimental investigation.
Genotype combinations at multiple loci can interact to generate extreme phenotypes. In the B6 × D2 cross, mice with homozygous DD genotypes at both of the QTL loci on chromosomes 4 and 11 are predicted to have low HDL cholesterol levels beyond those observed in either parental strain. This phenomenon of transgressive segregation has also been observed in the BXD recombinant inbred populations (8, 39). It is the complement of genetic buffering and both are population level phenomena that reflect the effects of different combinations of alleles at multiple loci on a trait. Different allelic combinations can lead to stable phenotypes, or they can shift a quantitative phenotype outside of its normal range under certain environmental conditions. The loss of allelic combination that promotes buffering can result in a disease state such as CVD induced by cholesterol imbalance.
GRANTS
This study was supported by National Institute of General Medical Sciences Grants GM-070683 and GM-076468.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank Drs. E. E. Schadt and A. J. Lusis for making the B6 × D2 data publicly available. We also thank Dr. Beverly Paigen and her laboratory for their efforts on the strain survey experiment.
REFERENCES
- 1.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature 397: 168–171, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Arnett DK, Baird AE, Barkley RA, Basson CT, Boerwinkle E, Ganesh SK, Herrington DM, Hong Y, Jaquish C, McDermott DA, O'Donnell CJ. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease: a scientific statement from the American Heart Association Council on Epidemiology and Prevention, the Stroke Council, and the Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation 115: 2878–2901, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science 232: 34–47, 1986 [DOI] [PubMed] [Google Scholar]
- 5.Cheverud JM, Vaughn TT, Pletscher LS, Peripato AC, Adams ES, Erikson CF, King-Ellison KJ. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome 12: 3–12, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354: 1264–1272, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Colinayo VV, Qiao JH, Wang X, Krass KL, Schadt E, Lusis AJ, Drake TA. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mamm Genome 14: 464–471, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Crawford DC, Nord AS, Badzioch MD, Ranchalis J, McKinstry LA, Ahearn M, Bertucci C, Shephard C, Wong M, Rieder MJ, Schellenberg GD, Nickerson DA, Heagerty PJ, Wijsman EM, Jarvik GP. A common VLDLR polymorphism interacts with APOE genotype in the prediction of carotid artery disease risk. J Lipid Res 49: 588–596, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11: 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J. Structural equation modeling with the sem package in R. Struct Eq Model 13: 465–486, 2006 [Google Scholar]
- 12.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis, 2nd ed.Boca Raton, FL: Chapman and Hall/CRC, 2004 [Google Scholar]
- 13.Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324, 1992 [DOI] [PubMed] [Google Scholar]
- 14.Hansen TF. The evolution of genetic architecture. Annu Rev Ecol Evol Syst 37: 123–157, 2006 [Google Scholar]
- 15.Jasnos L, Korona R. Epistatic buffering of fitness loss in yeast double deletion strains. Nat Genet 39: 550–554, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41: 56–65, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann EL, D'Abrera HJM. Nonparametrics: Statistical Methods Based on Ranks. New York: McGraw-Hill, 1988 [Google Scholar]
- 18.Li R, Tsaih SW, Shockley K, Stylianou IM, Wergedal J, Paigen B, Churchill GA. Structural model analysis of multiple quantitative traits. PLoS Genet 2: e114, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusis AJ. Atherosclerosis. Nature 407: 233–241, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler Thromb 14: 133–140, 1994 [DOI] [PubMed] [Google Scholar]
- 21.Nishina PM, Lowe S, Verstuyft J, Naggert JK, Kuypers FA, Paigen B. Effects of dietary fats from animal and plant sources on diet-induced fatty streak lesions in C57BL/6J mice. J Lipid Res 34: 1413–1422, 1993 [PubMed] [Google Scholar]
- 22.Ordovas JM, Corella D, Kaput J. Nutrient-gene interactions in lipoprotein metabolism - an overview. Forum Nutr 60: 102–109, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Paigen B. Genetics of responsiveness to high-fat and high-cholesterol diets in the mouse. Am J Clin Nutr 62: 458S–462S, 1995 [DOI] [PubMed] [Google Scholar]
- 24.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis 57: 65–73, 1985 [DOI] [PubMed] [Google Scholar]
- 25.Paigen K. A miracle enough: the power of mice. Nat Med 1: 215–220, 1995 [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro J, Bates D, DebRoy S, Sarkar D, team tRC. nlme: Linear and Nonlinear Mixed Effects Models (R package version 3.1–93), 2009 [Google Scholar]
- 27.Plump AS, Smith JD, Hayek T, Aalto-Setala K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell 71: 343–353, 1992 [DOI] [PubMed] [Google Scholar]
- 28.Rader DJ. Molecular regulation of HDL metabolism and function: implications for novel therapies. J Clin Invest 116: 3090–3100, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice SH. A general population genetic theory for the evolution of developmental interactions. Proc Natl Acad Sci USA 99: 15518–15523, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutherford SL. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays 22: 1095–1105, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37: 710–717, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302, 2003 [DOI] [PubMed] [Google Scholar]
- 33.Schwarz GE. Estimating the dimension of a model. Annal Stat 6: 461–464, 1978 [Google Scholar]
- 34.Shipley B. Cause and Correlation in Biology: A User's Guide to Path Analysis, Structural Equations, and Causal Inference, Cambridge: Cambridge University Press, 2000 [Google Scholar]
- 35.Shockley KR, Witmer D, Burgess-Herbert SL, Paigen B, Churchill GA. Effects of atherogenic diet on hepatic gene expression across mouse strains. Physiol Genomics 39: 172–182, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Meth Mol 224: 149–157, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Su Z, Ishimori N, Chen Y, Leiter EH, Churchill GA, Paigen B, Stylianou IM. Four additional mouse crosses improve the lipid QTL landscape and identify Lipg as a QTL gene. J Lipid Res 50: 2083–2094, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svenson KL, Smith RV, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol 102: 2369–2378, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, Phillips SJ. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome 10: 335–348, 1999 [DOI] [PubMed] [Google Scholar]
- 40.Wagner A. Robustness and Evoluability in Living Systems. Princeton, NJ: Princeton University Press, 2005 [Google Scholar]
- 41.Warden CH, Yi N, Fisler J. Epistasis among genes is a universal phenomenon in obesity: evidence from rodent models. Nutrition 20: 74–77, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40: 161–169, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]