Abstract
Background
We recently identified a compound, clemizole hydrochloride, that inhibits NS4B’s RNA binding and HCV replication. Although significant, clemizole’s antiviral effect is moderate (8μM EC50 against an HCV-2a clone). We hypothesized that combination of clemizole with other anti-HCV agents can increase the antiviral effect over that achieved with each drug alone, and could also decrease emergence of viral resistance.
Methods
Luciferase reporter-linked HCV replication assays were used to study the antiviral effects of drug combinations including clemizole. Data was analyzed using Loewe additivity and Bliss independence models for synergy, and resistance studies were performed using HCV colony formation assays.
Results
Clemizole’s antiviral effect was highly synergistic with the HCV protease inhibitors SCH503034 and VX950, without toxicity. In contrast, combinations of clemizole with either interferon, ribavirin, or the nucleoside (NM283) and non-nucleoside (HCV796) HCV polymerase inhibitors were additive. Furthermore, combination of clemizole with SCH503034 decreased the frequency of resistant mutants compared to treatment with either drug alone. Finally, no cross resistance to clemizole of SCH503034-resistant mutants (or vice versa) was observed.
Conclusions
Clemizole can yield high level synergy with the protease inhibitors class. Inclusion of clemizole in future anti-HCV cocktails can represent an attractive paradigm for increasing current virologic response rates.
Keywords: NS4B, RNA binding inhibitors, antiviral treatment, synergy, protease inhibitors, hepatitis C
Background
Over 150 million individuals are infected with hepatitis C virus (HCV) worldwide. Current standard of care (SOC), which consists of interferon-ribavirin combinations, is inadequate for the majority of patients [1]. The 9.6-kb positive single-stranded RNA HCV genome encodes a polyprotein which is cleaved into structural proteins (form the mature virus), and nonstructural (NS) proteins (facilitate viral genome replication) [2]. The NS3/4A protease and the NS5B RNA-dependent RNA polymerase have been the main focus of anti-HCV specific drug discovery efforts thus far. Indeed, several inhibitory compounds against these targets suppress viral replication in HCV-infected patients [3]. Monotherapy, however, was associated with the emergence of viral resistance andtreatment failure [3]. Furthermore, resistant mutations conferred resistance to additional inhibitors from the same class, excluding this class from these patients’ armamentarium [3]. While combining these emerging drugs with SOC regimens improves response rates [3], resistance and inadequate efficacy in non-responders to SOC remain important problems. Thus, there is an urgent need for drugs directed at novel HCV targets. Such drugs could be used in combination with SOC regimens or hopefully in future interferon-sparing cocktails.
The NS4B protein is a key player in HCV replication. It induces the formation of a novel intracellular membrane structure [4], which represents the site of HCV replication, and is required to assemble the other NS proteins within these membrane-associated replication complexes [5]. Disrupting NS4B function thus represents an attractive new anti-HCV strategy.
We have recently shown that an arginine rich-like motif within NS4B mediates binding to the 3′-terminus of the negative HCV strand and HCV RNA replication [6]. 18 pharmacological inhibitors of this activity were identified in a high-throughput screen using a novel microfluidics platform [6]. A lead compound, clemizole, was also found to inhibit HCV RNA replication [6]. This H1-histamine receptor antagonist has been used in humans (albeit for a different indication) and was well tolerated [7]. Although significant, clemizole’s antiviral effect is moderate with an EC50 of 8μM against HCV genotype 2a in the human hepatoma derived cell line, Huh7.5. Like other anti-HCV agents, genotype 1 clemizole-resistant mutants can be selected in vitro [6].
We hypothesized that combining clemizole with other anti-HCV agents can increase the antiviral effect achieved with one active drug alone and decrease emergence of viral resistance. Here we report that clemizole’s antiviral effect is highly synergistic with the emerging anti-HCV specific drugs, SCH503034 and VX950 targeting the HCV protease, without host cell toxicity. These results have exciting implications for novel strategies for combating HCV infections.
Methods
Compounds
Clemizole and ribavirin were purchased from Sigma and Interferon-α-B2 from PBL biomedical labs. SCH503034, VX950, NM283 and HCV796 were a gift from Leslie Holsinger (Virobay, Palo Alto). 10mM stocks were prepared and stored at −20°C. Clemizole and ribavirin were solubilzed in H20. SCH503034, VX950, NM283 and HCV796, were solubilzed in DMSO. Interferon was solubilized according to the manufacturer’s instructions.
Plasmids
FL-J6/JFH-5′C19Rluc2AUbi that consists of a full-length genotype 2a HCV genome and expresses Renilla luciferase was a gift from Dr. C.M. Rice [8]. Bart-79I was previously described [9]. Bart-79I-luc was made by replacing the neomycin-phospho-transferase gene by a Firefly luciferase gene in the Bart-79I plasmid.
Cell cultures and electroporation
Huh-7.5 cells were electroporated with in vitro transcribed FL-J6/JFH-5′C19Rluc2AUbi or Bart79I-luc RNAs, as described [6]. Cells were pooled and seeded in 96-well plates (~2–3×104 cells/well). Medium was replaced at 24hr and daily after. Cells were grown in 4 replicates in the presence of serial dilutions of the inhibitory compounds. Untreated cells with or without corresponding concentrations of DMSO were used as negative controls for DMSO and water-soluble compounds, respectively. After 72hr, cells were subjected to alamarBlue-based viability assays and luciferase assays.
Viability assays
Cells were incubated for 2hrs at 37ºC in the presence of either 10% alamarBlue reagent (TREK Diagnostic Systems) or CellTiter-Blue reagent (Promega). Fluorescence was detected using FLEXstationII 384 (Molecular Devices). Depending on the inhibitory compound’s solvent, water or DMSO, signal was normalized relative to untreated samples or samples grown in the presence of DMSO, respectively.
Luciferase assays
Viral RNA replication was determined using Renilla (for genotype 2a replicons) or Firefly (for genotype 1b replicons) luciferase assays (Promega). Cells were washed with PBS and shaked in lysis buffer. Following 15 minute incubation at −80°C and thawing, luciferase assay buffer containing the assay substrate was injected and luciferase activity was measured using a Berthold LB96V luminometer. Signal was normalized as described above. Experiments were repeated three times, each time with 4 replicates.
Focus formation assay
2×104 Huh7.5 cells were infected in triplicates with cell culture-grown HCV titered at 1.2×104 TCID50/ml, as described [10]. 2hrs after infection, cells were washed and treated daily with various concentrations of clemizole and SCH503034, either alone or in combination. After 72hrs, samples were subjected to viability assays, followed by fixation in 4% formaldehyde and permeabilization with saponin. HCV core protein was detected with primary anti-core monoclonal and secondary goat anti-mouse Alexa-594-conjugated antibodies. Foci were counted under an inverted microscope.
Colony formation assays
Huh7 cells electroporated with genotype 1b subgenomic HCV replicon (Bart-79I) [11] were treated in duplicates with various concentrations of clemizole and SCH503034, either alone or in combination. G418 was included to provide selective pressure on HCV replicon cells such that cells bearing wild-type replicons that are sensitive to the antiviral drugs are expected to die, while cells bearing resistant replicons are expected to grow and form colonies after 3 weeks. Plates were stained with crystal violet and the frequency of resistance was determined (number of colonies/number of input cells).
Selection of resistant mutants
Established HCV replicon-harboring cells [11] were passaged in the presence of neomycin and increasing concentration of either clemizole (1–16μM) or SCH503034 (0.25–2.5μM) in 5 replicates. Colonies that grew in the presence of the compounds were pooled, passaged 15–20 times and the replicating HCV RNA was subjected to sequence analysis[9]. Whole cell RNA electroporations were performed as described [6].
Analysis of combination data
Combination data were analyzed using the Loewe additivity and Bliss independence drug interaction models [12, 13]. CalcuSynTM (Biosoft, , Cambridge, UK) was used to quantify differences between observed effects and predicted ones. Drugs were mixed at fixed molar ratios matching their equipotent concentrations which were maintained during serial dilutions [12–14]. An isobologram was plotted [12, 13], demonstrating lines of theoretical additivity and experimental EC50, EC75, and EC90 values for the combination. Synergy, antagonism and additivity are indicated by values plotted to the left of the corresponding lines of additivity, their right or on these lines, respectively. Combination indices (CIs) at the EC50, EC70, and EC90 levels were also determined [15]. Indices<1 indicate synergy, indices=1 additivity and those>1 antagonism.
The MacSynergy II program (kindly provided by Prof. M.N. Prichard) was used to analyze data according to the Bliss independence model [16, 17]. The combination’s effect is determined by subtracting the experimental values from theoretical additive values [16]. A three-dimensional differential surface plot demonstrates synergy as peaks above a theoretical additive plane and antagonism as depressions below it [16]. Matrix data sets in four replicates were assessed at the 95% confidence level for each experiment [13, 16, 17]. Synergy (volume under the curve) and log volume were calculated. As suggested by Prichard et al. [17], such data sets should be interpreted as follows: volumes of synergy or antagonism at values of <25μM2% are insignificant, 25–50μM2%-minor but significant, 50–100μM2%-moderate and probably important in vivo, and >100μM2%-strong and likely to be important in vivo.
Statistical analysis
EC50s and CC50s were measured by fitting data to a three parameter logistic curve using the formula (where a, b and c represent minimum binding, maximum binding and logEC50 or LogCC50, respectively) (BioDataFit, Chang Bioscience).
Results
Clemizole’s antiviral effect is highly synergistic with SCH503034, a HCV protease inhibitor (PI)
We hypothesized that clemizole’s antiviral effect may be synergistic with emerging anti-HCV specific agents targeting other viral proteins. We first studied the antiviral activity of clemizole in combination with a PI currently studied in phase 2 trials, SCH503034 (Boceprevir) [18]. Following electroporation with J6/JFH (genotype 2a) HCV RNA genome harboring a luciferase reporter gene [8], Huh7.5 cells were grown in the presence of various concentrations of the individual compounds and their combinations. Luciferase assays and alamarBlue-based viability assays were performed at 72hr. Treatment with either compound alone resulted in a concentration-dependent inhibition of HCV replication. The average EC50 of clemizole alone was 8μM (p<0.05), as previously reported [6], with a CC50 of 35±0.5μM (p<0.05) (measured by alamarBlue-based assays and CellTiter-Blue assays). While EC50s between ~0.2μM to 0.574μM have been reported for SCH503034 [18, 19], and similarly determined by us for genotype 1b, the average EC50 using 2a genotype luciferase reporter gene assay was 0.8μM with a CC50>100μM (p<0.05) (Table 1).
Table 1. Inhibitory activity and cytotoxicity of the drugs used in this study.
Antiviral activity and cytotoxicityof the drugs used.
| Compound | EC50a,b | CC50c,d | ||||
|---|---|---|---|---|---|---|
| 2a genotype | 1b genotype | Measured by others (1b genotype) | 2a genotype | 1b genotype | Measured by others (1b genotype) | |
| clemizole | 8μM | 23±7μM | NA | 35±0.5μM | 40±5μM | NA |
| SCH-503034 | 0.8μM | 0.21±0.034μM | 0.21–0.57μM | >100μM | >100μM | >100μM |
| VX-950 | 0.3μM | ND | 0.35–0.56μM | ND | ND | 26.7–82μM |
| ITMN-191 | 0.08μM | 0.002μM | 0.002μM * | ND | ND | >50μM |
| Interferon** | 2.8±0.4 (u/ml) | ND | 1.8 (u/ml)*** | ND | ND | >10,000(u/ml) |
| Ribavirin | 14±3μM | ND | 15μM*** | >100μM | ND | NA |
| NM-283 | 1.16±0.2μM | ND | 1.23±0.52μM | ND | ND | >100μM |
| HCV-796 | 0.068±0.008μM | ND | 0.017±0.005μM* | ND | ND | >100μM |
Antiviral effect was determined using HCV J6-JFH (genotype 2a) and/or Bart79I (genotype 1b) luciferase reporter-linked replication assays after 3 days of treatment. EC50s were compared to EC50 values previously measured by others using conventional assays using genotype 1b replicons.
EC50 (half maximal effective concentration) - concentration of a drug which inhibits viral replication by 50%.
Cell viability was determined by an alamarBlue-based assay in parallel with the EC50 determinations.
CC50 (half maximal cytotoxic concentration) – concentration of a drug which causes a cytotoxic effect in 50% of the cells.
Numbers represent mean ± standard deviation.
Please, note the low EC50s for the polymerase inhibitor HCV796.
Interferon-α has important interactions with the adaptive and innate immune responses, however it also has potent antiviral activity. The latter is not believed to be mediated by a direct action on the virus or replication complex but rather, appears to act by inducing interferon-stimulated genes (ISGs), which establish a non-virus-specific antiviral state within the cell [38].
Measured in 2a genotype.
The two compounds’ combination resulted in a greater inhibition than either compound alone at all tested concentrations (Fig. 1A). For example, while SCH503034 alone at a concentration of 2.5μM decreased viral replication by ~1log, when combined with 5μM clemizole viral replication was inhibited by ~3logs. Furthermore, no significant cytotoxicity was measured with either compound alone or with the above combinations (unshown data). These results suggest that even relatively low concentrations of clemizole have a dramatic effect on viral replication when added to SCH503034.
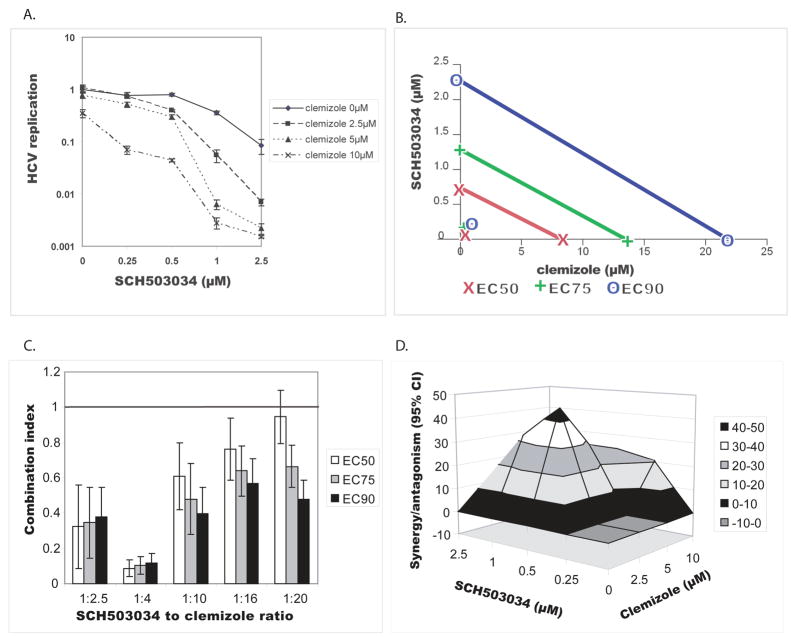
Fig. 1. Clemizole’s antiviral effect is highly synergistic with SCH503034, a HCV protease inhibitor.
Huh7.5 cells electroporated with a full-length J6/JFH HCV RNA genome harboring a luciferase reporter gene were grown in the presence of increasing concentrations of clemizole and SCH503034 either individually or in various combinations. Luciferase reporter-linked replication assays were performed at 72hr in parallel with alamarBlue-based viability assays. The experiment was performed 3 times each with 4 replicates. Four types of analyses are presented.
A. Dose response curves of SCH503034 in the absence or presence of increasing concentrations of clemizole. Error bars represent standard deviation.
B. Isobolograms (generated using CalcuSynTM). The lines denote the expected additive EC50, EC75, and EC90 values for the drug combination as calculated from the monotherapies. The experimental EC50, EC75, and EC90 values for the combination are marked by x, +, o, respectively. Note that these values plot to the far left of the corresponding lines, indicating synergy.
C. Combination Indices of SCH503034-clemizole combinations at the EC50, EC75 and EC90 measured at various drug ratios. A combination index greater than 1.0 indicates antagonism and a CI less than 1.0 indicates synergism. Error bars represent standard deviation.
D. Differential surface plot at the 95% confidence level (generated using MacSynergyII). The three-dimensional plot represents the differences between the actual experimental effects and the theoretical additive effects at various concentrations of the two compounds. Only statistically significant (95% confidence interval) differences between the two were considered at any given concentration. Peaks above the theoretical additive plane indicate synergy, whereas depressions below it indicate antagonism. The colors indicate the level of synergy/antagonism.
To formally characterize the SCH503034-clemizole combination’s interaction, a data sub-set where the drugs were mixed at a fixed molar ratio matching their equipotent concentrations (SCH503034 to clemizole ratio of 1:10), was analyzed using CalcuSynTM. As shown in the resulting isobolgram (Fig. 1B), the calculated EC50, EC75, and EC90 values for the SCH503034-clemizole combinations plotted far to the left of the corresponding lines of additivity, suggesting that the tested combinations are indeed synergistic [12, 13]. At an equipotent ratio of 1:10, the CIs at the EC50, EC70, and EC90 were 0.61, 0.479 and 0.397, respectively (Fig. 1C) [15]. Being below 0.9, these indices confirm that the interaction is synergistic. These CIs are similar in magnitude to the most potent synergistic interaction measured by others between HCV PIs and polymerase inhibitors [20]. While the interaction was found to be synergistic at any tested ratio, lowest CIs were measured at SCH503034 to clemizole ratio of 1:4 (Fig. 1C).
To confirm the nature of this interaction, to adjust for the various concentration ratios, and to better quantify the degree of the observed synergy, we further analyzed the data by a mathematical model, MacSynergy [16, 17]. 4 by 3 matrix data sets in four replicates were assessed at the 95% confidence level for each of the three experiments performed. The clemizole-SCH503034 combination had antiviral effects that were significantly more potent than the theoretical additive effects, supporting that this combination was indeed synergistic (Fig 1D). No evidence of antiviral antagonism was seen with any of the tested doses. The calculated synergy and log volume were 210μM2% and 19, respectively. According to the criteria suggested by Prichard et al. [17] (see methods), the clemizole-SCH503034 combination is considered strong and will likely be important in vivo. Importantly, since there was no cellular toxicity with either drug alone at the studied concentrations and no increase in cytotoxicity when used in combinations, the measured synergy is indeed specific and does not reflect synergistic toxicity.
The synergy of clemizole-SCH503034 combination is not genotype-specific
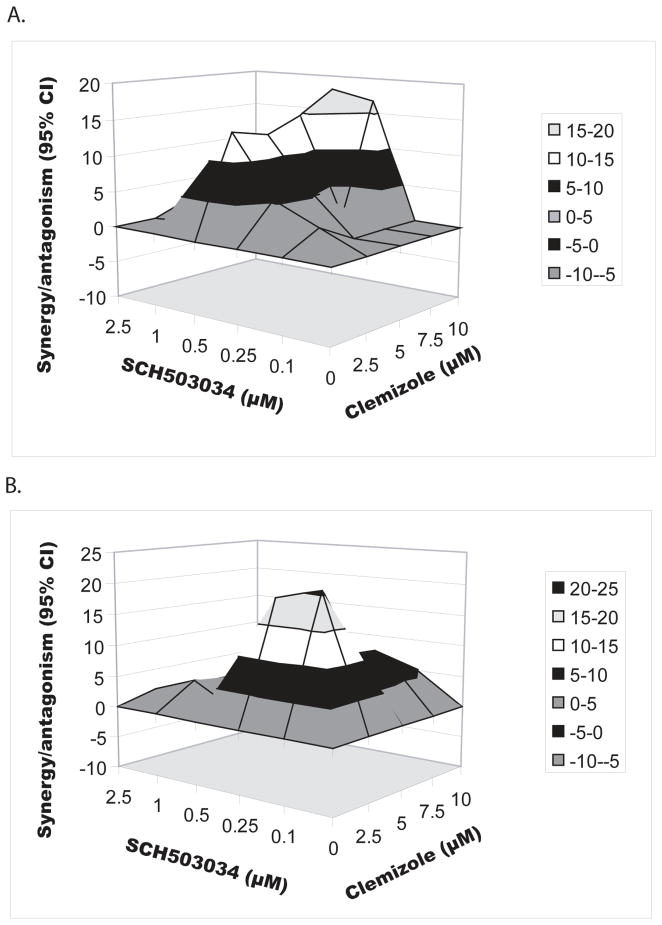
To determine whether the observed synergy of the clemizole-SCH503034 combination is genotype-specific, the experiments outlined above were repeated using a subgenomic genotype 1b Bart79I HCV replicon harboring a luciferase reporter gene [8]. In contrast to its effect in genotype 2a, very mild concentration-dependent inhibition of HCV replication was measured following 72hr treatment with clemizole alone, with an average EC50 of 24±6μM (p=0.01, R2=0.85) and an average CC50 of 40±5μM (p<0.05) (Table 1). It is possible that the lower sensitivity of this assay compared with the 2a luciferase reporter gene assay (resulting from lower level of genotype 1b replication compared with the genotype 2a clone) accounts for the difference in clemizole’s EC50s between genotypes. Alternatively, the difference might result from differential antiviral activity of clemizole against the two genotypes. Selection of clemizole-resistant mutants in 1b genotype replicon cells [6], suggests that clemizole does have an antiviral effect against genotype 1b. We thus favor the former possibility. Nevertheless, even low concentrations of clemizole surprisingly had a significant effect on genotype 1b viral replication when added to escalating concentrations of SCH503034, with a synergy volume of 100.04μM2% (MacSynergy) (Fig. 2A). Importantly, no cellular toxicity was measured at the concentrations used. These results suggest that the highly synergistic antiviral effect of combined clemizole-SCH503034 treatment is not genotype-specific. Since infection with genotype 1 HCV is the most common in the United States [21], and tends to be the least responsive to current SOC regimens [22], the synergistic antiviral effect of the clemizole-SCH503034 combination is important.
Fig 2. The clemizole-SCH503034 combination is synergistic across genotypes and in HCV-infected cells.
The antiviral effect of the clemizole-SCH503034 combination was determined using (A) genotype 1b luciferase reporter-linked replication assays, and (B) focus formation assays in cells infected with cell culture-grown HCV. Assays were performed following 72hr of treatment. Differential surface plot at the 95% confidence level are shown (generated using MacSynergyII). Peaks above the theoretical additive plane indicate synergy, whereas depressions below it indicate antagonism. The colors indicate the level of synergy/antagonism.
Clemizole-SCH503034 combination is synergistic in HCV-infected cells
To determine whether the clemizole-SCH503034 combination is synergistic in inhibiting direct viral replication (versus indirect assessments using luciferase reporter genes) we studied its antiviral effect by focus formation assays using cell culture-grown HCV [10]. While the average foci number in untreated wells was 46, lower numbers were counted with each drug alone in a dose-dependent manner. When combined, the two drugs resulted in substantially more potent antiviral effects than either compound alone. Importantly, neither drug alone nor the combinations showed cytotoxicity at the concentrations tested (unshown data). The synergy volume was 113μM2% (MacSynergy) (Fig. 2B). These results suggest that the highly synergistic antiviral effect of the clemizole-SCH503034 combination is also achieved in the context of viral infection.
The synergistic effect of NS4B RNA binding inhibitors and PIs combinations appears generalizable
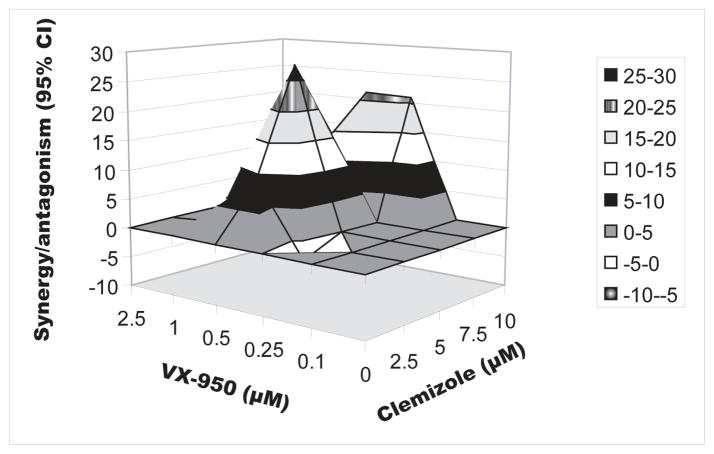
We hypothesized that the observed synergistic antiviral effect is also achieved when combining other NS4B RNA binding inhibitors with different HCV NS3 PIs. The antiviral effect of clemizole in combination with VX950 (Telaprevir), another PI [23], was thus determined. Genotype 2a luciferase reporter-linked assays and viability assays were performed as described above. The EC50 of VX950 alone was measured at ~300nM, similarly to prior reports [23, 24] (Table 1). In most concentrations tested, the combined drugs resulted in substantially more potent antiviral effects than the corresponding single agents (Fig. 3) with a synergy volume 97.51μM2% (MacSynergy). An insignificant antagonistic effect appeared in a single combination mixture with an antagonism volume of −2.83 μM2%. Importantly, neither drug alone nor the combinations showed cytotoxicity at the concentrations tested (unshown data). Furthermore, we have recently embarked on a clemizole derivatization program and identified a variety of such derivative molecules that have potency similar to, or greater than, clemizole (to be published elsewhere). When combined with SCH503034, one tested clemizole derivative demonstrated significant synergistic effects similar to the parental compound (unshown data). Taken together, these results suggest that the synergistic antiviral effect of the clemizole-SCH503034 combination may be generalizable and may reflect a broad synergism potential between the PI and NS4B RNA binding inhibitor classes of drugs. Since SCH503034 and VX950 are both ketoamide PIs, however, it remains to be determined whether combinations of the macrocyclic PIs, such as ITMN191 and BILN2061, with NS4B RNA binding inhibitors are similarly synergistic.
Fig. 3. The synergistic effect of NS4B RNA binding inhibitors and protease inhibitors combinations appears generalizable.
MacSynergy analysis of the antiviral effect of clemizole in combination with VX950 by luciferase reporter-linked replication assays after 72hr of treatment. A differential surface plot at the 95% confidence level is shown. Peaks above the theoretical additive plane indicate synergy, whereas depressions below it indicate antagonism. The colors indicate the level of synergy/antagonism.
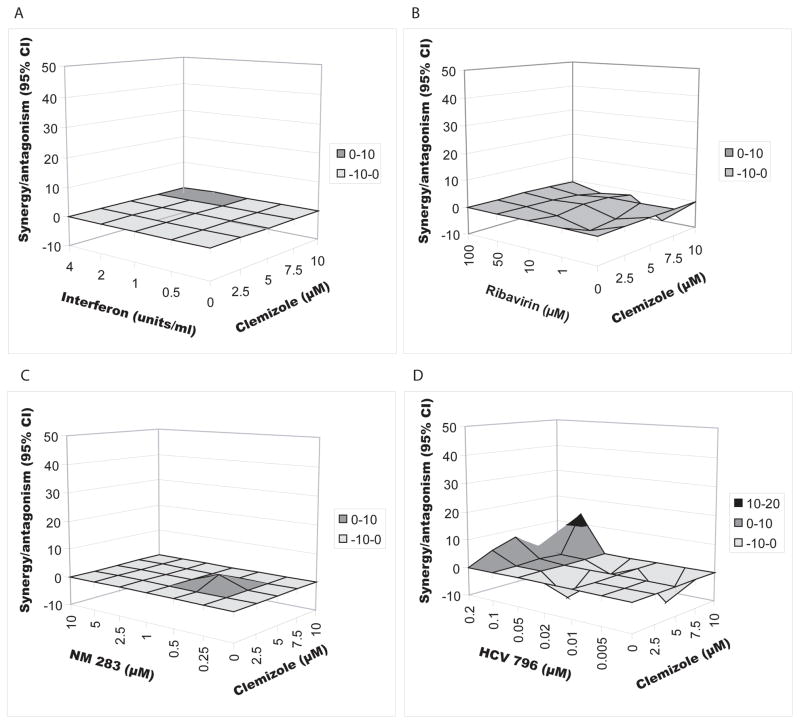
Combinations of clemizole with either interferon, ribavirin, a nucleoside analog or a non-nucleoside analog polymerase inhibitors are not synergistic but additive
We then studied the antiviral activity of clemizole in combination with either interferon, ribavirin, or two polymerase inhibitors [25, 26]: NM283 (valopicitabine), a nucleoside analog, or HCV796, a non-nucleoside analog. Genotype 2a luciferase reporter-linked assays were performed, as described above. EC50s of the individual compounds are shown in Table 1 [27]. The combination of clemizole with any of these compounds resulted in antiviral effects that were not significantly different from the theoretical additive effects (Fig. 4) (MacSynergy). Synergy volumes of 0.4μM2% and 3.57μM2%, and an antagonism volume of −15μM2% were measured for the combinations of clemizole with interferon, NM283 and ribavirin, respectively, indicating additivity. The synergy volume for the clemizole-HCV796 combination was 31.35μM2% with an antagonistic volume of −33.26μM2%, suggesting that overall this combination is largely additive too. Two-dimensional analysis using CalcuSyn yielded similar results (unshown data).
Fig. 4. Combinations of clemizole with either interferon, ribavirin, a nucleoside analog or a non-nucleoside analog polymerase inhibitors are not synergistic but additive.
MacSynergy analysis of the antiviral effect of clemizole in combination with interferon alpha (A), ribavirin (B), NM283 (C) and HCV796 (D) as measured by luciferase reporter-linked replication assays after 72hr of treatment. Differential surface plots at the 95% confidence level are shown. Peaks above the theoretical additive plane indicate synergy, whereas depressions below it indicate antagonism. The colors indicate the level of synergy/antagonism.
Clemizole-SCH503034 combinations significantly reduce the frequency of phenotypic resistance
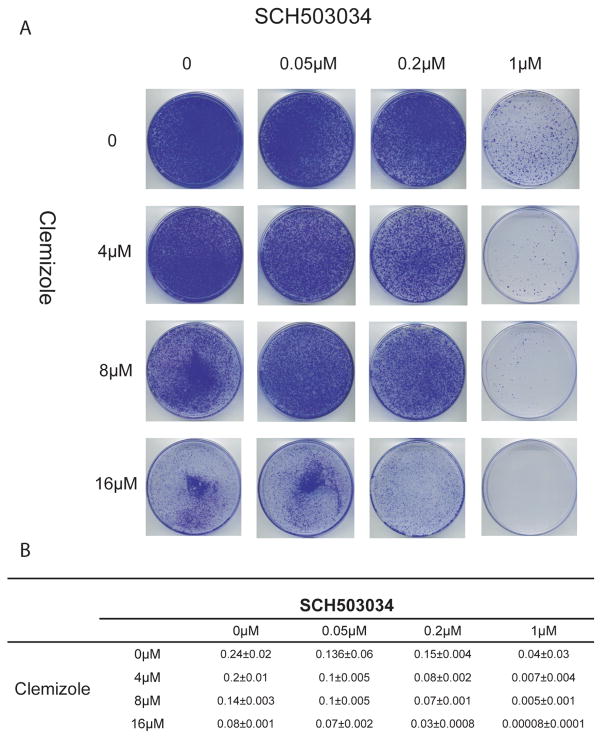
To test the hypothesis that clemizole-SCH503034 combinations can decrease the emergence of phenotypic viral resistance, we performed HCV colony formation assays in the presence of clemizole and/or SCH503034. There was an inverse correlation between the number of colonies and the compounds’ concentration (Fig. 5). The addition of clemizole even at a low concentration to SCH503034 significantly decreased the frequency of resistant colonies compared with SCH503034 alone. Other combinations of anti-HCV treatment were similarly shown by others to have such an effect on frequency of resistance [28] [29].
Fig. 5. Combinations of clemizole with SCH503034 significantly reduce the frequency of phenotypic resistance.
Huh7 cells electroporated de novo with a genotype 1b subgenomic HCV replicon were treated in duplicates with various concentrations of clemizole and SCH503034, either alone or in combinations, in the presence of G418 selection. Following 3 weeks of treatment, plates were stained with crystal violet.
A- Representative plates
B- Colonies were counted using Image-J (NIH) and their number was used to calculate the frequency of resistance (number of colonies/number of input cells).
To confirm that the emerged colonies indeed harbored mutations associated with resistance to the respective inhibitors, HCV RNA replicating in cells from pools of drug-resistant colonies was isolated and subjected to sequence analysis. As expected, replicons selected under SCH503034 pressure harbored mutations within the NS3 coding region, such as the A156T/V mutations previously shown to confer resistance to SCH503034 [28]. Similarly, previously described clemizole-resistant mutations were again selected within the NS4B coding region and the 3′-terminus of the negative viral genome in replicons extracted from cells treated with clemizole [6]. The effect of clemizole on the frequency of HCV resistance provides further rationale for its use in combination therapy with NS3 PIs.
There is no cross-resistance between clemizole and SCH503034
To confirm that there is no cross-resistance among these two classes of inhibitors, either SCH503034 or clemizole-resistant mutants were selected in HCV replicon-harboring cells and the HCV RNA was subjected to sequence analysis. None of the 5 independent SCH503034-treated pooled clones harbored replicons with mutations that mapped to the NS4B or the 3′ negative terminus. Similarly, no replicons that harbored mutations in NS3′s coding region were identified in 5 pooled clones treated with clemizole.
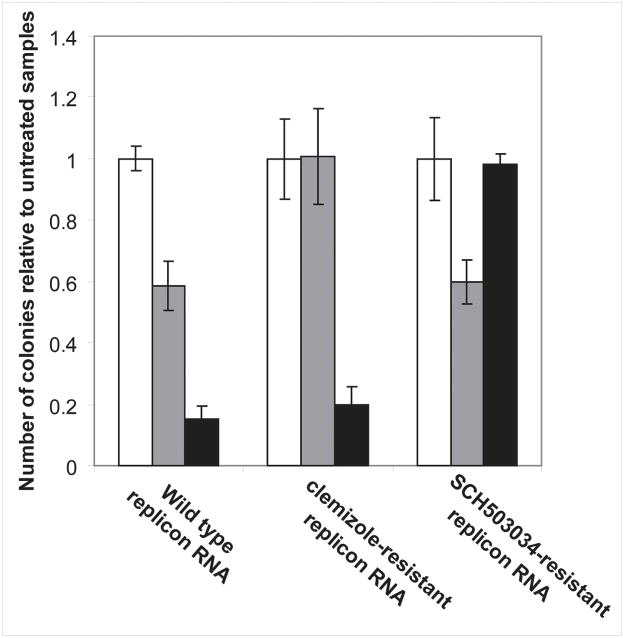
Lastly, Huh7.5 cells transfected with whole cell RNA extracted from a clemizole-resistant clone harboring the W55R mutation [6] were unaffected by 8μM clemizole, but remained sensitive to 2.5μM SCH503034 (Fig. 6). Reciprocally, decreased sensitivity to SCH503034 but not to clemizole was demonstrated in Huh7.5 cells transfected with whole cell RNA extracted from a SCH503034-resistant clone harboring the A156T mutation (Fig. 6). Cells transfected with whole cell RNA extracted from wild type replicon cells were sensitive to both drugs.
Fig 6. There is no cross-resistance between clemizole and SCH503034.
HCV replication in Huh7.5 cells electroporated with 50μg of whole cell RNA extracted from cells harboring either wild-type, the W55R NS4B mutant clone, or A156T NS3 mutant clone followed by growth in the absence (white bars), presence of 8μM clemizole (grey bars), or presence of 2.5μM SCH503034 (black bars). Results represent relative numbers of colonies obtained compared to each corresponding untreated control.
Discussion
As with HIV, effective pharmacologic control of HCV will likely best be achieved by a cocktail of drugs against independent virus-specific targets. Our results demonstrate that the antiviral effect of the recently discovered NS4B RNA binding inhibitor, clemizole [6] is highly synergistic with HCV PIs, and additive with interferon, ribavirin or HCV polymerase inhibitors. Importantly, combining clemizole with PIs doesn’t increase host cell toxicity. Moreover, the clemizole-SCH503034 combination decreases emergence of resistance without conferring cross-resistance.
Two major models, Loewe Additivity and Bliss Independence theory are used for analyzing interactions between drugs in combinations. While usually concordant, discordant results may be obtained when analyzing data in these two models [30, 31]. Both models defined the antiviral effect of the clemizole-SCH503034 combination as synergistic, excluding potential bias and validating the results. Furthermore, the magnitude of the clemizole-SCH503034 combination’s synergy was characterized as strong [17] further emphasizing its potential relevance in vivo. Clinical trials are needed to determine the maximally tolerated dose of clemizole (based on preclinical animal data, a no observed adverse events level (NOAEL) is estimated to be 100mg/kg/day [32], and the dose used for antihistamine therapy in humans was typically only ~2mg/kg/day) and pharmacokinetics in HCV patients in order to estimate what are the achievable serum and liver concentrations.
In addition to synergistically increasing the antiviral effect, the SCH503034-clemizole combination decreases the emergence of resistance. This is likely a result of the distinct mechanisms of action of these two drugs and the resulting different resistance profiles. The increased antiviral effect likely contributes as well. Moreover, each of these drugs can select for resistant mutants that have decreased fitness [6, 28]. Importantly, there was no evidence of cross-resistance between the two drugs. Last, the described synergistic antiviral effects may also permit a reduction in the dose or dosing frequency of individual agents, thereby minimizing potential toxicity and adverse effects. Taken together, these major advantages further emphasize clemizole’s potential as an important component of future combination regimens.
Synergy between antimicrobial drugs often implies a mechanistic interaction between the two targets [33, 34]. NS4B and NS3 have been shown to bind each other biochemically [35]. Furthermore, there is genetic evidence for their interaction [36]. We thus hypothesize that the interaction between these proteins, perhaps involving conformational changes, is the mechanism for the observed synergistic interaction between NS4B RNA binding inhibitors and PIs. Alternatively, it is possible that inhibiting both of these proteins affects two steps in a common pathway that is critical for viral replication thus resulting in synergy by a sequential blocking mechanism [37].
In summary, these results suggest that combination of even a moderate NS4B RNA binding inhibitor with PIs represents an attractive paradigm for increasing virologic response rates. Although we hypothesize that more potent inhibitors than clemizole can be obtained, because clemizole has already been extensively used in humans (albeit for a different indication) it may find immediate use as a critical component of next generation anti-HCV strategies.
Acknowledgments
Financial Support
This work was supported by NIH K08-AI079406-01 (to SE), a Stanford Digestive Disease Center (DDC) Pilot/Feasibility Award (to SE) provided support via NIH P30 DK56339 (“Molecular Pathogenesis of Digestive Diseases”), NIH RO1 DK066793 and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (to JSG), and by the Stanford SPARK program.
We thank Dr. Robert Prichard for kindly providing us the MacSynergy software and for helpful and insightful comments. We thank Leslie Holsinger (Virobay., Palo Alto) for kindly providing us some of the compounds used in this work. We also wish to thank the Stanford University SPARK Program.
List of Abbreviations
- HCV
Hepatitis C virus
- CI
Combination index
- NS
non-structural
- SOC
Standard of care
- PI
Protease inhibitor
- EC50
half maximal effective concentration
- CC50
half maximal cytotoxic concentration
Footnotes
Conference program
1. Einav S. Novel approaches to HCV therapy. In: “What’s hot in infectious diseases” session at the Western Regional Meeting, Ja2009, Carmel, CA.
2. Einav S, Hadas D. Sobol, Elizabeth Gehrig and Jeffrey S. Glenn. The Hepatitis C virus (HCV) NS4B RNA binding inhibitor, clemizole, is highly synergistic with HCV protease inhibitors. Infectious Disease Society of America (IDSA), Oct 2009, Philadelphia, PE (Program committee choice award).
Disclosure
JSG is a consultant to, receives research funding from, or has an equity interest in: Romark Laboratories, Roche, Genentech, Merck, and Eiger BioPharmaceuticals. SE and HDS have an equity interest in Eiger BioPharmaceuticals. EG has no commercial or other association that might pose a conflict of interest.
References
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Current topics in microbiology and immunology. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 3.Soriano V, Peters MG, Zeuzem S. New therapies for hepatitis C virus infection. Clinical infectious diseases. 2009;48:313–20. doi: 10.1086/595848. [DOI] [PubMed] [Google Scholar]
- 4.Egger D, Wölk B, Gosert R, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78:11393–400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Einav S, Gerber D, Bryson PD, et al. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nature Biotechnology. 2008;26:1019–27. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacques AAFV. Clinical evaluation of clemizole in allergic rhinitis. Int Rec Med. 1960:88–91. [PubMed] [Google Scholar]
- 8.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Einav S, Elazar M, Danieli T, Glenn JS. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol. 2004;78:11288–95. doi: 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 11.Elazar M, Cheong KH, Liu P, Greenberg HB, Rice CM, Glenn JS. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J Virol. 2003;77:6055–6061. doi: 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tallarida RJ. Drug synergism: its detection and applications. The Journal of pharmacology and experimental therapeutics. 2001;298:865–72. [PubMed] [Google Scholar]
- 13.Prichard MN, Shipman C. Analysis of combinations of antiviral drugs and design of effective multidrug therapies. Antivir Ther. 1996;1:9–20. [PubMed] [Google Scholar]
- 14.Korba BE. In vitro evaluation of combination therapies against hepatitis B virus replication. Antiviral Res. 1996;29:49–51. doi: 10.1016/0166-3542(95)00915-9. [DOI] [PubMed] [Google Scholar]
- 15.Tallarida RJ. The interaction index: a measure of drug synergism. Pain. 2002;98:163–8. doi: 10.1016/s0304-3959(02)00041-6. [DOI] [PubMed] [Google Scholar]
- 16.Prichard MN, Shipman C. A three-dimensional model to analyze drug-drug interactions. Antiviral Res. 1990;14:181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 17.Mark Neal Prichard KRA, Shipman Charled., Jr MacSynergy II Manual. 1993 [Google Scholar]
- 18.Malcolm BA, Liu R, Lahser F, et al. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother. 2006;50:1013–20. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartels DJ, Zhou Y, Zhang EZ, et al. Natural prevalence of hepatitis C virus variants with decreased sensitivity to NS3.4A protease inhibitors in treatment-naive subjects. J Infect Dis. 2008;198:800–7. doi: 10.1086/591141. [DOI] [PubMed] [Google Scholar]
- 20.Wyles DL, Kaihara KA, Vaida F, Schooley RT. Synergy of small molecular inhibitors of hepatitis C virus replication directed at multiple viral targets. J Virol. 2007;81:3005–8. doi: 10.1128/JVI.02083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau JYDG, Prescott LE, Maertens G, Lindsay KL, Qian K, Mizokami M, Simmonds P. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in the United States. Hepatitis Interventional Therapy Group. Ann Intern Med. 1996;124:868–76. doi: 10.7326/0003-4819-124-10-199605150-00002. [DOI] [PubMed] [Google Scholar]
- 22.Davis AUGLLJ. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26:122S–127S. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 23.Perni RB, Almquist SJ, Byrn RA, et al. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother. 2006;50:899–909. doi: 10.1128/AAC.50.3.899-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCown MF, Rajyaguru S, Le Pogam S, et al. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob Agents Chemother. 2008;52:1604–12. doi: 10.1128/AAC.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandra P, Raible D, Harper D, Speth J, Villano S, Bichier G. Antiviral activity of the non nucleoside polymerase inhibitor, HCV-796, in patients with chronic hepatitis C virus: preliminary results from a randomized, double-blind, placebo-controlled, ascending multiple dose study. Gastroenterology. 2006;130:A–748. [Google Scholar]
- 26.Chao CF, Fielman B, Myers M, Brown NA. First clinical results for a novel antiviral treatment for hepatitis C: a phase I/II dose escalation trial assessing tolerance, pharmacokinetics and antiviral activity on NM283, a novel antiviral treatment for hepatitis C. Gastroenterology. 2004;126:A681. [Google Scholar]
- 27.Kato T, Date T, Miyamoto M, et al. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J Clin Microbiol. 2005;43:5679–84. doi: 10.1128/JCM.43.11.5679-5684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 2006;70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Mathy JE, Ma S, Compton T, Lin K. Combinations of cyclophilin inhibitor NIM811 with hepatitis C Virus NS3-4A Protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob Agents Chemother. 2008;52:3267–75. doi: 10.1128/AAC.00498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dressler V, Müller G, Sühnel J. CombiTool--a new computer program for analyzing combination experiments with biologically active agents. Computers and Biomedical Research. 1999;32:145–60. doi: 10.1006/cbmr.1999.1509. [DOI] [PubMed] [Google Scholar]
- 31.Delaney WE, Yang H, Miller MD, Gibbs CS, Xiong S. Combinations of adefovir with nucleoside analogs produce additive antiviral effects against hepatitis B virus in vitro. Antimicrob Agents Chemother. 2004;48:3702–10. doi: 10.1128/AAC.48.10.3702-3710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkelstein M, Kromer C, Sweeney S, Delahunt C. Some aspects of the pharmacology of clemizole hydrochloride. J Am Pharm Assoc Am Pharm Assoc. 1960;49:18–22. [PubMed] [Google Scholar]
- 33.Matsuzaki K, Mitani Y, Akada KY, et al. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. 1998;37:15144–53. doi: 10.1021/bi9811617. [DOI] [PubMed] [Google Scholar]
- 34.Bell A. Antimalarial drug synergism and antagonism: mechanistic and clinical significance. FEMS microbiology letters. 2005;253:171–84. doi: 10.1016/j.femsle.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 35.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paredes AM, Blight KJ. A genetic interaction between hepatitis C virus NS4B and NS3 is important for RNA replication. J Virol. 2008;82:10671–83. doi: 10.1128/JVI.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black ML. Sequential Blockage as a theoretical basis for drug synergism. J Med Chem. 1963;6:145–53. doi: 10.1021/jm00338a014. [DOI] [PubMed] [Google Scholar]
- 38.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]