Abstract
Programmed genetic rearrangements in lymphocytes require transcription at antigen receptor genes to promote accessibility for initiating double-strand break (DSB) formation critical for DNA recombination and repair. Here, we showed that activated B cells deficient in the PTIP component of the MLL3 (mixed-lineage leukemia 3)–MLL4 complex display impaired trimethylation of histone 3 at lysine 4 (H3K4me3) and transcription initiation of downstream switch regions at the immunoglobulin heavy-chain (Igh) locus, leading to defective immunoglobulin class switching. We also showed that PTIP accumulation at DSBs contributes to class switch recombination (CSR) and genome stability independently of Igh switch transcription. These results demonstrate that PTIP promotes specific chromatin changes that control the accessibility of the Igh locus to CSR and suggest a nonredundant role for the MLL3-MLL4 complex in altering antibody effector function.
Mixed-lineage leukemia (MLL)–like complexes catalyze mono-, di-, and tri-methylation of histone H3 at lysine 4 (H3K4me), marks which all strongly correlate with active gene expression (1), but the direct role and specificity of these complexes during lymphocyte maturation are unknown. The PTIP (Pax interaction with transcription-activation domain protein-1, also known as Paxip1) protein stably associates with a subset of MLL-like complexes containing the MLL3 and MLL4 methyltransferases (2–4), suggesting a role for PTIP in transcriptional regulation. Embryonic lethality of Paxip1−/− mice at embryonic day 9.5 (5), however, has precluded any further elucidation of its biological function, with the exception of a role in adipogenesis (6).
Along with possible functions in transcriptional regulation, PTIP contains six BRCT (BRCA1 carboxy terminal) domains, implicating the protein in the DNA damage response (7). Indeed, upon treatment with ionizing radiation (IR), PTIP forms nuclear foci dependent on the γ-H2AX/MDC1/RNF8 damage response pathway and its own BRCT function, and also physically interacts with the DNA repair factor 53BP1 (7–9). PTIP-deficient cells are also hypersensitive to irradiation (9–11) and defective in homologous recombination (12). To what extent PTIP functions directly in double-strand break (DSB) repair, however, is unclear because loss of PTIP is also associated with defective proliferation (5) and a global decrease in H3K4me3 (3, 13), a chromatin mark of transcription initiation (14–16).
During V(D)J recombination in developing B cells in the bone marrow, recombination-activating gene 1/2 (RAG1/2)–mediated cleavage and DSB repair by the nonhomologous end-joining (NHEJ) pathway occur at accessible recombination signal sequence chromatin in antigen receptor genes and require actively transcribed gene elements (17). In response to ligand engagement from lipopolysaccharide (LPS), B cell receptor cross-linking, and/or cytokine stimulation, mature naïve immunoglobulin M (IgM)–expressing B cells in the periphery proliferate and become activated to undergo class-switch recombination (CSR) (17, 18). This second rearrangement event promotes recombination of an antigen recognition gene segment with different Igh constant regions to interact with different cell surface receptors for successful clearance of a pathogen (17, 18). During CSR, transcription of immunoglobulin heavy-chain (Igh) switch region chromatin is required for accessibility of AID (activation-induced cytidine deaminase) (19, 20), which leads to formation of DSBs both at the upstream switch (Sμ) region and one of the downstream switch regions (Sγ3, Sγ1, Sγ2b, Sγ2a, Sε, or Sα) (18). Subsequently, synapsis and DNA repair of the two switch regions are mediated by γ-H2AX/53BP1 and NHEJ proteins, leading to a recombined Igh locus (21). Given that DNA rearrangements in B lymphocytes require transcription and DNA repair at immunoglobulin genes and that PTIP is implicated in both of these processes, we investigated PTIP function in CSR.
Genome-wide H3K4me3 in LPS-stimulated PaxipΔ/Δ and control B cells
To determine how changes in histone H3K4 methylation regulate mature B cell differentiation, we used chromatin immunoprecipitation coupled with Illumina sequencing (ChIP-Seq) to compare genome-wide H3K4me3 profiles of resting murine B cells with B cells stimulated with LPS and antibody against IgD conjugated to dextran (αIgD-dextran, herein referred to as LPS stimulation), mitogens known to induce rapid proliferation and activation ex vivo. We observed that 1014 regions across the genome display more than a fourfold increase in H3K4me3 after LPS stimulation (fig. S1A and table S1). Only 13.2% of these LPS-induced H3K4me3 regions were localized within 4 kb of known transcriptional start sites, suggesting that LPS stimulation may initiate transcription at many uncharacterized genomic regions (fig. S1B).
Several genes whose transcripts are LPS inducible in B lymphocytes (18) displayed increased H3K4me3 upon LPS stimulation, including Il6, the Igh switch regions, and Aicda (encodes activation-induced cytidine deaminase) (fig. S1, C to E). In contrast, c-myc, which is induced during B cell activation but displays promoter-proximal polymerase II (Pol II) pausing (22), displayed similar H3K4me3 profiles in both resting and LPS-stimulated B cells (fig. S1E). These data support the notion that although c-myc may be poised for induced expression in mature B cells, Aicda, Il6, and the Igh switch regions are devoid of H3K4me3 until B cell activation, thereby comprising a different group of tightly regulated genes that have predominant control mechanisms that normally prevent initiation (15).
To investigate the genetic requirements and biological impact of LPS-induced H3K4me3, we crossed Paxipfl/fl mice with CD19-cre mice to generate B cell–specific PTIP knockout mice. Analysis of Cd19-cre Paxipfl/fl splenic B cells (herein referred to as PaxipΔ/Δ B cells) revealed reduced PTIP protein and transcript expression while displaying near-normal CD19+ B cell numbers and frequency of surface IgM+ cells (fig. S2, A to D). In contrast to primary fibroblasts and embryonic stem cells (5), PTIP-deficient B cells proliferated in a manner similar to that of controls in response to either LPS or LPS and interleukin-4 (IL-4) (fig. S2E). Together these data suggest that PTIP is largely dispensable for B cell development and cell cycle progression.
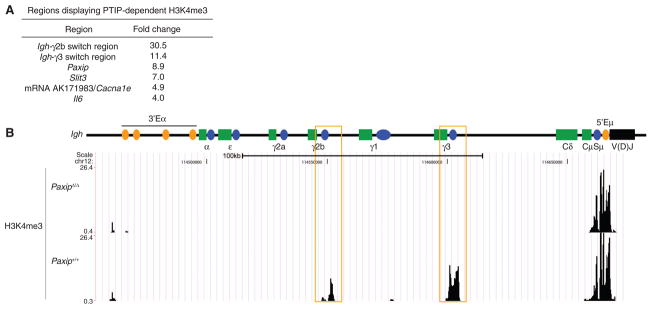
Normal proliferation of PaxipΔ/Δ B cells suggested the possibility of a more limited role for PTIP-mediated histone methylation in B cells compared to the global reduction of H3K4me3 observed in developing tissues (3, 13). We found that only six regions (Igh-γ2b, Igh-γ3, Slit3, Il6, Paxip1, and mRNA AK191783/Cacna1e) in PaxipΔ/Δ B cells displayed more than a fourfold decrease in H3K4me3 compared to controls, and 10 regions displayed more than a threefold decrease in H3K4me3 (Fig. 1A and fig. S3, A and B). No regions displayed consistently increased H3K4me3 in PaxipΔ/Δ B cells (fig. S3A). Consistent with PTIP deficiency in PaxipΔ/Δ cells, the Paxip gene itself also displayed reduced H3K4me3 (Fig. 1A) because of Cre-mediated deletion of its first exon (3). Taken together, our data demonstrate a selective role for PTIP in promoting histone methylation in activated B cells.
Fig. 1.
Genome-wide H3K4me3 changes in LPS-stimulated PaxipΔ/Δ B cells. (A) Table showing average fold change decrease of most affected regions from three independent H3K4me3 ChIP-Seq experiments with multiple mice. Values represent control/PaxipΔ/Δ ratios of sequence tag counts. (B) H3K4me3 ChIP-Seq profiles across the Igh constant region locus in B cells stimulated with LPS and α-IgD-dextran for 2 days. Data are representative of three independent experiments. In the illustration, green rectangles indicate constant (C) region exon segments, blue circles indicate switch (S) regions, orange ovals indicate enhancers (E), and the black rectangle indicates the antigen recognition V(D)J gene segment. The μ, δ, γ3, γ1, γ2b, γ2a, ε, and α isotypes correspond to immunoglobulins M, D, G3, G1, G2b, G2a, E, and A. LPS-induced switch regions are highlighted with orange boxes.
The Igh-γ2b and Igh-γ3 switch regions showed the greatest decrease in H3K4me3 from LPS-induced PaxipΔ/Δ B cells (Fig. 1, A and B). Moreover, PaxipΔ/Δ and control B cells stimulated with LPS and IL-4 displayed PTIP-dependent H3K4me3 at Igh-γ1 (fig. S4), indicating that PTIP also promotes H3K4me3 at other Igh switch regions under different conditions of B cell activation. Notably, the PTIP-dependent H3K4me3 at activated Igh switch regions occurs independently of AID-induced DNA damage (fig. S4), consistent with H3K4me3 associating with transcription rather than with DNA DSBs (14). PTIP deficiency, however, had no effect on H3K4me3 marking the Igh-μ enhancer (5′Eμ), μ switch repeat region (Sμ), or Igh-ε switch repeat region (Sε) under either stimulation condition (Fig. 1B and fig. S4), demonstrating the specificity of PTIP-mediated histone methylation. Normal H3K4me3 across the 5′Eμ region is consistent with the normal surface IgM expression observed on PaxipΔ/Δ B cells (fig. S2D).
Ig class-switching defects in PaxipΔ/Δ B cells
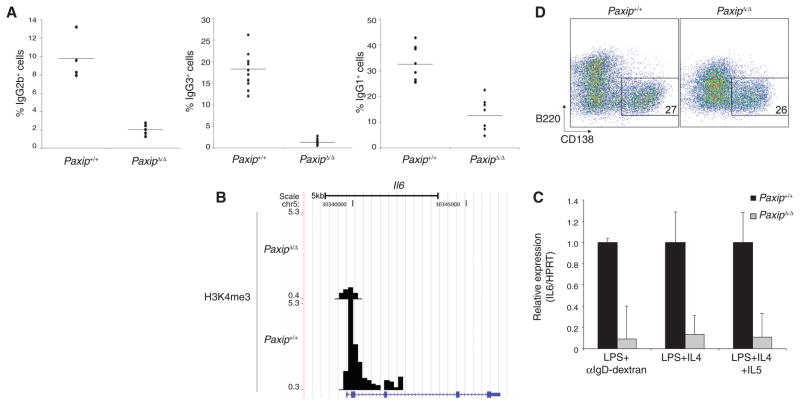
Aberrant regulation of Igh switch regions and Il6 have well-established implications for mature B cell function (18, 23). To understand the physiological relevance of PTIP-dependent H3K4me3 at Igh switch regions, we examined Ig class switching. Upon LPS stimulation, PaxipΔ/Δ B cells displayed 14- and 4.8-fold decreases in the frequency of IgG3 and IgG2b switching, respectively (Fig. 2A and fig. S5, A and B). PaxipΔ/Δ B cells also displayed a 2.6-fold decrease in IgG1 switching upon LPS and IL-4 stimulation (Fig. 2A and fig. S5, A and B). When CSR was assayed directly from genomic DNA of B cells stimulated with LPS and IL-4, PaxipΔ/Δ B cells displayed a similar defect in the level of Sμ-Sγ1 switch junctions (fig. S5C). As expected, PaxipΔ/Δ B cells showed normal cell proliferation (figs. S2E and S5, A and B) and cell survival (fig. S5D). We conclude that PTIP is critical for IgG3, IgG2b, and to a lesser extent, IgG1 class switching.
Fig. 2.
Ig CSR defects in PaxipΔ/Δ B cells. (A) Average percentage of IgG+ B cells from flow cytometric analyses of ex vivo stimulation (IgG2b+ and IgG3+ from LPS and α-IgD-dextran and IgG1+ from LPS and IL-4). Each dot represents an individual mouse, and the line represents the average (IgG2b+: 4.8-fold, P = 0.006; IgG3+: 14.1-fold, P = 0.00000005; IgG1+: 2.6-fold, P = 0.00003). (B) H3K4me3 ChIP-Seq profiles at the Il6 gene in B cells stimulated with LPS and α-IgD-dextran for 2 days. A UCSC (University of California Santa Cruz) gene annotation for Il6 is shown at the bottom. Data are representative of three independent experiments. (C) PaxipΔ/Δ-activated B cells display decreased Il6 transcript amounts. RT-qPCR analysis of Il6 transcripts from B cells stimulated for 3 days under the indicated conditions. (D) Flow cytometric analysis of B cells stimulated with LPS, IL-4, and IL-5 for 5 days and stained with anti-CD138 as a marker for plasma cells. Numbers indicate the percentage of total live CD138+ B220low B cells. Data are representative of two independent experiments.
IL-6 was further analyzed because it can function in both antibody secretion and the development and tumorigenesis of plasma cells (23) (Fig. 1A). We found that H3K4me3 at Il6 was both LPS inducible and PTIP dependent (fig. S1C and Fig. 2B) and that mRNA expression of Il6 was also impaired in PaxipΔ/Δ B cells (Fig. 2C). These defects, however, had no apparent effect on plasma cell differentiation ex vivo, as monitored by CD138 surface expression (Fig. 2D). Moreover, the IgG3 class-switching defect in PaxipΔ/Δ B cells was not rescued with exogenous recombinant IL-6 in the culture medium (fig. S6).
PTIP promotes Ig switch region transcription initiation
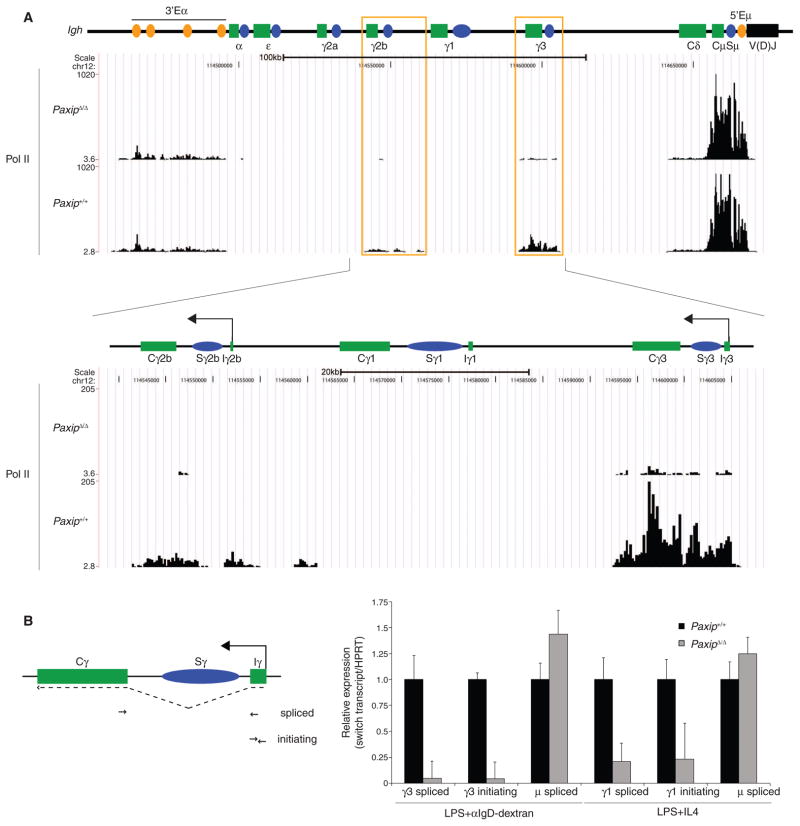
For most actively transcribed genes, H3K4me3 and Pol II accumulate within 2 kb of the transcription start sites (24). In contrast, H3K4me3 peaks at Igh switch regions are more broad, spanning up to 7 kb downstream of the germline transcript promoter, and include the mutagenic switch repeats and downstream of the switch region (Fig. 1B and figs. S4 and S7, A and B) (25). This broad H3K4me3 distribution at switch regions correlates with observed accumulation of Pol II at switch regions (Fig. 3A) (25, 26). Therefore, we considered whether PTIP might be important for elongation or splicing of switch transcripts. To investigate whether PTIP regulates transcription of Igh switch regions, we measured germline switch transcripts that had been spliced from the initiating (I) exon located 5′ of the switch region to the constant (C) exon located 3′ of the switch region from stimulated PaxipΔ/Δ and control B cells (Fig. 3B). IgM-expressing B cells express a μ sterile transcript, and we found that PaxipΔ/Δ B cells displayed near-normal μ switch transcript amounts in both conditions (Fig. 3B). This is consistent with the normal H3K4me3 patterns at Eμ and Sμ in the absence of PTIP (Fig. 1B and fig. S4). In contrast, both γ3 and γ1 spliced switch transcripts, important for IgG CSR, were severely impaired in PaxipΔ/Δ B cells under their respective stimulation conditions (Fig. 3B). Spliced transcripts at Igh-ε were unchanged in PaxipΔ/Δ B cells (fig. S7C), consistent with no change in H3K4me3 at this switch region in the absence of PTIP (fig. S4).
Fig. 3.
PTIP promotes Pol II association and transcription initiation at Igh switch regions. (A) Pol II ChIP-Seq profiles across the constant region locus in B cells stimulated with LPS and α-IgD-dextran for 2 days. Illustration is the same as in Fig. 1B. LPS-induced switch regions are highlighted with orange boxes. The lower panel shows the expanded profiles across γ2b, γ1, and γ3. Black arrows indicate sites of LPS- and α-IgD-dextran–induced transcription at γ3 and γ2b switch regions. “I” indicates initiating exon segment. (B) Left, illustration showing location of oligonucleotides used in RT-qPCR. Right, RT-qPCR analysis of spliced and initiating Igh switch transcripts from B cells stimulated under the indicated conditions for 3 days. Data are representative of two independent experiments.
To further delineate at which stage PTIP functions in promoting Igh-γ germline switch transcript expression, we also assayed for unspliced and initiating switch transcripts. PaxipΔ/Δ B cells showed similar deficiencies both in unspliced (fig. S7D) as well as in initiating switch transcripts (Fig. 3B) as were observed for spliced switch transcripts when we assayed across the IγCγ exon junction (Fig. 3B). We conclude that PTIP functions at a stage preceding pre-mRNA splicing to promote initiating Igh switch transcript expression.
In yeast, evidence suggests that initiating Pol II recruits the H3K4 methylase activity to mark transcription start sites (14). To test whether PTIP promotes Igh switch region transcripts at the level of Pol II association or downstream of pre-initiation complex assembly, we performed Pol II ChIP-Seq on PaxipΔ/Δ and control B cells stimulated with LPS. At Igh, although no major changes were observed at the μ, δ, or 3′ α enhancer (Eα) regions, Pol II association was severely impaired at both Igh-γ2b and γ3 regions (Fig. 3A). Furthermore, at the other sites displaying PTIP-dependent H3K4me3, including Il6 and Paxip itself, Pol II association was also impaired (fig. S8). We conclude that, upon LPS stimulation, PTIP is required for Pol II association at downstream switch regions along with a subset of the other regions displaying PTIP-dependent H3K4me3.
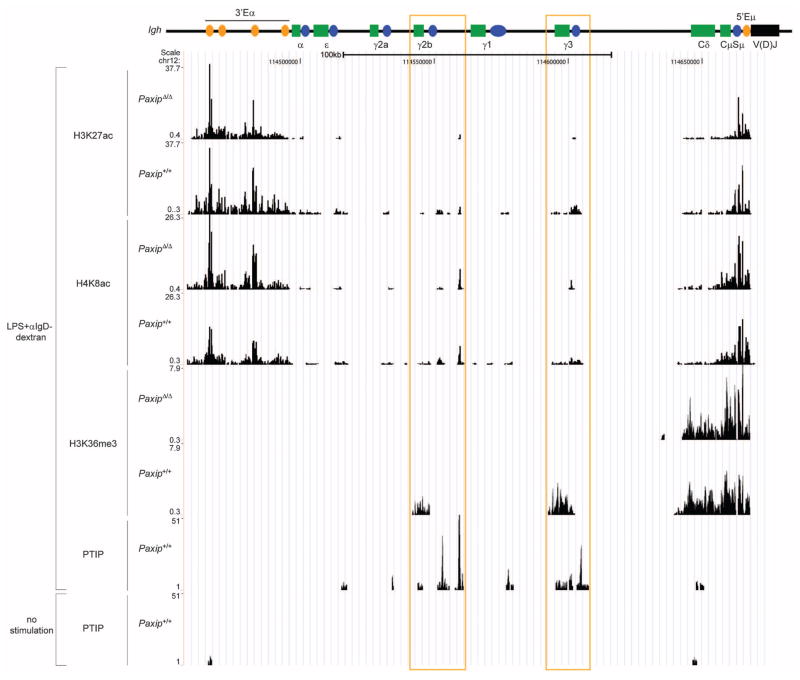
Histone acetylation facilitates decondensation of chromatin, which may promote transcriptional activation by increasing the accessibility of transcription factors and Pol II to promoters (27). To better understand the mechanism for PTIP-dependent Pol II association at Igh switch regions and other PTIP-affected regions, we performed ChIP-Seq for a number of histone acetylation marks (H2BK5ac, H3K9ac, H3K27ac, and H4K8ac) along with the H3K36me3 mark associated with transcription elongation (24). At Igh, our data indicate that PTIP is required for all of these histone modifications at activated switch regions, and, similar to H3K4me3 and Pol II, PTIP is dispensable for these modifications at the μ, δ, and 3′Eα regions (Fig. 4 and fig. S9). Thus, loss of H3K4me3 correlates with impaired histone acetylation. Although PTIP is also required for H3K4me2 at activated Igh switch regions (fig. S9), the H3K4me1 mark was present similarly across the Igh locus in both PaxipΔ/Δ and control B cells (fig. S9), indicating that PTIP is required for most but not all chromatin signatures of Igh switch region transcription. This PTIP dependency for other histone modifications largely holds true for all regions displaying PTIP-dependent H3K4me3 (fig. S8). Taken together, our results indicate that loss of PTIP correlates with impaired H3K4me2/3, histone acetylation, and Pol II association and suggest that chromatin accessibility at Igh switch regions may be regulated by a hierarchy of histone modifications.
Fig. 4.
PTIP directly localizes to Igh upon LPS stimulation and promotes histone acetylation and H3K36me3 at activated Igh switch regions. The indicated histone modification (H3K27ac, H4K8ac, and H3K36me3) and PTIP ChIP-Seq profiles across the Igh constant region locus in B cells either stimulated with LPS and α-IgD-dextran for 2 days or with no stimulation. Significant PTIP association is presented as fold change enrichment of control/PaxipΔ/Δ island tag counts. PTIP ChIP-Seq data in stimulated cells are representative of two independent experiments. Illustration is the same as in Fig. 1B. LPS-induced switch regions are highlighted with orange boxes.
To test whether PTIP-dependent H3K4me3 in activated B cells is directly mediated by the PTIP-associated methyltransferase complex, we performed genome-wide association analysis of PTIP and the shared MLL-like complex component, ASH2, using ChIP-Seq. Substantial enrichment of PTIP was observed at 9647 islands in LPS-stimulated control B cells, and localization was substantially enriched at promoter regions (fig. S10, A and B). By comparison, ASH2 binding in LPS-stimulated B cells was observed at 27,806 islands and was also enriched at promoter regions (fig. S10, A and B). Although the majority (78.6%) of LPS-induced H3K4me3 that we identified earlier (Fig. 1A and table S1) also displayed ASH2 binding (fig. S10C), only 27.8% show direct PTIP binding (fig. S10C), suggesting that PTIP function in gene expression during B cell activation may be very limited. Notably, nearly all (94.7%) of the LPS-induced H3K4me3 sites showing PTIP binding also displayed ASH2 binding (fig. S10C). These data are consistent with PTIP associating with an MLL-like complex, which colocalizes with H3K4me3 marks at transcription start sites (1–4). These data also suggest that PTIP may be important for targeting the MLL3-MLL4 complex to a subset of genomic loci. Indeed, substantial PTIP and ASH2 ChIP-Seq colocalization was observed at five of the six regions displaying PTIP-dependent H3K4me3, including Igh-γ3, Igh-γ2b, Il6, Slit3, and Paxip itself (Fig. 4 and figs. S8 and S11A).
By comparing resting and stimulated B cells, we also found that PTIP and ASH2 localization at Igh switch regions was dependent on LPS stimulation (Fig. 4 and fig. S11A), suggesting that the PTIP-associated complex is actively recruited to the Igh locus during B cell activation. Consistent with these data, localization of PTIP and another shared MLL-like complex component, RBBP5, was also observed near the Igh-γ1 transcription start site in LPS- and IL-4–stimulated B cells using ChIP-qPCR (fig. S11B). Furthermore, RBBP5 localization at Igh-γ1 was dependent on PTIP (fig. S11B), suggesting that PTIP is required for targeting the methyltransferase complex to Igh switch regions. As a specificity control, we did not observe enrichment of PTIP by ChIP-qPCR (quantitative polymerase chain reaction) near the Igh-γ1 transcription start site in LPS- and IL-4–stimulated mouse embryonic fibroblasts (MEFs) (fig. S11C). We conclude that PTIP-dependent H3K4me3 at Igh-γ3, Igh-γ2b, and Igh-γ1 in stimulated B cells results from direct association of the PTIP-associated H3K4 methyltransferase complex nearby their transcription start sites.
PTIP accumulation at DNA breaks contributes to CSR independently of its role in transcription
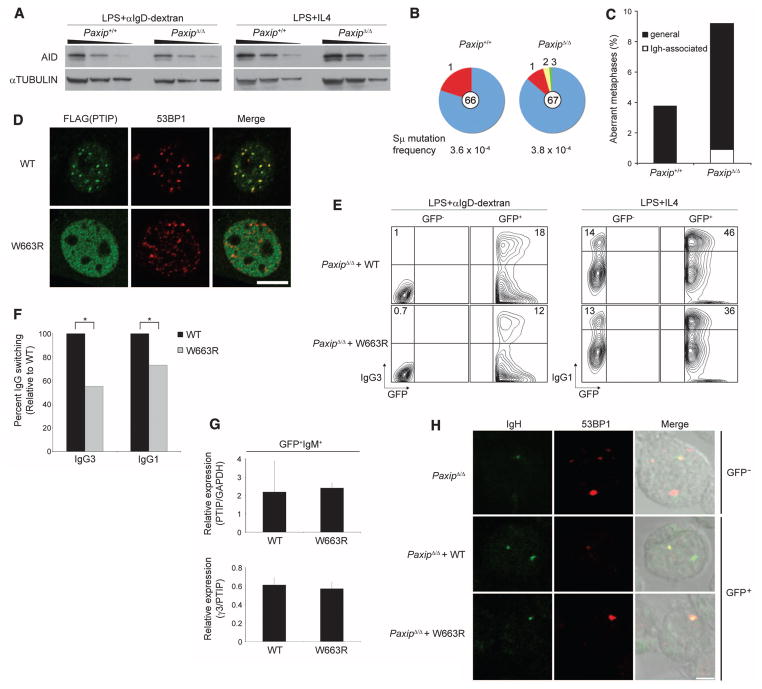
AID expression is required for mutations at Igh switch regions and for CSR (28–31); however, AID protein expression was similar in PaxipΔ/Δ and control B cells (Fig. 5A), consistent with normal H3K4me3 and Pol II at the Aicda locus in PaxipΔ/Δ B cells (fig. S1E). Similarly, we observed no major differences in H3K4me3, Pol II association, or expression of genes implicated in CSR, suggesting that disruption of Paxip does not cause indirect expression defects in known CSR-related factors (fig. S12, A and B). Moreover, a similar frequency of mutations was observed in PaxipΔ/Δ B cells in a region immediately 5′ of the Sμ repeat sequence (Fig. 5B), suggesting that AID targeting and mutation at Sμ occurs independently of PTIP.
Fig. 5.
PTIP foci formation contributes to Ig class-switching and genome stability. (A) Western blot of AID and α-TUBULIN from B cells stimulated for 3 days. (B) Sμ mutation analysis of genomic DNA from CFSE (carboxyfluorescein succinimydyl ester)–labeled B cells sorted for IgM and five cell divisions after stimulation with LPS for 4 days. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. Frequency of mutations per base pair sequenced and the total number of independent sequences analyzed are indicated below and in the center of each chart, respectively. Data are representative of two independent experiments. (C) Genomic instability analysis of metaphase spreads from B cells stimulated with LPS for 3 days (two mice of each genotype, Paxip+/+: n = 261; PaxipΔ/Δ: n = 218). Abnormalities specifically associated with the Igh locus (Igh-associated) or with all other chromosomes (general) are shown. (D) Immunofluorescent images of retrovirally infected MEFs exposed to 10-Gy irradiation with 4 hours recovery showing PTIP and 53BP1 foci formation. Bar, 20 μm. (E) Flow cytometric analysis of retrovirally infected PaxipΔ/Δ stimulated B cells stained 3 days after infection with IgG antibodies. Numbers indicate the percentages of total live IgG+ cells in GFP− (no retroviral expression) or GFP+ (retroviral expression) gates. (F) Average percentage of IgG switching from infection experiments. Data are from four independent experiments for each condition (*P < 0.05). (G) RT-qPCR of Paxip (top) and Igh-γ3 switch (bottom) transcripts from retrovirally infected GFP+IgM+ sorted PaxipΔ/Δ B cells stimulated with LPS. (H) Representative images of sorted B cells stimulated with LPS and IL-4 for 2 days and stained with α53BP1 antibody (red) before FISH detection of the Igh-Cα region (green). Among the total number of cells examined, those that contained 53BP1 and Igh-Cα foci were analyzed [GFP− PaxipΔ/Δ, 100 cells; wild-type (WT) control GFP+, 89 cells; W663R mutant GFP+, 97 cells]. Coincidence of a 53BP1 focus with one or both Igh-Cα alleles was detected in 42% of GFP− PaxipΔ/Δ, 30% of WT control GFP+, and 43% of W663R mutant GFP+ cells. Bar, 5 μm.
PTIP-mediated changes in Igh chromatin structure could also affect NHEJ of broken switch regions. To directly assay for alterations in DNA repair, we analyzed switch recombination junctions from successfully class-switched IgG1-positive B cells. We found that Sμ-Sγ1 junctions were indistinguishable between PaxipΔ/Δ and control, showing no differences in the amount of donor/acceptor homology at the junctions, suggesting that classical NHEJ is proficient in PaxipΔ/Δ B cells (fig. S13). Nevertheless, B cells with CSR-related DNA repair deficiencies show Igh-associated DNA breaks and general genomic instability (21, 32, 33). Indeed, we found that 8.3% of metaphases from PaxipΔ/Δ B cells showed general genomic instability outside of the Igh locus, compared to 3.8% in control cells, marked by chromosome breaks, chromatid breaks, and translocations (Fig. 5C). Furthermore, 0.9% of PaxipΔ/Δ metaphases displayed Igh-associated instability compared with none in control cells (Fig. 5C). We conclude that PTIP suppresses both general and Igh-associated genomic instability in B cells. DNA breaks and translocations at Igh may arise from AID-induced mutations at the μ, γ3, and γ2b regions because these are the transcriptionally active switch loci upon LPS stimulation (17, 18, 21). Given that AID is targeted to the upstream Sμ region (Fig. 5B) while transcription (and presumably accessibility) of downstream (Sγ) switch regions is impaired (Fig. 3, A and B), we presume that the observed Igh instability in PTIP-deficient cells is the result of unresolved AID-dependent breaks at Sμ.
53BP1 accumulates at sites of AID-induced Igh breaks and promotes DNA end-joining during CSR (21). We further examined a role for PTIP in DNA repair of CSR-associated breaks because PTIP contains six BRCT domains and can physically interact with 53BP1. A mutation of Trp663 to Arg (W663R) within BRCT domain 3 (BRCT3) of PTIP abolishes irradiation-induced foci formation of PTIP (8, 11), demonstrating the critical importance of this residue in targeting PTIP to DNA DSBs. To test whether accumulation of PTIP at sites of AID-induced DNA damage plays a role in Ig class switching, we generated retroviral expression constructs for B cell infection containing either control or W663R mutant PTIP and IRES-GFP (green fluorescent protein) to label the infected cells. After verifying that our ectopically expressed PTIP protein accumulated at sites of DSBs and the W663R mutant retrovirus expressed a DNA damage foci-defective PTIP mutant (Fig. 5D), we infected stimulated PaxipΔ/Δ B cells with retrovirus expressing either wild-type control or W663R mutant and measured class switching. First, comparing switching in GFP+ and GFP− cells infected with control retrovirus indicated that both IgG3 and IgG1 class-switching defects observed in PaxipΔ/Δ B cells are fully rescued (Fig. 5E), demonstrating that ectopic PTIP expression is functional. In contrast, the W663R mutant retrovirus restored IgG3 and IgG1 class switching only to 52% and 66%, respectively, of switching restored by control retrovirus (Fig. 5, E and F). This difference in the efficiency of rescue is not caused by impaired PTIP or γ3 spliced switch transcript amounts (Fig. 5G). These data indicate that the defect in class switching observed in W663R mutant B cells, unlike that in PaxipΔ/Δ B cells, is not due to aberrant germline switch transcription.
Mutation of W663 or any of the four C-terminal BRCT domains abolishes IR-induced 53BP1 coprecipitation with PTIP (8, 11). To determine whether disruption of Paxip or W663 impairs 53BP1 accumulation at Igh in B cells undergoing CSR, we infected PaxipΔ/Δ B cells with either W663R mutant or control retrovirus and processed the cells for immunocytochemistry–fluorescence in situ hybridization (FISH) to visualize protein and DNA simultaneously (30). W663R mutant and control B cells showed a similar percentage of 53BP1/Igh colocalizing (Fig. 5H), suggesting that defective CSR in the W663R mutant was not caused by impaired 53BP1 accumulation at Igh. These data support our observations (fig. S14) and those of others (11) that PTIP is dispensable for IR-induced 53BP1 foci formation in MEFs.
Discussion
Observations from mice with Igh switch promoter deletions and subsequent biochemical studies (17–20) have together demonstrated that switch-region transcription targets AID activity to Igh and is required for AID-dependent DSB formation and class switching (fig. S15, step 3). Histone modifications at the Igh locus and interactions with the transcription machinery have been suggested to be important for AID accessibility and targeting (25, 34, 35), but there has been little genetic evidence to support this notion. Our experiments call attention to PTIP as a critical factor for directly mediating histone modifications and promoting transcription initiation of germline Igh switch regions to target AID activity and also functioning, subsequently, in the repair of AID-induced DNA breaks.
PTIP promotes Igh switch transcription initiation
We propose that the downstream Igh constant region locus, as a cluster of gene segments that lack H3K4me3, Pol II, and transcription initiation, is an example of a tightly repressed gene cluster (15) that, upon B cell stimulation, requires PTIP to alleviate regulatory mechanisms that ensure proper targeting and stability of the switch regions (36). Because disruption of PTIP does not appear to alter MLL3-MLL4 complex integrity (2), our data suggest that PTIP confers specificity for recruitment of MLL3-MLL4 activity to particular genomic regions. Based on the observations that PTIP can directly interact with the PAX2 transcription factor (37) and mediate recruitment of H3K4me activity to a PAX2-dependent promoter (3), we speculate that PTIP-mediated H3K4me results from direct interaction with DNA-binding transcription factors (fig. S15, step 1) (36). In this model, upon PTIP-associated MLL3-MLL4 complex recruitment, H3K4me3 is catalyzed at transcription start sites (fig. S15, step 2). The H3K4me3 mark could then recruit acetylases and chromatin remodelers that alter chromatin structure at transcription start sites, promoting recruitment and/or stabilization of Pol II to initiate transcription (fig. S15, step 2). Nevertheless, we cannot exclude an alternative model in which PTIP stabilizes a transcription factor–DNA complex at the Igh locus leading to more efficient transcription initiation by Pol II, which in turn promotes H3K4 di- and trimethylation.
PTIP functions in DNA repair during class switching
γ-H2AX amplifies the DNA damage signal, in part, by accumulating the E3 ubiquitin ligase RNF8 to promote H2A/H2AX ubiquitination critical for subsequent accumulation of PTIP to DSBs (9, 11). Because CSR-associated DSBs are marked by γ-H2AX foci (30) and both H2AX- and RNF8-deficient mice also exhibit mild defects in CSR due to DSB repair defects (38–40), we speculate that, after AID-dependent DSB formation (fig. S15, step 3), PTIP may act downstream of H2AX and RNF8 to facilitate synapsis and/or repair efficiency of CSR-associated DNA breaks (fig. S15, step 4). Although DNA damage–induced accumulation of PTIP promotes efficient Ig class switching, this DNA repair function, however, cannot entirely account for the severe class-switching defects observed in PaxipΔ/Δ B cells, which arise to a larger extent from impaired transcription initiation of downstream Igh switch regions.
Supplementary Material
Acknowledgments
We thank G. Gutierrez-Cruz, V. Sartorelli, M. Kruhlak, K. Cui, and Q. Tang for technical assistance. This work was supported by funding from the Intramural Research Program of the NIH, National Cancer Institute, and Center for Cancer Research to A.N.; the Division of Intramural Research at the National Heart, Lung, and Blood Institute to K.Z.; and an NIH Director’s Challenge Award on “Epigenomic regulation of mammalian development and differentiation” to A.N., K.Z., and R.C. M.C.N. is a Howard Hughes Medical Institute investigator. The authors declare no competing financial interests and apologize for not being able to cite all primary references due to space limitations. All sequence data have been deposited in the Gene Expression Omnibus database (accession number GSE20852) at http://ncbi.nlm.nih.gov/geo/.
Footnotes
References and Notes
- 1.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Cho YW, et al. J Biol Chem. 2007;282:20395. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel SR, Kim D, Levitan I, Dressler GR. Dev Cell. 2007;13:580. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Issaeva I, et al. Mol Cell Biol. 2007;27:1889. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho EA, Prindle MJ, Dressler GR. Mol Cell Biol. 2003;23:1666. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YW, et al. Cell Metab. 2009;10:27. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manke IA, Lowery DM, Nguyen A, Yaffe MB. Science. 2003;302:636. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 8.Munoz IM, Jowsey PA, Toth R, Rouse J. Nucleic Acids Res. 2007;35:5312. doi: 10.1093/nar/gkm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Prindle MJ, Dressler GR, Yu X. J Biol Chem. 2009;284:18078. doi: 10.1074/jbc.M109.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jowsey PA, Doherty AJ, Rouse J. J Biol Chem. 2004;279:55562. doi: 10.1074/jbc.M411021200. [DOI] [PubMed] [Google Scholar]
- 11.Gong Z, Cho YW, Kim JE, Ge K, Chen J. J Biol Chem. 2009;284:7284. doi: 10.1074/jbc.M809158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Takenaka K, Takeda S. Genes Cells. 2010;15:243. doi: 10.1111/j.1365-2443.2009.01379.x. [DOI] [PubMed] [Google Scholar]
- 13.Fang M, et al. Development. 2009;136:1929. doi: 10.1242/dev.026559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shilatifard A. Annu Rev Biochem. 2006;75:243. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 15.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. Cell. 2007;130:77. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh TY, Cuddapah S, Cui K, Zhao K. Proc Natl Acad Sci USA. 2006;103:15782. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Adv Immunol. 2005;86:43. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 18.Stavnezer J, Guikema JE, Schrader CE. Annu Rev Immunol. 2008;26:261. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Nat Immunol. 2003;4:452. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhuri J, et al. Nature. 2003;422:726. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 21.Nussenzweig A, Nussenzweig MC. Cell. 2010;141:27. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lis J. Cold Spring Harb Symp Quant Biol. 1998;63:347. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 23.Potter M. Immunol Rev. 2003;194:177. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Schones DE, Zhao K. Curr Opin Genet Dev. 2009;19:127. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. J Exp Med. 2009;206:1817. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopal D, et al. J Exp Med. 2009;206:1237. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Carey M, Workman JL. Cell. 2007;128:707. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Muramatsu M, et al. Cell. 2000;102:553. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 29.Revy P, et al. Cell. 2000;102:565. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 30.Petersen S, et al. Nature. 2001;414:660. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue K, Rada C, Neuberger MS. J Exp Med. 2006;203:2085. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco S, et al. Mol Cell. 2006;21:201. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Callén E, et al. Cell. 2007;130:63. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Nambu Y, et al. Science. 2003;302:2137. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 35.Kuang FL, Luo Z, Scharff MD. Proc Natl Acad Sci USA. 2009;106:5288. doi: 10.1073/pnas.0901368106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Materials and methods are available as supporting material on Science Online.
- 37.Lechner MS, Levitan I, Dressler GR. Nucleic Acids Res. 2000;28:2741. doi: 10.1093/nar/28.14.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandran S, et al. Proc Natl Acad Sci USA. 2010;107:809. [Google Scholar]
- 39.Santos MA, et al. J Exp Med. 2010;207:973. doi: 10.1084/jem.20092308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, et al. J Exp Med. 2010;207:983. doi: 10.1084/jem.20092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.