Abstract
The only clinically validated assay available to determine HIV tropism is Trofile, an assay that possesses some limitations. Our first aim was to develop a new phenotypic tropism test (TROCAI [tropism coreceptor assay information]) and to categorize results generated by this test according to the virological response to a short-term exposure to the CCR5 receptor antagonist maraviroc (maraviroc clinical test). Our second aim was to compare TROCAI results to those obtained by Trofile enhanced sensitivity (ES) and to different genotypic algorithms. TROCAI assayed HIV tropism in 33 HIV-infected patient viral isolates obtained from a modified coculture, followed by multiple infection cycles of indicator cells. TROCAI obtained a reportable result in all patients with viral loads of >500 HIV RNA copies/ml and in 3/6 patients with <500 HIV RNA copies/ml (30/33 patients, 91.9%). Patients who responded to maraviroc had an X4-using virus proportion in indicator cell supernatant of 0 to 0.41%. Hence, we used the threshold of 0.5% to categorize TROCAI results as R5 (<0.5%) or dual/mixed (>0.5%). The concordance between TROCAI and Trofile (ES) was 22/24 (91.6%), and with genotypic approaches it was 22/26 (84.6%). TROCAI results, which were categorized in this study by the maraviroc clinical test, could be used as a test in addition to those currently used to select patients for treatment with CCR5 antagonists.
Since the first CCR5 receptor antagonist drug, maraviroc (MRV), was commercialized, tropism assays have become a leading topic in HIV research. According to actual guidelines (12), MRV is part of a regimen for pretreated and viremic treated patients whose viruses have been classified as R5 tropic by using a validated tropism test.
Different genotypic and phenotypic tools have been used to assay HIV tropism (2). Despite the improvements, genotypic algorithms are not sensitive enough, and their predictive values have limitations (7, 14, 16). Another genotypic approach is ultradeep sequencing; however, this is currently expensive and not applicable to daily clinical practice (1). Two different commercially available phenotypic assays allow the determination of viral tropism: the tropism recombinant test (TRT) (VIRalliance, France) (24) and the original phenotypic test Trofile (Monogram Biosciences, South San Francisco, CA) (27). Trofile is the only clinically validated tropism test; however, it has some limitations, such as (i) high cost, (ii) prolonged time to obtain a confirmed result, (iii) availability (samples need to be sent to South San Francisco, CA), (iv) a considerable proportion of “nonreportable” results, (v) the requirement that samples contain more than 1,000 HIV RNA copies/ml, and (vi) limited access in developing countries. Furthermore, discordance in two consecutive Trofile results (separated by approximately 1 month) has been observed in ∼10% of patients before being exposed to CCR5 inhibitors (15, 23). These limitations may exclude some patients from using MRV as part of a therapeutic program. Therefore, it is important to develop resources other than Trofile (enhanced sensitivity [ES]) to test HIV tropism.
In this study, we developed a new phenotypic HIV-1 tropism assay (TROCAI [tropism coreceptor assay information]) based on the production of viral isolates from patients and multiple rounds of infection of indicator cells, modifying a technique previously reported by our group (22). In parallel, we have shown and recently extended the findings that the virological response to a short-term exposure to MRV could be used to predict the indication for CCR5 receptor antagonists, the maraviroc clinical test (MCT) (10, 11). This scenario gave us the unique opportunity to compare TROCAI results with those obtained after MCT.
The first aim of this work was to describe a new phenotypic tropism test (TROCAI) and to categorize TROCAI results by using MCT. Our second aim was to compare TROCAI results with those obtained by the most commonly used tests to assay HIV tropism, Trofile ES and different genotypic algorithms.
MATERIALS AND METHODS
Patients.
The study was performed at the Infectious Diseases Service of Virgen del Rocio University Hospital (Seville, Spain). From 1 July 2008 through 1 October 2009, 42 patients were included in the MCT (10). Thirty-three had peripheral blood mononuclear cell (PBMC) samples available and were included in the present study. Briefly, MCT inclusion criteria were (i) persistently detectable viral load of >40 HIV RNA copies/ml during the previous 6 months, (ii) no highly active antiretroviral therapy (HAART) modification in the preceding 6 months, (iii) no HAART reintroduction after previous supervised treatment interruption (STI), (iv) no previous treatment with CCR5 receptor antagonists, and (v) future therapeutic options other than MRV.
Patients or guardians (for those patients under 18 years old) provided written, informed consent, and the ethical committee of the hospital approved the study.
Quantification of HIV RNA.
HIV-1 RNA was measured in fresh plasma samples from patients and in frozen samples of cell-free supernatants by quantitative PCR (Cobas Ampliprep/Cobas TaqMan HIV-1 test; Roche Molecular Systems, Basel, Switzerland) according to the manufacturer's instructions. This assay has a detection limit of 40 HIV RNA copies/ml.
Determination of HIV-1 coreceptor usage. (i) MCT.
Patients began an 8-day exposure to MRV. The response was considered positive if a significant viral load reduction, defined as a reduction of ≥1 log10 HIV RNA copies/ml, or an undetectable viral load (<40 HIV RNA copies/ml) was achieved at day 8 (10).
(ii) Trofile (ES).
Circulating virus was quantified in patient plasma samples containing more than 1,000 HIV RNA copies/ml according to the manufacturer's instructions (27) by using the PhenoSense HIV entry assay for coreceptor tropism (Monogram Biosciences Inc., South San Francisco, CA). This version of Trofile claims to have greater sensitivity, detecting 0.3% of X4 strains (20).
(iii) Genotypic methods.
V3 loop amplification and sequencing were performed as previously described (25). V3 sequences were interpreted using 4 different bioinformatic genotypic predictors of tropism (PSSM, geno2pheno, C4.5, and SVM), as previously reported (3), which are freely available at 3 websites: geno2pheno at http://coreceptor.bioinf.mpi-inf.mpg.de/index.php, PSSM at http://fortinbras.us/cgi-bin/fssm/fssm.pl, and C4.5 and SVM at http://genomiac2.ucsd.edu:8080/wetcat/tropism.html. In all cases, HIV-1 variants were classified as R5 or dual/mixed tropic (D/M), with the latter including pure X4 and dual/mixed viruses. When two or more genotypic predictors obtained a D/M result, the tropism was considered D/M.
(iv) TROCAI.
TROCAI is based on the production of viral isolates from patients through improvement of a coculture technique previously published by our group (22) and multiple rounds of infection of indicator cells. TROCAI consists of two main steps as follows.
(a) Primary HIV-1 isolation.
Patient PBMCs were isolated from fresh whole-blood samples by density gradient centrifugation (BD Vacutainer CPT) prior to MRV administration and frozen until needed. To produce viral stocks, patient PBMCs were thawed and cocultured with HIV-hepatitis C virus (HCV)-uninfected donor PBMCs that had been pretreated with 5 μg/ml of phytohemagglutinin (PHA; Boehringer Ingelheim). Cells were cultured in RPMI 1640 medium (Biochrom AG) supplemented with 10% fetal calf serum (FCS; Biochrom AG), 20 U/ml of interleukin 2 (IL-2; R&D Systems), 2 mM glutamine, 100 μg/ml streptomycin, and 100 U/ml penicillin (Sigma-Aldrich). Cultures were maintained until they were considered positive (see below). Cell-free supernatants were harvested three times a week and stored at −80°C. Cultures were fed once per week with PHA-stimulated HIV-uninfected donor PBMCs.
Cultures were considered positive when at least two consecutive viral load measurements were >1 × 106 HIV RNA copies/ml. The first cell-free supernatant with an HIV viral load above this value was then used for further infection assays. In some patients with a plasma viral load below 1,000 HIV-1 RNA copies/ml, CD8+ T cells were depleted in both infected and uninfected PBMCs by using magnetic bead separation (Miltenyi Biotec, Auburn, CA) to favor viral replication.
(b) Determination of HIV-1 tropism.
Tropism was determined using U87 cell lines (U87-CD4+CCR5+ and U87-CD4+CXCR4+) cultured in Dulbecco's modified Eagle's medium (DMEM high glucose; PAA Laboratories, Germany) supplemented with 5% fetal calf serum (FCS; Biochrom AG), 2 mM glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin (Sigma-Aldrich), and 2 μg/ml puromycin and 300 μg/ml Geneticin G418 (Sigma).
U87 cell lines were infected with 1 × 106 HIV RNA copies of cell-free virus supernatant overnight. Medium was changed at day 3 and tested for HIV-1 RNA copies/ml on day 6 as described above.
The measurements of HIV RNA copies/ml in the supernatant of the cultures of X4 and R5 cell lines at day 6 showed the tropism of viral isolates from the patients. Results are presented as the proportion of virus isolates using the X4 or R5 coreceptor. We used the X4-tropic virus strain NL4.3 and the R5-tropic virus strain BaL as positive controls.
Statistical analysis.
Statistical analyses were performed using the Statistical Package for the Social Sciences software (SPSS 16.0; SPSS Inc., Chicago, IL). Differences between groups were analyzed with Mann-Whitney U tests for continuous variables and with chi-square tests for categorical variables. All continuous variables were expressed as medians (interquartile range [IQR]), and categorical variables were expressed as percentages. All differences with a P value of <0.05 were considered statistically significant.
RESULTS
Viral production of PBMC cocultures.
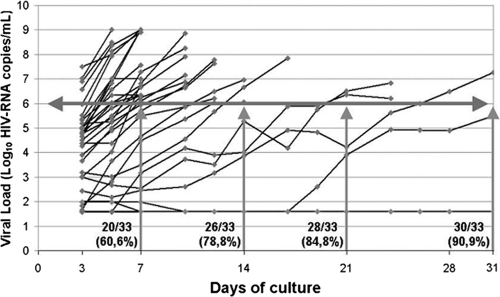
Baseline characteristics of the patients are shown in Table 1. Viral production is shown in Fig. 1. We observed that 30/33 (90.9%) cultures were positive within 1 month. We obtained positive cultures in 27/27 (100%) of patients with viral loads above 500 HIV RNA copies/ml and in 3/6 (50%) of patients with viral loads below 500 HIV RNA copies/ml.
TABLE 1.
Baseline characteristics of patients (n = 33)
| Patient characteristic | Valuec |
|---|---|
| Age (yr) | 42.1 (32.5-45.4) |
| Male sex, n (%) | 23 (69.7) |
| HCV coinfection,an (%) | 13 (39.4) |
| CD4+ (no. of cells/mm3) | 223 (112-365) |
| Risk group, n (%) | |
| IDUb | 19 (57.6) |
| Sexual transmission | 7 (21.3) |
| Vertical transmission | 5 (15.2) |
| Blood transfusion | 2 (6.1) |
| Stage C (CDC), n (%) | 17 (51.5) |
| Plasma HIV-1 RNA (no. of copies/ml), n (%) | |
| <500 | 6 (18.2) |
| 500-999 | 3 (9.1) |
| >1,000 | 24 (72.7) |
Positive PCR for hepatitis C virus.
IDU, intravenous drug users.
Values other than number (percent) are expressed as medians (interquartile range [IQR]).
FIG. 1.
Viral dynamics during coculture. The horizontal gray line represents the threshold of positive cultures (>1 × 106 HIV RNA copies/ml). Vertical gray lines show the positive cultures and cumulative frequencies after 7, 14, 21, and 31 days of culture. Black lines represent the viral production from each patient. Cultures were stopped if more than two consecutive measurements were above 1 × 106 HIV RNA copies/ml.
Of the nine cultures from patients with viral loads under 1,000 HIV RNA copies/ml, the first two were positive without CD8+ T-cell depletion. However, the next two were negative, so we performed CD8+ T-cell depletion in these samples, obtaining positive cultures in both. In the remaining five cultures, CD8+ T cells were depleted, with two being positive and the remaining three being negative, having viral loads under 500 HIV RNA copies/ml. Therefore, CD8+ T-cell depletion was assayed in 7/9 patients with viral loads of <1,000 HIV RNA copies/ml, and a positive culture was obtained in 4 of these patients (57.1%).
Categorization of TROCAI results by using MCT.
MCT was used to categorize TROCAI results in two ways: D/M and R5 tropic. This was done by correlating the proportion of X4/R5 virus in U87 cell line supernatant with the response to MRV short-term exposure. In this way, as shown in Table 2, all patients with positive MCT results showed a TROCAI result of ≤0.41% X4 strains and were classified as R5 with the exception of patients 20, 21, and 22, whose cocultures were negative. All patients with a negative MCT showed a TROCAI result of ≥0.6% X4 strains and were classified as D/M. Indeed, all patients with a positive MCT (except the three patients mentioned above) showed ≤2.52 × 103 HIV RNA copies/ml in U87-CD4+CXCR4+ cell supernatant, whereas all patients negative for MCT showed ≥5.91 × 104 HIV RNA copies/ml in U87-CD4+CXCR4+ cell supernatant with the exception of a single patient, number 24.
TABLE 2.
Coreceptor assay resultsa
| Patient no. | MCT | TROCAI |
Trofile | Genotype | PBVL | ||||
|---|---|---|---|---|---|---|---|---|---|
| VL U87-X4 | VL U87-R5 | % X4 | % R5 | Categorization | |||||
| 1 | + | 0 | 9.28 × 106 | 0 | 100 | R5 | R5 | R5 | 421,000 |
| 2 | + | 0 | 4.31 × 106 | 0 | 100 | R5 | R5 | R5 | 7,350 |
| 3 | + | 0 | 1.27 × 104 | 0 | 100 | R5 | R5 | R5 | 16,100 |
| 4 | + | 0 | 5.23 × 104 | 0 | 100 | R5 | R5 | R5 | 28,000 |
| 5 | + | 0 | 5.68 × 106 | 0 | 100 | R5 | D/M | R5 | 15,500 |
| 6 | + | 0 | 5.53 × 107 | 0 | 100 | R5 | ND | NS | 810 |
| 7 | + | 0 | 3.49 × 104 | 0 | 100 | R5 | ND | R5 | 496 |
| 8 | + | 2.19 × 102 | 4.61 × 107 | 0 | 100 | R5 | R5 | R5 | 114,000 |
| 9 | + | 4.38 × 102 | 1.00 × 109 | 0 | 100 | R5 | R5 | NS | 1,660 |
| 10 | + | 5.17 × 102 | 3.90 × 107 | 0 | 100 | R5 | R5 | R5 | 10,500 |
| 11 | + | 6.87 × 102 | 2.53 × 107 | 0 | 100 | R5 | ND | R5 | 835 |
| 12 | + | 7.06 × 102 | 7.19 × 108 | 0 | 100 | R5 | R5 | R5 | 22,500 |
| 13 | + | 1.62 × 103 | 7.50 × 107 | 0 | 100 | R5 | R5 | R5 | 47,000 |
| 14 | + | 1.99 × 103 | 6.41 × 107 | 0 | 100 | R5 | R5 | R5 | 14,100 |
| 15 | + | 2.52 × 103 | 2.43 × 107 | 0.01 | 99.99 | R5 | ND | R5 | 437 |
| 16 | + | 1.05 × 102 | 4.76 × 105 | 0.02 | 99.98 | R5 | R5 | R5 | 50,900 |
| 17 | + | 9.40 × 102 | 1.89 × 106 | 0.05 | 99.95 | R5 | R5 | R5 | 167,000 |
| 18 | + | 7.94 × 102 | 6.75 × 105 | 0.12 | 99.88 | R5 | R5 | NA | 6,060 |
| 19 | + | 1.55 × 103 | 3.75 × 105 | 0.41 | 99.59 | R5 | R5 | R5 | 42,700 |
| 20 | + | ND | ND | ND | ND | NC | ND | R5 | 457 |
| 21 | + | ND | ND | ND | ND | NC | ND | NA | 294 |
| 22 | + | ND | ND | ND | ND | NC | ND | NA | 87 |
| 23 | − | 5.91 × 104 | 9.81 × 106 | 0.6 | 99.4 | D/M | D/M | D/M | 32,900 |
| 24 | − | 1.28 × 103 | 1.64 × 105 | 0.77 | 99.23 | D/M | ND | D/M | 827 |
| 25 | − | 2.70 × 107 | 1.11 × 109 | 2.37 | 97.63 | D/M | R5 | D/M | 40,000 |
| 26 | − | 7.47 × 105 | 2.75 × 107 | 2.65 | 97.35 | D/M | D/M | D/M | 192,000 |
| 27 | − | 2.21 × 105 | 2.89 × 106 | 7.1 | 92.9 | D/M | D/M | R5 | 1,000,000 |
| 28 | − | 1.49 × 106 | 6.20 × 106 | 19.38 | 80.62 | D/M | D/M | R5 | 74,300 |
| 29 | − | 7.98 × 106 | 1.84 × 106 | 81.26 | 18.74 | D/M | D/M | R5 | 33,000 |
| 30 | − | 4.12 × 106 | 6.67 × 105 | 86.07 | 13.93 | D/M | D/M | D/M | 5,110 |
| 31 | − | 3.36 × 106 | 1.95 × 104 | 99.42 | 0.57 | D/M | ND | NA | 271 |
| 32 | − | 1.44 × 108 | 4.69 × 106 | 96.85 | 3.15 | D/M | D/M | D/M | 147,000 |
| 33 | − | 5.05 × 107 | 6.37 × 105 | 98.75 | 1.25 | D/M | D/M | R5 | 306,000 |
VL U87-X4, viral load (number of HIV RNA copies/ml) in U87-CD4+CXCR4+ cell supernatant; VL U87-R5, viral load (number of HIV RNA copies/ml) in U87-CD4+CCR5+ cell supernatant; PBVL, plasma baseline viral load; NC, negative coculture; NS, no sample; ND, not determined (<1,000 HIV RNA copies/ml); NA, not amplifiable. Discordances between tropism predictions are shown in boldface.
Patient 24 showed only 1.28 × 103 HIV RNA copies/ml in U87-CD4+CXCR4+ cell supernatant; however, when the X4/R5 proportion was considered, this patient was categorized as D/M by TROCAI.
Interestingly, virus production from patient 24 cultures reached only 2.98 × 105 HIV RNA copies/ml, and this amount of virus was not enough to infect U87 cell lines. Therefore, we tested a new strategy to infect U87 cell lines in this patient, using spinoculation to increase viral adsorption on the cell membrane without affecting viral entry (21) and obtaining a positive result. As a control, we performed spinoculation in another patient with positive culture and obtained the same percentages of X4 and R5 strains with and without spinoculation (data not shown).
Correlation among TROCAI, Trofile (ES), and genotypic analysis. (i) TROCAI and Trofile (ES).
Trofile (ES) could report results only for 24/33 (72.7%) patients, whereas TROCAI reported results for 30/33 (90.9%) patients. The remaining patients had viral loads under 1,000 HIV RNA copies/ml, which could not be reported by the Trofile (ES) assay. A global concordance of 22/24 (91.6%) between TROCAI and Trofile (ES) was observed. Two nonconcordant results were found (patients 5 and 25). Patient 5 was reported as D/M by Trofile (ES) but was positive by MCT. TROCAI showed a result consisting of 100% R5 strains. On the other hand, patient 25 was reported as R5 by Trofile (ES) but was negative by MCT. In this case, we found 2.4% X4 strains and 97.6% R5 strains using TROCAI, and the result was therefore considered D/M.
(ii) TROCAI and genotype analysis.
Genotypic approaches reported results for 27/31 (87.1%) patients. The V3 loop of 4 patients could not be amplified, and for 2 other patients, no samples were available. A global concordance of 22/26 (84.6%) between TROCAI and genotype was observed in available serum samples (Table 2).
In testing the four algorithms separately, the highest correlation with TROCAI was found using geno2pheno (88.5%), followed by PSSM (80.8%) and then by SVM (80.8%), and the lowest was found with C4.5 (76.9%) (Table 3).
TABLE 3.
Predictions of the four genotypic approachesa
| Patient no. | Sample | Prediction by: |
|||||
|---|---|---|---|---|---|---|---|
| G2P | PSSM | SVM | C4.5 | MCT | TROCAI | ||
| 1 | Serum | CCR5 | CCR5 | D/M | CCR5 | + | R5 |
| 2 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| V-Iso | D/M | D/M | D/M | CCR5 | + | R5 | |
| 3 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| V-Iso | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| P-DNA | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| 4 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| V-Iso | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| P-DNA | CCR5 | CCR5 | D/M | CCR5 | + | R5 | |
| 5 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| V-Iso | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| P-DNA | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| 6 | Serum | No sample | No sample | No sample | No sample | + | R5 |
| 7 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 8 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 9 | Serum | No sample | No sample | No sample | No sample | + | R5 |
| 10 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 11 | Serum | D/M | CCR5 | CCR5 | CCR5 | + | R5 |
| 12 | Serum | CCR5 | D/M | CCR5 | CCR5 | + | R5 |
| 13 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| P-DNA | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| 14 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 15 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| P-DNA | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 | |
| 16 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 17 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 18 | Serum | Not amplified | Not amplified | Not amplified | Not amplified | + | R5 |
| 19 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | R5 |
| 20 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | + | NA |
| 21 | Serum | Not amplified | Not amplified | Not amplified | Not amplified | + | NA |
| 22 | Serum | Not amplified | Not amplified | Not amplified | Not amplified | + | NA |
| 23 | Serum | D/M | D/M | D/M | D/M | − | D/M |
| 24 | Serum | D/M | D/M | D/M | CCR5 | − | D/M |
| 25 | Serum | D/M | D/M | D/M | D/M | − | D/M |
| 26 | Serum | D/M | D/M | CCR5 | CCR5 | − | D/M |
| V-Iso | D/M | D/M | CCR5 | CCR5 | − | D/M | |
| P-DNA | D/M | D/M | CCR5 | CCR5 | − | D/M | |
| 27 | Serum | CCR5 | CCR5 | CCR5 | CCR5 | − | D/M |
| V-Iso | CCR5 | CCR5 | CCR5 | CCR5 | − | D/M | |
| P-DNA | D/M | D/M | CCR5 | CCR5 | − | D/M | |
| 28 | Serum | CCR5 | CCR5 | D/M | CCR5 | − | D/M |
| V-Iso | CCR5 | CCR5 | CCR5 | CCR5 | − | D/M | |
| 29 | Serum | D/M | CCR5 | CCR5 | CCR5 | − | D/M |
| 30 | Serum | D/M | D/M | D/M | D/M | − | D/M |
| P-DNA | D/M | D/M | D/M | D/M | − | D/M | |
| 31 | Serum | Not amplified | Not amplified | Not amplified | Not amplified | − | D/M |
| 32 | Serum | D/M | D/M | D/M | D/M | − | D/M |
| 33 | Serum | D/M | CCR5 | CCR5 | CCR5 | − | D/M |
G2P, geno2pheno algorithm; PSSM, PSSM algorithm; C4.5, C4.5 algorithm; SVM, SVM algorithm; MCT, maraviroc clinical test; V-Iso, virus isolates; P-DNA, proviral DNA; NA, not available.
To explore possible tropism changes among serum, virus isolates, and proviral DNA, genotype analysis was also performed under these three conditions in the 10 available patient samples. Interestingly, despite the fact that the same tropism result was observed in most of the samples, some changes were found in the amino acid sequence, with a mean change of 2.1 amino acids (aa) per sequence between serum and viral isolates and of 1.1 aa per sequence between serum and proviral DNA (see Table S1 in the supplemental material).
Furthermore, we found discordances between samples in 2/10 patients (see Table S1 in the supplemental material). For patient 27, the genotype result for serum and virus isolates was R5, concordant with TROCAI, but in proviral DNA, the result was considered D/M. For patient 2, reported as R5 by TROCAI, the genotype was concordant with TROCAI in serum but not in virus isolates, producing a D/M result.
Viral tropism evolution during viral isolate production.
One important concern was that viral evolution during multiple infection cycles could alter HIV tropism (24). To clarify this issue, we repeated TROCAI in three virus isolates from the same patient obtained with a delayed viral production from three consecutive days of coculture (days 26, 28, and 31). The TROCAI results were found to be similar in each of the three samples. The amount of virus found in both X4 and R5 cell line supernatants gradually increased, but no change was observed in the proportions of the X4/R5 virus variants (Table 4).
TABLE 4.
Results of TROCAI in relation to viral evolution during coculturea
| Patient no. | Day | VL U87-X4 | VL U87-R5 | % X4 | % R5 | TROCAI result |
|---|---|---|---|---|---|---|
| 11 | 26 | 6.87 × 102 | 2.53 × 107 | 0.00 | 100.00 | R5 |
| 11 | 28 | 2.62 × 103 | 2.25 × 107 | 0.01 | 99.99 | R5 |
| 11 | 31 | 4.18 × 104 | 1.00 × 109 | 0.00 | 100.00 | R5 |
VL U87-X4, viral load (number of HIV RNA copies/ml) in U87-CD4+CXCR4+ cell supernatant; VL U87-R5, viral load (number of HIV RNA copies/ml) in U87-CD4+CCR5+ cell supernatant; day, number of days of coculture; R5, CCR5-using virus.
DISCUSSION
In this study, we have developed a novel assay to measure HIV tropism. The results obtained using TROCAI displayed a high degree of concordance with the widely used Trofile (ES) assay, thus demonstrating the potential of TROCAI for HIV tropism testing. Furthermore, we were able to obtain a reportable result in a larger number of patients using the TROCAI test than with the Trofile (ES) assay.
The first aim of this study was to describe a new phenotypic tropism test (TROCAI) and to categorize TROCAI results by using MCT. MCT provided us the unique opportunity to associate the response to a short-term exposure to MRV with the proportion of X4 virus in U87 cell line supernatant obtained in TROCAI. Using MCT, we were able to classify subjects with >0.5% X4-using variants assayed with TROCAI as D/M. This limit has to be taken with caution, and a greater sample size is needed to firmly establish a more definitive cutoff point. Further studies are needed to explain why patients with a small number of X4 strains in the X4 cell line supernatant (even less than 1%) do not respond to MRV exposure in our system. It will also be very interesting to test TROCAI at different time points throughout the course of HIV infection. The study of tropism evolution will help to clarify unanswered questions regarding HIV tropism and the related clinical implications.
Once TROCAI results were categorized, our second aim was to compare TROCAI results with those obtained by Trofile (ES) and different genotypic algorithms. In our study, the global concordance between TROCAI and Trofile (ES) was 22/24 (91.6%). Only two discordant results were found between TROCAI and Trofile (ES) (patients 5 and 25). However, both were clearly categorized using MCT in light of the TROCAI results, and in both patients, genotypic approaches were also concordant with TROCAI (Table 2). According to these results, both patients would have been treated incorrectly if only Trofile (ES) results had been considered.
In a study comparing first-generation Trofile and different phenotypic assay results, correlations similar to that found in the present study between TROCAI and the second-generation Trofile (ES) were shown (13). In a recent study also comparing Trofile (ES) with other phenotypic tropism test results, similar correlations were found between Trofile (ES) and TROCAI (5). However, in this study, clonal HIV-1 variants using limiting dilution biological cloning were used, and these viruses were obtained from only two patients and not in consecutive plasma and PBMC samples (5). On the other hand, correlations found between Trofile (ES) and bioinformatics algorithms in our study are similar to those found in other studies (6, 16, 18, 19). In 8/10 (80%) patients genotyped from serum, proviral DNA, and virus isolates, the same tropism was predicted, supporting the absence of differences in the tropism among different viral sources (26). One of the discordant result sets was that for patient 2, for which genotypic approaches reported a D/M result in the virus isolate; however, the TROCAI result was clearly R5 (0% X4 and 100% R5) (Table 2).
One important aspect of TROCAI is that the rate of positive cultures obtained was 90.9%. However, this rate was reached using frozen PBMCs and is higher than recently reported in studies using both frozen (52%) (13) and fresh (66.4%) (8) PBMCs. Furthermore, we believe that this rate could be increased by introducing improvements to the culture conditions (4, 9, 17, 28). In our study, only 5/33 cultures were negative after a period of 3 weeks. We maintained these five cultures for an additional 2 weeks, at which time 2 cultures became positive. It is worth noting that 26/30 (86.6%) of the TROCAI results were obtained in less than 3 weeks.
One potential limitation of our assay is the traditional notion that extended virus culturing during virus stock production may lead to adaptation of the quasispecies to the new environment, which may not represent the original in vivo HIV population (24). Nonetheless, the TROCAI results do not seem to change after this process (Table 4). This result supports the idea that multiple cycles of infection do not change virus tropism, at least during the time period studied. Furthermore, this result suggests that the proportion of X4/R5 virus is a more reliable tropism result than is the number of HIV RNA copies/ml found in the X4 cell line supernatant.
In summary, TROCAI results, obtained after categorization of the experimental data using a clinical response to MRV, showed a high concordance with Trofile (ES) and genotypic approaches. TROCAI increased the number of reportable results compared with Trofile (ES). For all of these reasons, we suggest that TROCAI could be an additional test to assay HIV tropism.
Supplementary Material
Acknowledgments
This work was supported by Redes Telemáticas de Investigación Cooperativa en Salud (RETICS; 2006, Red de SIDA RD06/0006/0021, 2007-2010) and Fondo de Investigacion Sanitaria (PS09-01595). A.G.-S. has a grant from Fundación Reina Mercedes. E.R.-M. has a grant from Fondo de Investigaciones Sanitarias (CP08/00172). A.G.-P. has a grant from Junta de Andalucía P06-CTS-01579. S.F.-M. has a grant from Fondo de Investigaciones Sanitarias (FIS06/00176).
We are grateful to all of the patients who participated in this study; to José Manuel Lara Ruiz from the Immunology Service for his technical support; to Marien Gutierrez, Magdalena Rodriguez, and Paco Cano for their clinical support; to Maria Tavares, Catherin Hogan, and Luke Noon for critical reading of the manuscript; and to Oscar Jacome for his logistic support. Samples from patients were kindly provided by the HIV BioBank integrated in the Spanish AIDS Research Network (RIS).
There are no conflicts of interest to declare.
Footnotes
Published ahead of print on 13 October 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Archer, J., M. S. Braverman, B. E. Taillon, B. Desany, I. James, P. R. Harrigan, M. Lewis, and D. L. Robertson. 2009. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 23:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, E. A., R. W. Doms, E. M. Fenyö, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 3.Chueca, N., C. Garrido, M. Alvarez, E. Poveda, J. de Dios Luna, N. Zahonero, J. Hernández-Quero, V. Soriano, C. Maroto, C. de Mendoza, and F. García. 2009. Improvement in the determination of HIV-1 tropism using the V3 gene sequence and a combination of bioinformatic tools. J. Med. Virol. 81:763-767. [DOI] [PubMed] [Google Scholar]
- 4.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coakley, E., J. D. Reeves, W. Huang, M. Mangas-Ruiz, I. Maurer, A. M. Harskamp, S. Gupta, Y. Lie, C. J. Petropoulos, H. Schuitemaker, and A. B. van 't Wout. 2009. Comparison of the HIV-1 tropism profiles in clinical samples by the Trofile and MT-2 cell assays. Antimicrob. Agents Chemother. 53:4686-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delobel, P., M. T. Nugeyre, M. Cazabat, C. Pasquier, B. Marchou, P. Massip, F. Barre-Sinoussi, N. Israël, and J. Izopet. 2007. Population-based sequencing of the V3 region of env for predicting the coreceptor usage of human immunodeficiency virus type 1 quasispecies. J. Clin. Microbiol. 45:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Mendoza, C., K. Van Baelen, E. Poveda, E. Rondelez, N. Zahonero, L. Stuyver, C. Garrido, J. Villacian, V. Soriano, and the Spanish HIV Seroconverter Study Group. 2008. Performance of a population-based HIV-1 tropism phenotypic assay and correlation with V3 genotypic prediction tools in recent HIV-1 seroconverters. J. Acquir. Immune Defic. Syndr. 48:241-244. [DOI] [PubMed] [Google Scholar]
- 8.Demeter, L. M., R. J. Bosch, R. W. Coombs, S. Fiscus, J. Bremer, V. A. Johnson, A. Erice, J. B. Jackson, S. A. Spector, K. M. Squires, M. A. Fischl, M. D. Hughes, and S. M. Hammer. 2002. Detection of replication-competent human immunodeficiency virus type 1 (HIV-1) in cultures from patients with levels of HIV-1 RNA in plasma suppressed to less than 500 or 50 copies per milliliter. J. Clin. Microbiol. 40:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 10.Genebat, M., E. Ruiz-Mateos, J. A. León, A. González-Serna, I. Pulido, I. Rivas, S. Ferrando, B. Sanchez, M. A. Muñoz, and M. Leal. 2009. Correlation between Trofile test and virological response to a short-term maraviroc exposure in HIV-infected patients. J. Antimicrob. Chemother. 64:845-849. [DOI] [PubMed] [Google Scholar]
- 11.Genebat, M., E. Ruiz-Mateos, I. Pulido, A. González-Serna, A. García-Pergañeda, G. Méndez, M. C. Romero-Sánchez, S. Ferrando-Martínez, and M. Leal. 2010. Long-term immunovirological effect and tolerability of a maraviroc-containing regimen in routine clinical practice. Curr. HIV Res. 8:482-486. [DOI] [PubMed]
- 12.Hammer, S. M., J. J. Eron, Jr., P. Reiss, R. T. Schooley, M. A. Thompson, S. Walmsley, P. Cahn, M. A. Fischl, J. M. Gatell, M. S. Hirsch, D. M. Jacobsen, J. S. Montaner, D. D. Richman, P. G. Yeni, P. A. Volberding, and the International AIDS Society-USA. 2008. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA 300:555-570. [DOI] [PubMed] [Google Scholar]
- 13.Hosoya, N., Z. Su, T. Wilkin, R. M. Gulick, C. Flexner, M. D. Hughes, P. R. Skolnik, F. Giguel, W. L. Greaves, E. Coakley, and D. R. Kuritzkes. 2009. Assessing HIV-1 tropism: a comparison of assays using replication competent virus versus plasma-derived pseudotyped virions. J. Clin. Microbiol. 47:2604-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, W., J. Toma, S. Fransen, E. Stawiski, J. D. Reeves, J. M. Whitcomb, N. Parkin, and C. J. Petropoulos. 2008. Coreceptor tropism can be influenced by amino acid substitutions in the gp41 transmembrane subunit of human immunodeficiency virus type 1 envelope protein. J. Virol. 82:5584-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landovitz, R. J., J. B. Angel, C. Hoffmann, H. Horst, M. Opravil, Long J., W. Greaves, and G. Fätkenheuer. 2008. Phase II study of vicriviroc versus efavirenz (both with zidovudine/lamivudine) in treatment-naive subjects with HIV-1 infection. J. Infect. Dis. 198:1113-1122. [DOI] [PubMed] [Google Scholar]
- 16.Low, A. J., W. Dong, D. Chan, T. Sing, R. Swanstrom, M. Jensen, S. Pillai, B. Good, and P. R. Harrigan. 2007. Current V3 genotyping algorithms are inadequate for predicting X4 coreceptor usage in clinical isolates. AIDS 21:F17-F24. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz, M., M. Vesanen, K. Tenner-Racz, Y. Cao, J. M. Binley, A. Talal, A. Hurley, X. Jin, M. R. Chaudhry, M. Yaman, S. Frankel, M. Heath-Chiozzi, J. M. Leonard, J. P. Moore, P. Racz, D. F. Nixon, and D. D. Ho. 1999. The effect of commencing combination antiretroviral therapy soon after human immunodeficiency virus type 1 infection on viral replication and antiviral immune responses. J. Infect. Dis. 179:527-537. [DOI] [PubMed] [Google Scholar]
- 18.Poveda, E., E. Seclén, M. del Mar González, F. García, N. Chueca, A. Aguilera, J. J. Rodríguez, J. González-Lahoz, and V. Soriano. 2009. Design and validation of new genotypic tools for easy and reliable estimation of HIV tropism before using CCR5 antagonists. J. Antimicrob. Chemother. 63:1006-1010. [DOI] [PubMed] [Google Scholar]
- 19.Raymond, S., P. Delobel, M. Mavigner, M. Cazabat, C. Souyris, K. Sandres-Sauné, L. Cuzin, B. Marchou, P. Massip, and J. Izopet. 2008. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS 22:F11-F16. [DOI] [PubMed] [Google Scholar]
- 20.Reeves, J. D., E. Coakley, C. J. Petropoulos, and J. M. Whitcomb. 2009. An enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: a review of analytical and clinical studies. J. Viral Entry 3:94-102. [Google Scholar]
- 21.Ruiz-Mateos, E., A. Pelchen-Matthews, M. Deneka, and M. Marsh. 2008. CD63 is not required for production of infectious human immunodeficiency virus type 1 in human macrophages. J. Virol. 82:4751-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Mateos, E., A. Rubio, A. Vallejo, R. De la Rosa, A. Sanchez-Quijano, E. Lissen, and M. Leal. 2004. Thymic volume is associated independently with the magnitude of short- and long-term repopulation of CD4+ T cells in HIV-infected adults after highly active antiretroviral therapy (HAART). Clin. Exp. Immunol. 136:501-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schürmann, D., G. Fätkenheuer, J. Reynes, C. Michelet, F. Raffi, J. van Lier, M. Caceres, A. Keung, A. Sansone-Parsons, L. M. Dunkle, and C. Hoffmann. 2007. Antiviral activity, pharmacokinetics and safety of vicriviroc, an oral CCR5 antagonist, during 14-day monotherapy in HIV-infected adults. AIDS 21:1293-1299. [DOI] [PubMed] [Google Scholar]
- 24.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallejo, A., E. Ruiz-Mateos, S. Molina-Pinelo, N. Soriano-Sarabia, B. de Felipe, S. Gutierrez, A. Sánchez-Quijano, E. Lissen, and M. Leal. 2006. Immunovirologic characteristics of human immunodeficiency virus-infected patients consisting mainly of injecting drug users on highly active antiretroviral treatment with prolonged virologic failure. Viral Immunol. 19:759-767. [DOI] [PubMed] [Google Scholar]
- 26.Verhofstede, C., L. Vandekerckhove, V. V. Eygen, E. Demecheleer, I. Vandenbroucke, B. Winters, J. Plum, D. Vogelaers, and L. Stuyver. 2009. CXCR4-using HIV type 1 variants are more commonly found in peripheral blood mononuclear cell DNA than in plasma RNA. J. Acquir. Immune Defic. Syndr. 50:126-136. [DOI] [PubMed] [Google Scholar]
- 27.Whitcomb, J. M., W. Huang, S. Fransen, K. Limoli, J. Toma, T. Wrin, C. Chappey, L. D. Kiss, E. E. Paxinos, and C. J. Petropoulos. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong, J. K., M. Hezareh, H. F. Günthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.