Abstract
This study examined in vivo and in vitro colonization by Aggregatibacter actinomycetemcomitans, an organism highly associated with aggressive periodontitis. Thirteen volunteers (5 were A. actinomycetemcomitans positive for buccal epithelial cells [BECs] and teeth, 5 were A. actinomycetemcomitans positive for teeth only, and 3 were A. actinomycetemcomitans-negative controls) had two mandibular stents fabricated. Each stent contained 3 removable hydroxyapatite (HA) tooth surrogates. One HA square was removed from a stent at 5 time points over 7 h to assess the transfer of A. actinomycetemcomitans from teeth or BECs to HA. Streptococcus, Actinomyces, A. actinomycetemcomitans, and total anaerobic counts were evaluated on each square over time. In vitro experiments evaluated binding, desorption, transfer, and reattachment of A. actinomycetemcomitans wild-type and mutant strains to BECs and saliva-coated HA (SHA). Streptococcus and Actinomyces formed 80% of the cultivable flora on HA in all subjects. Transfer of A. actinomycetemcomitans to HA was not seen in subjects with A. actinomycetemcomitans on teeth only. All 5 subjects with A. actinomycetemcomitans on BECs showed transfer of A. actinomycetemcomitans to HA. In vitro, A. actinomycetemcomitans desorbed from BECs and transferred to SHA. A. actinomycetemcomitans binding to SHA was irreversible and did not transfer to BECs. The adhesin Aae showed specificity for BECs. Fimbrial mutants showed the greatest reduction in binding to SHA. A. actinomycetemcomitans migrated from BECs to HA in vivo and to SHA in vitro; however, A. actinomycetemcomitans movement from teeth and SHA to BECs did not occur. In vivo, A. actinomycetemcomitans colonized HA within 6 h and thus can be considered an early colonizer. BECs are a likely reservoir for A. actinomycetemcomitans tooth colonization.
Aggregatibacter actinomycetemcomitans has frequently been associated with the etiology of localized aggressive periodontitis (LAP) (41). More recently, longitudinal studies of both humans and animals have added to this evidence and made the association between A. actinomycetemcomitans and LAP even more compelling (10, 14). Furthermore, virulence factors associated with A. actinomycetemcomitans and studied on a molecular level appear to be consistent with the pathogenesis described for LAP (11, 12, 17). Taken together, these data suggest that A. actinomycetemcomitans could play an important role in the initiation and progression of LAP.
A. actinomycetemcomitans resides in the oral cavity, is a member of the Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella (HACEK) group of “pathogenic” microorganisms, and has been found on six continents (21). While population-based data are far from extensive, A. actinomycetemcomitans has been found in a small subset of individuals in the North and South American continents as well as in Europe, with a lower prevalence in Caucasian individuals and a higher prevalence in individuals from Asia and Africa (2, 3, 15, 16, 28, 29). These demographic patterns raise questions as to how and why certain groups of individuals are more prone to A. actinomycetemcomitans colonization. This global distribution could result from infection due to random but more frequent exposure to A. actinomycetemcomitans by individuals or groups of individuals on one continent than on another (21). Alternatively, A. actinomycetemcomitans colonization could be dependent on the specificity of the outer membrane proteins (OMPs) of the infecting bacteria and their interaction and coupling with surface receptors of host cells (1, 37). If exposure is the driving force for attachment, then colonization would occur in a nonspecific fashion and could be dependent upon the level of A. actinomycetemcomitans in the environment. If specificity is the driving force, then one would expect host selection to dictate A. actinomycetemcomitans attachment and that only those bacteria that interact with appropriate tissue receptors would be colonized by A. actinomycetemcomitans (37).
Several groups have been working on the identification and characterization of adherence factors that allow A. actinomycetemcomitans to colonize the oral cavity (8, 24, 35). In vitro studies have suggested that soft tissue binding occurs in a specific manner and is mediated by the autotransporter adhesins ApiA and Aae (11, 24, 40). In contrast, binding of A. actinomycetemcomitans to tooth surfaces appears to occur in a nonspecific manner and is dominated by OMPs such as Flp (19, 31). There is no general agreement as to whether tooth or tissue surfaces form the primary site for A. actinomycetemcomitans colonization in the oral cavity (8, 23). In a longitudinal study of LAP, we confirmed reports by others and identified buccal epithelial cells (BECs) as a primary site for A. actinomycetemcomitans colonization in the oral cavities of healthy A. actinomycetemcomitans-positive individuals (10). However, definitive proof showing that buccal cells can seed tooth surfaces is lacking.
Because A. actinomycetemcomitans is typically found in the mouth in <20% of the population (41), studying the role of A. actinomycetemcomitans colonization of clean tooth surfaces in the general population has presented a difficult challenge. As a result, studies that have examined the relationship of A. actinomycetemcomitans to early events in the colonization process in humans are rare (26). However, one study of monkeys, though limited in scope, has shown that A. actinomycetemcomitans can colonize teeth within a 5- to 8-h period after tooth cleaning (22). In that study, the main goal was to determine the influence of a carbohydrate diet on Streptococcus mutans colonization of thoroughly cleaned teeth, and identification of A. actinomycetemcomitans on tooth surfaces was discovered by chance. However, because these teeth were cleaned, the likely source of A. actinomycetemcomitans was the primate mucosa (22). Recent studies have shown that A. actinomycetemcomitans can be found in predentate children, suggesting that the mucosa can serve as a potential reservoir for tooth colonization (33).
In an effort to help resolve questions related to early tooth and tissue colonization in humans, we explored the possibility of examining the colonization patterns of a number of A. actinomycetemcomitans-positive participants already enrolled in an ongoing longitudinal study (9). Since we had access to these subjects and had knowledge of their carriage of A. actinomycetemcomitans, we felt that we could recruit an appropriate subject population to study A. actinomycetemcomitans colonization of tooth surrogates in vivo (9). Overall, examination of the colonization patterns of A. actinomycetemcomitans can lead to a better understanding of the role of OMPs in A. actinomycetemcomitans binding, which could lead to potentially useful preventive and therapeutic strategies for LAP. The studies we report herein were designed to begin to address the following two important biological questions related to A. actinomycetemcomitans colonization of the oral cavity. Is A. actinomycetemcomitans an early colonizer of hydroxyapatite (HA) tooth surrogate surfaces? Do BECs serve as an effective reservoir for A. actinomycetemcomitans transfer to HA tooth surrogates?
MATERIALS AND METHODS
Study population.
In the ongoing longitudinal study, subjects were screened for the presence of A. actinomycetemcomitans and for their periodontal, oral, and general health. A group of A. actinomycetemcomitans-positive subjects with healthy periodontia and a cohort of A. actinomycetemcomitans-negative subjects were recalled at 6-month intervals to assess their periodontal status. Results of the clinical, microbiological, and host aspects of the longitudinal study have been reported elsewhere (9, 10). Based on limited data related to early colonization of tooth surfaces by A. actinomycetemcomitans, as shown in one monkey study (22) and one human study (26), we estimated that 5 subjects with A. actinomycetemcomitans on buccal cells and teeth or with A. actinomycetemcomitans on buccal sites alone and 5 subjects who were A. actinomycetemcomitans positive for teeth alone would be sufficient to supply pilot data for exploration of this biological process. As such, we reviewed the charts of 147 A. actinomycetemcomitans-positive subjects derived from a panel of over 1,000 subjects screened who participated in a longitudinal study of A. actinomycetemcomitans-induced periodontal disease (10). From this group, we selected 10 subjects who were A. actinomycetemcomitans positive for buccal and tooth sites, 5 who were positive for buccal sites alone, and 8 who were A. actinomycetemcomitans positive for tooth sites only. The initial selection was based on the availability of subjects for participation in this study, which required three separate visits (mandibular impressions, fitting of custom stents, and an observational period during the colonization study). Of those approached, none in the group that was A. actinomycetemcomitans positive for buccal sites alone were either available or willing to volunteer for the study, while 5 who were A. actinomycetemcomitans positive for tooth sites alone and 5 who were A. actinomycetemcomitans positive for tooth and buccal sites were available and volunteered to participate. We then recruited three A. actinomycetemcomitans-negative subjects, who served as controls. Thus, in total, 13 volunteers (10 who were A. actinomycetemcomitans positive and 3 who were A. actinomycetemcomitans negative) participated in the study.
Among the 10 A. actinomycetemcomitans-positive subjects, 5 of 5 subjects in the group that had A. actinomycetemcomitans in buccal and tooth sites had a maximum of one 5-mm pocket. These subjects had a mean age of 15.05 ± 0.45 years, and all five were female; one was Hispanic, while the other four were African-American. The five subjects in the group that had A. actinomycetemcomitans on tooth sites alone had one pocket with a range of 5 to 6 mm. These subjects had a mean age of 15.93 ± 0.25 years, and four were female; four were African-American, and one was of Hispanic heritage. The three subjects in the control group were all A. actinomycetemcomitans negative, all demonstrated good oral and medical health, and none had a 5-mm pocket. Two of the controls were female (Caucasian), and one was male (African-American), with a mean age of 20.05 ± 1.5 years. Potential participants were excluded if they were smokers, were on antibiotic therapy within the last 3 months, were either pregnant or lactating, or required medication for any systemic condition.
To participate, all volunteers and/or their legal guardians gave consent, using a form that was reviewed and approved by the Institutional Review Board (IRB) of the University of Medicine and Dentistry of New Jersey (UMDNJ), which had also reviewed and approved the study protocol. Both parental consent and the child's assent were received prior to subject participation in the study.
In vivo experimental procedures.
Each participant had a mandibular alginate impression taken to produce an individual stone model, which was used to design a custom acrylic mandibular stent. The stent extended from the second molar to the first premolar, just below the gingival margin and just above the incisal edge of the occlusal surface of the teeth. Both left- and right-sided stents were made. Each stent contained four recessed areas into which HA squares could be inserted and retained using soft wax. In this manner, only the surface of the HA square was exposed to accumulating plaque. Three HA squares were placed in each of the stents (left and right). HA squares and their stents were made as previously described (7).
The stents were placed into the subjects' mouths early in the morning, and one square each was removed at 5 time points over a 7-h period to assess colonization. Thus, squares were removed 5 min, 2 h, 4 h, 6 h, and 7 h after placement. At each time point, an HA square was removed, placed into 1 ml of phosphate-buffered saline (PBS), and sonicated for 30 s with a Branson sonicator equipped with a cup horn attachment (model 200; Branson, Co., Danbury, CT) set at an output of 4, with a 50% pulsed duty cycle and low power output. Bacterial cells released into the supernatant were plated on the following media: blood agar for enumeration of total aerobes; mitis salivarius agar for enumeration of Streptococcus (13); CFAT agar for enumeration of Actinomyces (42); and A. actinomycetemcomitans growth medium (AAGM), composed of Trypticase soy broth supplemented with 6 g of yeast extract, 8 g of glucose, and 4 g of sodium bicarbonate in 1 liter of deionized water (used as broth), and AAGMBV (AAGM supplemented with 75 μg bacitracin ml−1 [Sigma] and 5 μg vancomycin ml−1) (used as agar) for enumeration of A. actinomycetemcomitans (6). For each plating, a 100-μl aliquot of the supernatant was taken in duplicate, spotted, and then spread over the agar for separation. Samples were diluted from 100 to 10−6 in PBS for plating on each type of agar. For total anaerobic colony counts, blood agar plates were incubated at 37°C for 3 to 5 days under anaerobic conditions. For enumeration of Streptococcus and Actinomyces, plates were incubated at 37°C for 3 to 5 days under anaerobic conditions. For A. actinomycetemcomitans, plates were incubated at 37°C for 3 to 5 days in a CO2 environment. Plates with 30 to 100 colonies were counted, and CFU per ml were determined after the appropriate period of incubation. A. actinomycetemcomitans was initially identified by its distinctive colonial morphology on selective media and then confirmed by its catalase positivity and by PCR analysis (6).
Bacterial strains and growth conditions for in vitro studies.
A. actinomycetemcomitans strain IDH 781 was used for the in vitro studies described below. In vitro studies were designed to investigate attachment of A. actinomycetemcomitans to or desorption of A. actinomycetemcomitans from saliva-coated HA (SHA) and/or BECs, which were used as the host colonizing surfaces. A. actinomycetemcomitans strains were grown in AAGM broth or AAGMBV agar as described above (6). Strain IDH 781 was made nalidixic acid resistant by being subjected to increasing concentrations of nalidixic acid as described previously (11). Strains carrying mutations previously created in key OMPs that are known to affect binding of A. actinomycetemcomitans included strains JK 1046 (an aae mutant), JK 1047 (an flp mutant), and JK 1051 (an apiA mutant) (11, 40). These strains were used to determine the effects of the mutations on binding to BECs or SHA, as described below. In binding experiments, mutated strains were compared to each other and to IDH 781, the wild-type unaltered parental strain, and results obtained from these experiments were calculated as ratios of bound cells to unbound cells for SHA and/or BECs, in CFU/ml, as described below. Table 1 describes the origins of the mutated strains and the nalidixic acid resistance marker used to identify and label the parent A. actinomycetemcomitans strain, which was named IDH 781N (for nalidixic acid resistance). The mutated strains were used in SHA and BEC binding experiments only. The wild-type strain was used in binding, desorption, and reattachment/transfer experiments. The protocols used for these studies are described below.
TABLE 1.
Bacterial strains used for this study
| Strain | Relevant characteristica | Reference |
|---|---|---|
| IDH 781N | Spontaneous Nalr variant of IDH 781 | 11 |
| JK 1046 | IDH 781N aae::R6Kγ ori/KAN; Kmr | 11 |
| JK1047 | IDH 781N flp-1::Tn903 kan; Kmr | 11 |
| JK1051 | IDH 781N apiA::R6Kγ ori/KAN; Kmr | 40 |
Nalr, nalidixic acid resistant; Kmr, kanamycin resistant.
SHA binding assay.
For attachment assays, 50 mg of HA beads (Macrosorb C; Gallard-Schlesinger Inc., Carle Place, NY) were equilibrated in buffered KCl and then coated with saliva as described previously (5). In brief, approximately 30 ml of whole unstimulated saliva was collected and pooled for use in the assay. Three individuals of 25 to 45 years old who were in good medical and dental health volunteered to contribute 10 ml of saliva for use in the study. These individuals all signed the appropriate consent forms approved by the UMDNJ IRB. The collected saliva samples were pooled, separated into equal aliquots, frozen, and thawed for preparation for coating the HA beads. For HA treatment, the thawed saliva was clarified by centrifugation at 10,000 × g for 30 min. One milliliter of clarified saliva was added to 50 mg of HA for 2 h at 37°C for equilibration. The beads were subjected to centrifugation at 12,000 × g for 10 min and were then washed 3 times with buffered KCl to prepare SHA for incubation with bacteria as described above.
One-milliliter aliquots of the adjusted stock suspension of the wild-type bacteria and the various IDH 781 attachment knockouts were grown to achieve an approximate optical density at 590 nm of 0.9, which is equivalent to 1 × 109 bacteria/ml. Clumping cells were disrupted by sonication for 15 s at low power output as described below (5, 6). For the binding assay, 200 μl of bacteria was added to 50 mg of SHA beads. The beads and bacteria were rotated at 60 cycles per minute for 1 h at 37°C. After the beads had been allowed to settle for 30 min, 100 μl of the supernatant was removed, and the supernatant was plated on agar to estimate the number of unbound cells. To estimate the quantity of bound cells, the remaining supernatant was removed, and the settled beads containing the bound bacteria were washed, resuspended in buffered KCl, and subjected to pulse sonication for 30 s, using a Branson 200 sonicator with a cup horn attachment, as described previously, in order to dissociate cells that remained bound to SHA so that they could be enumerated. One hundred microliters of the buffered KCl supernatant derived from the sonicated beads was plated in duplicate to estimate the number of IDH 781 cells bound to SHA, in CFU/ml, as previously described (5, 6). The quantity of bound to unbound cells was calculated as a ratio and presented as a whole number (i.e., a ratio of 256/1 is presented as 256).
BEC binding assay.
To evaluate bacterial binding to epithelial cells, IDH 781 cells were prepared as described above to achieve an A590 of 0.9. Host cells were obtained from medically healthy subjects with no periodontal disease who signed consent forms approved by the UMDNJ IRB. BECs were obtained by gentle but firm scraping of the side of the cheek with a wooden tongue depressor, and cells thus obtained were suspended in PBS. Five to seven such scrapings supplied a sufficient volume of cells to yield 5 × 104 cells per ml, as estimated by microscopic examination using a hemocytometer. To be sure that BECs were not contaminated by A. actinomycetemcomitans, buccal cells were collected from A. actinomycetemcomitans-negative subjects, and BECs thus obtained were checked by DNA analysis to corroborate that they were free of A. actinomycetemcomitans. The binding assay has been shown to be useful for the evaluation of attachment to intact but nonviable cells. We have shown previously that 95% of BECs collected in this manner have intact morphology, do not harbor A. actinomycetemcomitans, and are permeable to trypan blue (40). Gentamicin treatment is known to kill bacterial cells on the periphery of mammalian cells and allowed us to determine that A. actinomycetemcomitans does not penetrate BECs when added under the conditions described for this assay (40).
For the bacterial component of the binding assay, a starting dose of A. actinomycetemcomitans that was equivalent to 1 × 109 cells/ml was used. A 250-μl aliquot of IDH 781 grown in this manner was added to 250 μl of BECs in a 2-ml polypropylene microcentrifuge tube to achieve a ratio of 103 to 104 bacterial cells per BEC. Attachment of IDH 781 to BECs was assessed as previously described (11). Briefly, tubes containing a mixture of host and bacterial cells were gently rotated for 60 min at 37°C to allow for active interaction of bacterial cells and BECs. Following the 60-min incubation period, the bacterial cell-BEC mixture was placed into a 15-ml centrifuge tube containing 10 ml of a 5% Ficoll 400 gradient, which was then subjected to centrifugation at 800 × g in a swinging bucket rotor for 10 min to separate IDH 781 bound to BECs from unbound IDH 781. In this manner, BECs alone and BECs with bound bacteria passed through the gradient, while individual bacteria unbound to BECs remained suspended in the Ficoll. After removal of the Ficoll, the pellet containing the BECs and BECs with bound bacteria was resuspended in PBS and subjected to vortex agitation. A 100-μl aliquot of the suspension was serially diluted and plated on AAGMBV agar. The agar plates were incubated at 37°C in a 10% CO2 atmosphere for 3 days and counted to enumerate bacteria bound to BECs, in CFU/ml, which was then converted to CFU/BEC (11).
SHA and BEC desorption protocol.
In preparation for desorption protocols, SHA, BECs, and IDH 781 were prepared as described previously. After being washed 3 times in PBS, SHA-coated beads were distributed into six microcentrifuge tubes for incubation with IDH 781. BECs were prepared as described above and then placed into a 50-ml centrifuge tube containing 3 ml of PBS. The tube was then vortexed vigorously for 30 s to disperse the BECs for distribution into six microcentrifuge tubes for incubation with IDH 781. A. actinomycetemcomitans strain IDH 781 was prepared as described for incubation with SHA and BECs.
Tubes containing SHA or BECs were then mixed with A. actinomycetemcomitans in PBS plus 10% sodium bicarbonate which had been adjusted to an A590 of 0.9. The six BEC and six SHA tubes mixed with IDH 781 were then incubated at 37°C while rotating for 2 h to allow for A. actinomycetemcomitans binding. After 2 h, two SHA tubes were analyzed for levels of bound and unbound IDH 781 present, as previously described, and 2 BEC tubes were analyzed for CFU/BEC. The remaining tubes were gently washed 3 times with PBS to remove unbound cells, and then these four SHA and/or four BEC samples were resuspended in 1 ml of PBS and incubated at 37°C while rotating at 60 cycles per minute. At 0, 15, 30, 60, 120, and 180 min, 100-μl aliquots were removed from the supernatants of both SHA and BEC tubes and analyzed for levels of IDH 781 that desorbed from either SHA or BECs (by plating on AAGMBV agar). With removal of each supernatant, 100 μl of PBS was readded to the sampled tubes to maintain the volume for later samplings. AAGMBV plates were incubated for 72 h at 37°C in 10% CO2, and then plate counts were performed to determine CFU/ml for cells desorbed from either SHA or BECs.
Transfer of A. actinomycetemcomitans from BEC to SHA and vice versa. (i) Preparation of A. actinomycetemcomitans IDH 781 culture.
A. actinomycetemcomitans IDH 781 was inoculated into a petri dish containing 15 ml of AAGM broth and incubated for 3 days at 37°C in 10% CO2. The A. actinomycetemcomitans biofilm culture that accumulated on the surface of the petri dish was scraped from the bottom of the dish, dispersed by trituration, and adjusted to an A590 of 0.9. A 100-μl aliquot of the dispersed sample was serially diluted in PBS and plated on AAGMBV agar for enumeration. The A590 reading was equivalent to 1 × 109 cells/ml.
(ii) Preparation of BECs.
BECs were scraped from the buccal mucosa of an A. actinomycetemcomitans-negative volunteer by use of a sterile wooden tongue depressor, as described above, to obtain 5 × 104 BECs/ml, as determined by a hemacytometer.
(iii) Preparation of SHA.
Hydroxyapatite squares of 3 mm by 3 mm by 1 mm were fabricated. Each HA square was placed in a microcentrifuge tube containing 1 ml of clarified saliva obtained from an A. actinomycetemcomitans-negative individual and incubated for 30 min at 37°C while rotating at 60 rpm. HA squares thus treated were washed 3 times in 1 ml of PBS to develop an SHA square for testing.
(iv) Transfer of IDH 781 from BECs to SHA.
One milliliter of a PBS solution containing 5 × 104 BECs/ml was added to four microcentrifuge tubes (Fig. 1A, tubes 1 to 4). One milliliter of IDH 781 cells containing 1 × 109 cells/ml was added to each tube, the mixture was triturated to disperse bacteria and BECs, and incubation was continued for 30 min at 37°C with rotation at 60 rpm. After incubation, the contents of tubes 1 and 2 were applied to a 15-ml tube containing 10 ml of 5% Ficoll 400 for separation of BECs from unattached A. actinomycetemcomitans IDH 781, as described previously. The pellet was resuspended in PBS, serially diluted, and plated on AAGMBV agar for enumeration of A. actinomycetemcomitans attached to BECs.
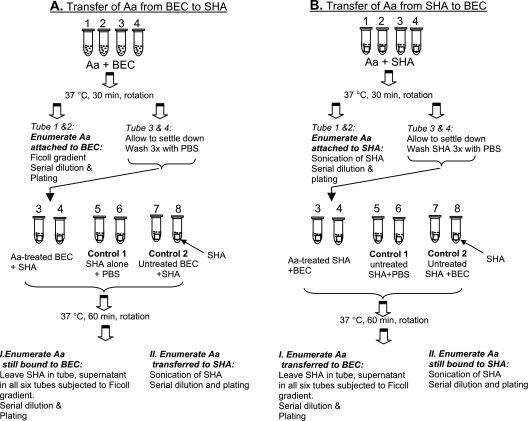
FIG. 1.
Protocol for transfer of Aggregatibacter to oral tissues. (A) Transfer from BECs to SHA. (First row) Tubes 1 to 4, BECs and A. actinomycetemcomitans (Aa) were incubated for 30 min while rotating. (Second row) After incubation, tubes 1 and 2 were used to calculate the level of A. actinomycetemcomitans bound to BECs by using the Ficoll method, and the level was reported as the number of A. actinomycetemcomitans cells/BEC. Tubes 3 and 4 were subjected to low-speed centrifugation to precipitate BECs with bound A. actinomycetemcomitans. Precipitates were washed three times with PBS to remove unbound A. actinomycetemcomitans. (Third row) Tubes 3 and 4, BECs with bound A. actinomycetemcomitans resuspended in 1 ml of PBS. One SHA square was added to each tube to determine the transfer of A. actinomycetemcomitans from BECs to SHA. Tubes 5 and 6, one SHA square alone suspended in PBS with no BECs (control 1); tubes 7 and 8, BECs with no A. actinomycetemcomitans added, suspended in 1 ml of PBS to which was added one SHA square (control 2). Tubes 3 to 8 were incubated with rotation for 60 min. All fluid was removed, leaving the SHA square in the tube. I, supernatant was subjected to Ficoll separation for enumeration of A. actinomycetemcomitans cells; II, SHA square was subjected to sonication for removal and enumeration of SHA-bound A. actinomycetemcomitans. (B) Transfer from SHA to BECs. (First row) Tubes 1 to 4, SHA was incubated with A. actinomycetemcomitans for 30 min while rotating. (Second row) SHA squares from tubes 1 and 2 were removed and subjected to sonication to detach A. actinomycetemcomitans bound to SHA for enumeration. Tubes 3 and 4, containing SHA with attached A. actinomycetemcomitans, were washed to remove unbound A. actinomycetemcomitans. (Third row) One milliliter of PBS was added to tubes 3 and 4. BECs were added to assess the transfer of A. actinomycetemcomitans from SHA to BECs. Tubes 5 and 6 contained one untreated SHA square each (control 1). Tubes 7 and 8 had one SHA square that was not exposed to A. actinomycetemcomitans, to which BECs that were not exposed to A. actinomycetemcomitans were also added, acting as a second control. Supernatant fluid was removed from tubes 3 to 8 and subjected to Ficoll separation to assess A. actinomycetemcomitans. SHA squares remained in the tubes. Tubes 3 to 8 with SHA remaining were subjected to sonication to determine the amount of A. actinomycetemcomitans attached to the SHA.
Tubes 3 and 4 were subjected to low-speed centrifugation at 500 × g for 5 min to pellet the BECs. The pelleted BECs were then resuspended in 1 ml of PBS. To determine transfer of IDH 781 from BECs to SHA, one SHA square was added to microcentrifuge tube 3 and one SHA square was added to tube 4. Each tube contained IDH 781-treated BECs. Controls consisted of two microcentrifuge tubes, each with SHA and no BECs (tubes 5 and 6), and two microcentrifuge tubes containing untreated BECs with SHA squares added (tubes 7 and 8).
All six microcentrifuge tubes (tubes 3 to 8) were incubated at 37°C for 60 min while rotating at 60 rpm. After 60 min, the contents of the tubes were allowed to settle, and the supernatants from all tubes were removed and applied to 5% Ficoll 400 to assess the level of A. actinomycetemcomitans remaining bound to BECs after the experimental period of 60 min. After the Ficoll was decanted, the BECs (tubes 3, 4, 7, and 8 had BECs; tubes 5 and 6 had SHA only) that were pelleted were resuspended in 1 ml of PBS, serially diluted in PBS, and then plated on AAGMBV agar to assess the level of IDH 781 bound to BECs. The SHA squares in the remaining 6 tubes were moved to new tubes, washed three times with PBS, and then resuspended in 1 ml of PBS. The tubes containing the SHA (tubes 3 to 8) were then subjected to 2 min of sonication, using a Branson 200 sonicator (output = 1, duty cycle = 50%), to remove any attached bacteria. The supernatant resulting from the sonication was removed, serially diluted, and plated on AAGMBV agar for enumeration of IDH 781 that had adhered to SHA squares and was removed by sonication. All experiments were done twice in triplicate.
(v) Transfer of IDH 781 from SHA to BECs.
One SHA square was placed into each of four microcentrifuge tubes (Fig. 1B). To tubes 1 to 4, 1 ml of a suspension of IDH 781 in PBS at a concentration of 1 × 109 bacteria/ml was added, triturated, and then allowed to incubate for 30 min at 37°C while rotating at 60 rpm. After 30 min, SHA squares were moved from one tube to a second tube and washed three times with PBS, and then SHA was removed from tubes 1 and 2 and subjected to 60 s of sonication, using a Branson 200 sonicator as described above, to detach SHA-bound A. actinomycetemcomitans. The supernatant thus obtained was removed, serially diluted, and plated on AAGMBV agar for enumeration of A. actinomycetemcomitans that had adhered to the SHA squares. In tubes 3 and 4, SHA squares with A. actinomycetemcomitans attached after washing were resuspended in 1 ml of PBS to which 1 ml of BECs at a concentration of 5 × 104 BECs/ml in PBS had been added. Controls consisted of tubes 5 and 6, which contained untreated SHA alone with no BECs added, and tubes 7 and 8, with untreated SHA and untreated BECs. All tubes were incubated at 37°C for 60 min while rotating at 60 rpm. After 60 min, the tubes were left undisturbed to allow SHA to settle, and the suspensions containing BECs in tubes 3, 4, 7, and 8 were applied to 5% Ficoll 400 and subjected to low-speed centrifugation as described above. The Ficoll was decanted, and the BECs with attached A. actinomycetemcomitans or without A. actinomycetemcomitans were resuspended in 1 ml of PBS, serially diluted, and then plated on AAGMBV agar for enumeration of A. actinomycetemcomitans. The remaining SHA squares in each of the 6 tubes were moved to new tubes, gently washed three times with PBS, and then resuspended in 1 ml PBS. The tubes were then subjected to 60 s of sonication, using a Branson 200 sonicator as described above, to remove attached A. actinomycetemcomitans. The supernatant thus obtained was removed, serially diluted, and plated on AAGMBV agar for enumeration of A. actinomycetemcomitans that had adhered to the SHA square. All experiments were done twice in triplicate.
Data analysis.
In vivo data were analyzed by a chi-square test using 5 time points over the 7-h period to determine the frequencies of A. actinomycetemcomitans recovery in the three groups. Significance was set at P values of ≤0.05. The in vitro mutational analysis, which compared wild-type IDH 781 to three OMP mutants, generated parametric data which were analyzed by analysis of variance (ANOVA). All other in vitro experiments were considered descriptive in nature.
RESULTS
In vivo study.
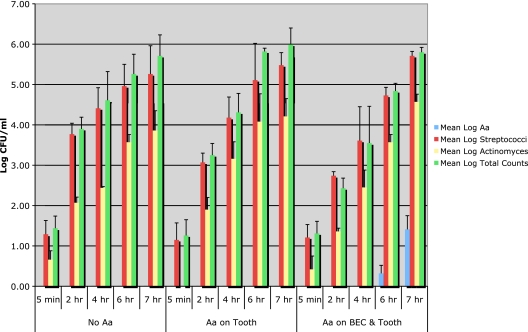
The microbiologic results obtained in the in vivo experiments are summarized in Fig. 2. At each of the sampling times (from 5 min to 7 h) for all 13 subjects, Streptococcus represented at least 80% of the total mean log counts, while Actinomyces species represented approximately 10% of the total level of Streptococcus. Thus, the level of Actinomyces species was consistently about 1 log lower than that of Streptococcus throughout the study period. Total anaerobic counts started at a level of 1 log 5 min after HA placement and showed a consistent increase to 6 log by 7 h after placement. These results are consistent with cultural studies of early plaque formation and confirm data showing that Streptococcus and Actinomyces are early pioneer colonizers of tooth surfaces (26, 34). When examined as relative percentages of total counts, Actinomyces and Streptococcus counts were higher for the group of subjects who showed A. actinomycetemcomitans on both BECs and teeth than for the other two groups. Actinomyces was shown to form about 10 to 18% of the total flora in the first 4 h of plaque accumulation for this group of subjects (see Table S1 in the supplemental material). No A. actinomycetemcomitans colonization of the HA tooth surrogates was found in control subjects who did not harbor A. actinomycetemcomitans in the screening assay or in subjects who had A. actinomycetemcomitans in tooth or pocket sites only. A. actinomycetemcomitans was detected on the 4-h HA square for one subject who had A. actinomycetemcomitans on buccal cells in vivo in the screening assay. The remaining four subjects harboring A. actinomycetemcomitans on their buccal cells at screening showed A. actinomycetemcomitans on HA squares within 6 h after stent placement. A. actinomycetemcomitans was seen to increase by 1 log or more in all 5 subjects when 6-h counts were compared to 7-h counts. The relative percentage of A. actinomycetemcomitans was <1% of the total flora but increased almost 4-fold from the 6-h to the 7-h time point (see Table S1 in the supplemental material). This colonization occurred only in those subjects who showed A. actinomycetemcomitans on their buccal cells, not in subjects with A. actinomycetemcomitans on their tooth surfaces or in pockets. The data derived from this in vivo study support the concept that buccal cells can serve as a reservoir for A. actinomycetemcomitans tooth colonization.
FIG. 2.
In vivo colonization of Aggregatibacter on tooth surrogates in humans. Bars show data obtained 5 min, 2 h, 4 h, 6 h, and 7 h after removal of hydroxyapatite squares from subjects' mouths for cultural assessment of total anaerobic counts and levels of Streptococcus, Actinomyces, and A. actinomycetemcomitans. Colony counts for control subjects (no A. actinomycetemcomitans) are shown in the left panel, while colony counts for the 5 subjects with A. actinomycetemcomitans in both buccal and tooth/pocket sites are shown in the right panel. Streptococcus, Actinomyces, and total anaerobic counts were high for all groups, while A. actinomycetemcomitans was seen only in subjects with A. actinomycetemcomitans at screening in both buccal and tooth or pocket sites, not in the control subjects or subjects with A. actinomycetemcomitans in tooth or pocket sites only. A. actinomycetemcomitans was found in all 5 subjects with A. actinomycetemcomitans in both buccal and tooth sites and increased by 1 log when 6-h squares were compared to 7-h squares.
In vitro experiments to determine A. actinomycetemcomitans mutant strain binding. (i) SHA binding.
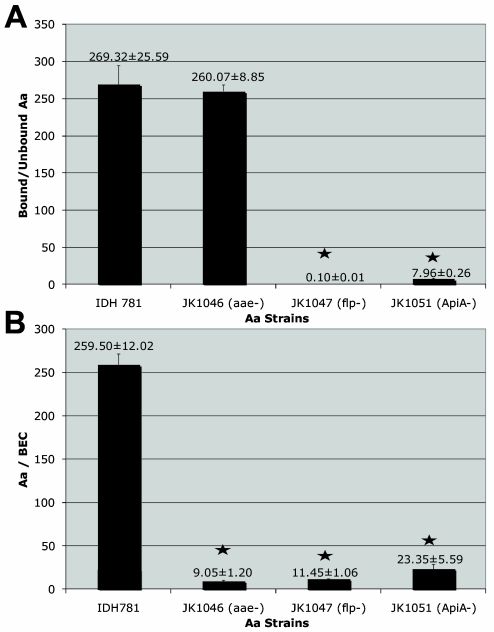
Wild-type IDH 781 bound to SHA at a ratio of 269 ± 25.6 bound to unbound cells, while the aae mutant strain (JK 1046) bound at a similar level, and thus a ratio of 260 ± 8.9 bound to unbound cells was found for SHA. In contrast, the apiA mutant strain (JK 1051) bound to SHA at a ratio of 8.0 ± 0.26 bound to unbound cells, and the flp mutant strain (JK 1047) bound at a negligible level of 0.01 ± 0.01 bound to unbound cells. In summary, both wild-type IDH 781 and JK 1046 bound to SHA at a significantly higher level than did JK 1047 (flp mutant) and JK 1051 (apiA mutant) (P ≤ 0.0001) (Fig. 3A).
FIG. 3.
(A) In vitro binding of A. actinomycetemcomitans to SHA. The IDH 781 wild-type strain bound at a ratio of 269.32 ± 25.59 bound to unbound cells. Binding of JK 1046 (the aae mutant strain) was not appreciably reduced compared to that of the wild-type strain. Binding of all other mutant strains was significantly reduced (P < 0.001). Thus, JK 1057 (the flp mutant strain) and JK 1051 (the apiA mutant strain) showed significantly reduced binding to SHA. (B) In vitro binding of A. actinomycetemcomitans to BECs. The IDH 781 wild-type strain bound at a level of 259.5 ± 12.02 CFU/BEC. Binding of all other mutant strains was reduced significantly (P < 0.001).
(ii) BEC binding.
Wild-type IDH 781 bound to BECs at a level of 259.5 ± 12.02 CFU/BEC. This binding was significantly higher than that of any of the other strains tested (P < 0.0001). The next highest binding occurred in strain JK 1051 (apiA mutant), which expressed both Aae and Flp. JK 1051 bound to BECs at a level of 23.4 ± 5.6 CFU/BEC, while JK 1046 (aae mutant) bound at a level of 9.0 ± 1.2 CFU/BEC and the flp mutant strain (JK 1047) bound to BECs at a similar level, 11.5 ± 1.06 CFU/BEC (Fig. 3B).
Desorption of A. actinomycetemcomitans from host surfaces. (i) SHA desorption.
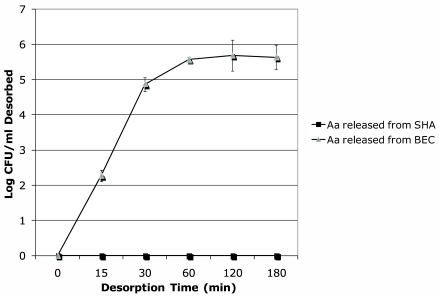
IDH 781, the wild-type A. actinomycetemcomitans strain, was used to evaluate attachment and desorption in all in vitro assays. In desorption experiments that followed attachment as described above, 1 h after completion of the attachment experiments the number of IDH 781 cells found in the supernatant solution was minimal and was calculated as 4.1 × 102 CFU/ml (data not shown). With respect to desorption, IDH 781 was never found to increase in the SHA supernatant over the 3-h desorption period, remained bound to SHA, and therefore would be unavailable for binding to “other” surfaces (Fig. 4).
FIG. 4.
Desorption of A. actinomycetemcomitans from SHA and BECs. There was no desorption of IDH 781 from SHA that could be detected over the entire 3-h experimental period. In contrast, desorption of IDH 781 from BECs began almost immediately and reached a plateau within 1 h after completion of the attachment phase of the experiment.
(ii) BEC desorption.
In the desorption phase of the experiment that followed the attachment phase, IDH 781 detachment from BECs began almost instantaneously after the attachment phase was halted, and thus a plateau was reached within 1 h of the initial assessment. At 3 h postincubation, about two-thirds of the IDH 781 cells initially attached to BECs had desorbed (6 × 105 IDH 781 CFU bound initially, and only 1.8 × 102 CFU were still attached after 3 h). These results indicate that more A. actinomycetemcomitans IDH 781 cells were found in the BEC supernatant than bound to BECs, and thus a large number of A. actinomycetemcomitans cells would be available for transfer and binding to “other” surfaces (Fig. 4).
Transfer experiments. (i) A. actinomycetemcomitans transfer from BECs to SHA.
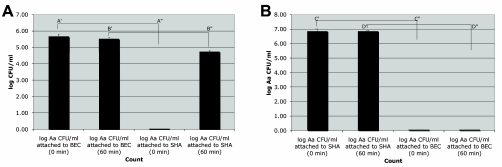
Untreated BECs with SHA (control tubes 7 and 8) and the untreated SHA (control tubes 5 and 6) showed no A. actinomycetemcomitans. In tubes 1 and 2, IDH 781 achieved a level of 4.9 × 105 total cells bound to BECs (Fig. 5A; A′ = time zero) (5.69 log10 cells). In tubes 3 and 4, BECs containing bound IDH 781 were coincubated with one aseptic, untreated SHA square (3 mm by 3 mm by 1 mm). At time zero, the SHA square had no bacteria (Fig. 5A; A" = time zero) (0 IDH 781 bacteria). After 1 h of coincubation, BECs were found to contain 3.5 × 105 IDH 781 cells/ml (Fig. 5A; B′ = 60 min) (5.54 log10 IDH 781 cells), while the SHA square contained 5.6 × 104 IDH 781 cells (Fig. 5A; B" = 60 min) (4.75 log10 IDH 781 cells).
FIG. 5.
Transfer of A. actinomycetemcomitans from BECs to SHA and vice versa. (A) Transfer from BECs to SHA. Columns A, time zero, there were 5 × 104 IDH 781 cells bound to BECs (log10 = 5.69) and no A. actinomycetemcomitans cells bound to the sterile SHA square; columns B, 60 min of coincubation, 3.5 × 105 IDH 781 cells were bound to BECs (log10 = 5.54) and 5.6 × 104 IDH 781 cells were found to be transferred to the SHA square (log10 = 4.75). (B) Transfer from SHA to BECs. Column C′, time zero, there were 7.2 × 106 IDH 781 cells bound to the SHA square (log10 = 6.86), and no IDH 781 cells were seen on the BECs at this time; column D′, 60 min of coincubation with BECs, IDH 781 levels were similar to those at time zero on the treated SHA surface (D), while no IDH 781 cells were shown to be transferred to the BECs (column D").
(ii) A. actinomycetemcomitans transfer from SHA to BECs.
Control tubes 5 and 6, with untreated SHA and no BECs, and control tubes 7 and 8, with untreated SHA and untreated BECs, showed no IDH 781. The SHA squares were prepared by incubating approximately 1 × 109 IDH 781 cells/ml with an aseptic SHA square for 30 min, which resulted in a level of 7.2 × 106 IDH cells/SHA square, as seen in tubes 1 and 2 (Fig. 5B; C′ = time zero) (6.86 log10 IDH 781 cells). SHA squares 3 and 4, the IDH 781-treated squares, had BECs added at a concentration of 5.1 × 104 BECs in 1 ml of PBS. The BECs measured at this time had no bacteria (Fig. 5B; C" = time zero) (IDH 781 cells/BEC). After 1 h of coincubation in tubes 3 and 4, the SHA contained 7.1 × 106 IDH 781 cells/square (Fig. 5B; D′ = 60 min) (6.85 log10 cells), and no IDH 781 was detected on the BECs assessed (Fig. 5B; D" = 60 min) (IDH 781 binding to BECs = 0). Thus, no transfer of IDH 781 from the SHA square to the BECs occurred in these in vitro experiments (Fig. 5B; D" = 60 min).
DISCUSSION
The present studies set out to address two questions concerning A. actinomycetemcomitans colonization. First, could A. actinomycetemcomitans be considered an early colonizer of tooth surfaces? We defined early colonization as the de novo appearance of A. actinomycetemcomitans on an uninfected, standard-sized, tooth-related surface (HA) within a 6-h period (22). Since it is known that early colonizers of teeth are bacteria from the Streptococcus and Actinomyces genera, we decided to evaluate colonization of A. actinomycetemcomitans in the context of these pioneer colonizers (34). Data taken from our in vivo study suggest that A. actinomycetemcomitans can be considered an early colonizer, as demonstrated by the observations that all 5 subjects who had A. actinomycetemcomitans on their buccal cells showed A. actinomycetemcomitans on tooth surrogates within 6 hours and that counts increased by 1 log over the next hour. While we cannot rule out the fact that A. actinomycetemcomitans could have been associated with the pioneer colonizers Streptococcus and Actinomyces, we feel confident that A. actinomycetemcomitans, based on our definition, can be considered an early tooth colonizer. Although the lack of interbacterial association still needs to be shown in vivo, our in vitro data indicate that A. actinomycetemcomitans does not require other microbes to attach to and avidly colonize tooth-like surfaces (Fig. 3 to 5). In spite of this remaining question, the data from this study are consistent with other reports indicating that A. actinomycetemcomitans can colonize both primate and human enamel within a 6-hour period after tooth prophylaxis (22, 26). As mentioned previously, these prior studies reported colonization of A. actinomycetemcomitans as a chance happening (22, 26). Neither of the previous reports attempted to speculate why A. actinomycetemcomitans, considered a poor colonizer, was found on tooth surfaces or how this finding could be interpreted in light of the known sequence of events in the microbial development of dental plaque (27, 34). Our findings suggest that the ability of A. actinomycetemcomitans to colonize tooth surfaces has been underestimated in the past, probably because A. actinomycetemcomitans is not present in most individuals, thus making it difficult to assess its status in early plaque formation.
The second question posed in this study addressed the role of the buccal mucosa as a reservoir for tooth colonization. In the design of the in vivo study, we contemplated collecting saliva at baseline and at other time points. We decided not to include saliva collection in this pilot study because saliva is typically considered a fluid that supports person-to-person as well as intraoral transmission but is not considered a stable intraoral bacterial niche as a result of constant swallowing (26, 33, 39). In contrast, buccal sites and tooth sites are considered to be stable reservoirs for many oral bacteria (4, 10, 36). We surmise, based on our data, that saliva acted as a fluid medium that helped to transfer A. actinomycetemcomitans from buccal to HA sites. Moreover, as a result of the pilot data generated in this study, we feel that collection of saliva in future studies may help to better define its role in intraoral transfer and its relationship to the early stages of disease.
In the in vivo study, we showed that A. actinomycetemcomitans colonized naïve tooth surrogates in cases where A. actinomycetemcomitans was found on BECs at screening but did not colonize tooth surrogates when A. actinomycetemcomitans was not found on BECs but was found on teeth alone. Thus, our in vivo data suggest that A. actinomycetemcomitans can be transferred from BECs to HA but not from teeth to HA. Our in vitro data support this observation and indicate that buccal cells can provide a reservoir for A. actinomycetemcomitans colonization as well as a source for transfer of A. actinomycetemcomitans to tooth surfaces. Previously, we proposed, based on the literature and with no experimental data, that BECs could provide a likely oral reservoir for recolonization of tooth surfaces after therapy (8). This hypothesis was based on the facts that A. actinomycetemcomitans was seen on and in BECs (4, 30, 36) and that A. actinomycetemcomitans bound to BECs in vitro, demonstrated saturable kinetics, and reached equilibrium quite readily, as seen in previous in vitro studies (8). The data obtained from this study support these concepts and suggest that binding of A. actinomycetemcomitans to buccal cells is reversible, and thus that the organism is available for interaction with other cells. Our interpretation is not meant to imply that A. actinomycetemcomitans could not move from one tooth to another over time, but our results do suggest that movement from tooth surface to tooth surface is less likely to occur than movement from buccal to tooth surfaces. To our knowledge, a direct comparison of desorption of A. actinomycetemcomitans from BECs and from SHA and examination of its subsequent transfer to oral tissues have not been done prior to the experiments reported herein (5, 6).
Our in vitro data support the concept that A. actinomycetemcomitans is readily released from BECs, since 1 h after A. actinomycetemcomitans attachment to BECs was achieved, A. actinomycetemcomitans was found to be contained within the supernatant fluid (11). Once A. actinomycetemcomitans was desorbed from BECs, it was then in a position to be transferred to SHA surfaces (Fig. 5). Thus, A. actinomycetemcomitans binding to BECs is a dynamic process, and as a result, BECs can provide a surface for binding, release, and transfer of A. actinomycetemcomitans from one surface to another. In a converse manner, the aggregative behavior and tenacious adherence to SHA of A. actinomycetemcomitans do not permit it to be released into the supernatant fluid. As a result of this persistent binding to SHA, no detectable desorption of A. actinomycetemcomitans could be seen, and transfer from SHA to BECs did not occur. These in vitro data provide a biological rationale that supports the in vivo observation indicating that those subjects with A. actinomycetemcomitans on their BECs could subsequently colonize SHA in vivo and suggest that BECs can provide a reservoir for A. actinomycetemcomitans in the oral cavity. Along these lines, our results support previous claims that the oral mucosal surfaces can serve as a reservoir for A. actinomycetemcomitans and other Gram-negative microbes (30, 36).
With regard to A. actinomycetemcomitans binding to SHA, the in vitro data suggest that A. actinomycetemcomitans bound to tooth-like surfaces (mediated predominantly by Flp) is aggregative, linear, and static (Fig. 3A). Furthermore, the aggregative behavior demonstrated by A. actinomycetemcomitans and attributed to Flp appears to allow A. actinomycetemcomitans to pile onto the initial tooth surface in vivo to form a tooth-associated early-colonizing biofilm (8). In the mutational studies, flp mutation had the greatest effect on A. actinomycetemcomitans binding to SHA, while aae mutation had the greatest effect on binding to BECs (Fig. 3B). Both Flp and ApiA appeared to have an effect on binding to both SHA and BECs, while Aae appeared to affect only binding to BECs. In the case of ApiA-mediated binding and autoaggregation, ApiA effects have been shown to occur exclusively when A. actinomycetemcomitans is present at a high density, as seen in the in vitro studies we reported (in this study, a density of 1 × 109 cells/ml was used) (11, 40). Since most infections occur at a low density of infecting cells, ApiA is likely to have a minimal effect in the early stages of LAP. In contrast, Flp and Aae are effective at low cell densities and are thus more likely to influence the early stages of LAP. Taken together, evidence indicating that A. actinomycetemcomitans can be (i) transmitted from mother to child (25, 33, 39) in predentate children, (ii) detected on buccal sites as the predominant colonization site in the oral cavity in healthy children (4, 19), and (iii) detected primarily in Old World primates and humans speaks to the possibility that A. actinomycetemcomitans adhesins that demonstrate specificity at low cell densities, such as Aae, could be critical for initial colonization of soft tissue sites in the oral cavity (8). Since Aae appears to be specific for buccal epithelia obtained from humans and Old World primates at low cell densities (11, 40), we suggest that specificity may outweigh powerful nonspecific attachment factors such as Flp in terms of A. actinomycetemcomitans transmission and its initial host selection; however, this hypothesis still needs to be proven.
As shown in previous in vitro studies, movement of A. actinomycetemcomitans away from the initial tooth-related biofilm would take some time (18, 20). The data from the current in vitro and in vivo experiments support clinical studies showing that A. actinomycetemcomitans binds avidly to tooth surfaces and is difficult to eliminate (32, 38). These data also suggest that therapeutic approaches directed at the physical removal of A. actinomycetemcomitans from tooth surfaces without consideration of soft tissue reservoirs can fall short of full eradication of A. actinomycetemcomitans, since A. actinomycetemcomitans appears to recolonize after standard therapy (8).
In conclusion, in this paper we have shown that the presence of A. actinomycetemcomitans on buccal cells in vivo can lead to colonization of initially aseptic tooth surrogates within 4 to 7 h, thereby supporting the argument that A. actinomycetemcomitans may use buccal cells as a reservoir for initial attachment and eventual movement to nonshedding tooth surfaces. This conclusion does not rule out the fact that other mucosal surfaces can also take part in this dynamic process, but we have focused on the buccal epithelium in this study. Moreover, because A. actinomycetemcomitans colonizes the tooth surface within a period as early as 4 to 6 h, A. actinomycetemcomitans may be considered among the early colonizers of tooth surfaces under the right circumstances (mucosal reservoir) and in vulnerable individuals (those who harbor A. actinomycetemcomitans). The significance of this dynamic process warrants further consideration in the context of the aggressive mucosal infection localized aggressive periodontitis.
Supplementary Material
Acknowledgments
This work was supported in part by Public Health Research grants DE-017968 and DE-016306 from the National Institute of Dental and Craniofacial Research.
We thank Maribasappa Karched for his thoughtful suggestions.
Footnotes
Published ahead of print on 29 September 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Beachey, E. H. 1981. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surfaces. J. Infect. Dis. 143:475-481. [DOI] [PubMed] [Google Scholar]
- 2.Bueno, L. C., M. P. A. Meyer, and J. M. DiRienzo. 1998. Relationship between conversion of localized juvenile periodontitis-susceptible children from health to disease and Actinobacillus actinomycetemcomitans leukotoxin promoter structure. J. Periodontol. 69:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortelli, J. R., S. C. Cortelli, S. Jordan, V. I. Haraszthy, and J. J. Zambon. 2005. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J. Clin. Periodontol. 32:860-866. [DOI] [PubMed] [Google Scholar]
- 4.Eger, T., L. Zoller, H. P. Muller, S. Hoffmann, and D. Lobinsky. 1996. Potential diagnostic value of sampling oral mucosal surfaces for Actinobacillus actinomycetemcomitans in young adults. Eur. J. Oral Sci. 104:112-117. [DOI] [PubMed] [Google Scholar]
- 5.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 6.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 7.Fine, D. H., D. Furgang, L. M. Steinberg, A. M. Olshan, B. E. Kohut, J. F. Coelho, and D. S. Harper. 2000. A model for clinical evaluation of anti-microbial effects of agents on plaque colonization. Am. J. Dent. 13:153-158. [PubMed] [Google Scholar]
- 8.Fine, D. H., J. B. Kaplan, S. C. Kachlany, and H. C. Schreiner. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol. 2000 42:114-157. [DOI] [PubMed] [Google Scholar]
- 9.Fine, D. H., K. Markowitz, D. Furgang, K. Fairlie, J. Ferrrandiz, C. Nasri, M. McKiernan, R. Donnelly, and J. Gunsolley. 2009. Macrophage inflammatory protein 1-a: a salivary biomarker of bone loss in a longitudinal cohort study of children at risk for aggressive periodontal disease? J. Periodontol. 80:106-113. [DOI] [PubMed] [Google Scholar]
- 10.Fine, D. H., K. Markowitz, D. Furgang, K. Fairlie, J. Ferrandiz, C. Nasri, M. McKiernan, and J. Gunsolley. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: a longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45:3859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fine, D. H., K. Velliyagounder, D. Furgang, and J. B. Kaplan. 2005. The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and Old World primates. Infect. Immun. 73:1947-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fives-Taylor, P. M., D. H. Meyer, K. P. Mintz, and C. Brissette. 1999. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000 20:136-167. [DOI] [PubMed] [Google Scholar]
- 13.Gold, O. G., H. V. Jordan, and J. van Houte. 1973. A selective medium for Streptococcus mutans. Arch. Oral Biol. 18:1357-1364. [DOI] [PubMed] [Google Scholar]
- 14.Haubek, D., O.-K. Ennibi, P. Poulsen, M. Vaeth, and M. Kilian. 2008. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet 371:237-242. [DOI] [PubMed] [Google Scholar]
- 15.Haubek, D., K. Poulsen, J. Westergaard, G. Dahlen, and M. Kilian. 1996. Highly toxic clone of Actinobacillus actinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J. Clin. Microbiol. 34:1576-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, T., T. Nishihara, D. R. Demuth, and I. Ishikawa. 1999. A novel insertion sequence increases the expression of leukotoxicity in Actinobacillus actinomycetemcomitans clinical isolates. J. Periodontol. 70:1261-1268. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, B., M. Wilson, L. Sharp, and J. M. Ward. 2002. Actinobacillus actinomycetemcomitans. J. Med. Microbiol. 51:1013-1020. [DOI] [PubMed] [Google Scholar]
- 18.Izano, E. A., I. Sadovskaya, H. Wang, E. Vinogradov, C. Ragunath, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2008. Poly-N-acetylglucosamine mediates biofilm formation and resistance in Aggregatibacter actinomycetemcomitans. Microb. Pathog. 44:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. Flp-1, first representative of a new pilin gene subfamily, is required for nonspecific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan, J. B., M. F. Meyenhoff, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilian, M., E. V. G. Frandsen, D. Haubek, and K. Poulsen. 2006. The etiology of periodontal disease revisited by population genetic analysis. Periodontol. 2000 42:158-179. [DOI] [PubMed] [Google Scholar]
- 22.Kilian, M., and G. Rolla. 1975. Initial colonization of teeth in monkeys as related to diet. Infect. Immun. 14:1022-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. England, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsuzawa, H., R. Asakawa, T. Kawai, K. Ochiai, T. Fujikawara, M. A. Taubman, M. Ohara, H. Kurihara, and M. Sugai. 2002. Identification of six major outer membrane proteins from Actinobacillus actinomycetemcomitans. Gene 288:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Lamell, W. C., A. L. Griffin, D. L. McClellan, and E. J. Leys. 2000. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J. Clin. Microbiol. 38:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, J., E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, and F. G. Oppenheim. 2004. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97:1311-1318. [DOI] [PubMed] [Google Scholar]
- 27.Loesche, W. J., and S. A. Syed. 1978. Bacteriology of human experimental gingivitis: effect of plaque and gingivitis score. Infect. Immun. 21:830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macheleidt, A., H.-P. Muller, T. Eger, M. Putzker, and L. Zoller. 1999. Absence of an especially toxic clone among isolates of Actinobacillus actinomycetemcomitans recovered from army recruits. Clin. Oral Invest. 3:161-167. [DOI] [PubMed] [Google Scholar]
- 29.Mombelli, A., R. Gmur, N. P. Lang, E. Corbet, and J. Frey. 1999. Actinobacillus actinomycetemcomitans in Chinese adults. Serotype distribution and analysis of leukotoxin gene promoter locus. J. Clin. Periodontol. 26:505-510. [DOI] [PubMed] [Google Scholar]
- 30.Muller, H. P., A. Heinecke, A. Fuhrmann, T. Eger, and L. Zoller. 2001. Intraoral distribution of Actinobacillus actinomycetemcomitans in young adults with minimal periodontal disease. J. Periodontal Res. 36:114-123. [DOI] [PubMed] [Google Scholar]
- 31.Planet, P. J., S. C. Kachlany, D. H. Fine, R. DeSalle, and D. H. Figurski. 2003. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34:193-198. [DOI] [PubMed] [Google Scholar]
- 32.Renvert, S., M. Wikstrom, G. Dahlen, J. Slots, and J. Egelberg. 1990. Effect of root debridement on the elimination of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis from periodontal pockets. J. Clin. Periodontol. 17:345-350. [DOI] [PubMed] [Google Scholar]
- 33.Ringler, J., M. Houpt, H. C. Schreiner, M. Mohan, and D. H. Fine. 2009. The prevalence of selected oral bacteria in children as determined by real-time PCR. M.S. thesis. NJDS, UMDNJ, Newark, NJ.
- 34.Ritz, H. L. 1967. Microbial population shifts in developing human dental plaque. Arch. Oral Biol. 12:1561-1568. [DOI] [PubMed] [Google Scholar]
- 35.Rose, J. E., D. H. Meyer, and P. M. Fives-Taylor. 2003. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 71:2384-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudney, K. J., R. Chen, and J. G. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selwood, R., R. A. Gibbons, G. W. Jones, and J. M. Rutter. 1975. Adhesion of enteropathogenic Escherichia coli to pig intestinal brush borders: the existence of two pig phenotypes. Med. Microbiol. 8:405-411. [DOI] [PubMed] [Google Scholar]
- 38.Takamatsu, N., K. Yano, T. He, M. Umeda, and I. Ishikawa. 1999. Effect of initial periodontal therapy on the frequency of detecting Bacteroides forsythus, Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. J. Periodontol. 70:574-580. [DOI] [PubMed] [Google Scholar]
- 39.Tanner, A., P. M. Milgrom, R. Kent, Jr., S. A. Mokeem, R. C. Page, S. A. Liao, C. A. Riedy, and J. Bruss. 2002. Similarity of the oral microbiota of pre-school children with that of their caregivers in a population-based study. Oral Microbiol. Immunol. 17:379-387. [DOI] [PubMed] [Google Scholar]
- 40.Yue, G., J. B. Kaplan, D. Furgang, K. G. Manfield, and D. H. Fine. 2007. A second Aggregatibacter actinomycetemcomitans autotransporter adhesin exhibits specificity for buccal epithelial cells in humans and Old World primates. Infect. Immun. 75:4440-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 42.Zylbert, L. J., and H. V. Jordan. 1982. Development of a selective medium for detection and enumeration of Actinomyces viscosus and Actinomyces naeslundii in dental plaque. J. Clin. Microbiol. 15:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.