Abstract
Saksenaea is a monotypic genus belonging to the order Mucorales and capable of producing severe human infections. Through a polyphasic study based on analysis of the sequences of the internal transcribed spacer (ITS) region, domains D1 and D2 of the 28S rRNA gene, and the elongation factor 1α (EF-1α) gene, as well as by evaluation of relevant morphological and physiological characteristics of a set of clinical and environmental strains, we have demonstrated that Saksenaea vasiformis is a complex of species. We propose as new species Saksenaea oblongispora, characterized by oblong sporangiospores and unable to grow at 42°C, and Saksenaea erythrospora, characterized by large sporangiophores and sporangia and by ellipsoid sporangiospores, biconcave in the lateral view. Itraconazole, posaconazole, and terbinafine were active against all isolates included in the study, while amphotericin B, voriconazole, and the echinocandins showed low activity.
The genus Saksenaea S. B. Saksena, belonging to the subphylum Mucoromycotina, was first isolated from a forest soil in India (38). Saksenaea vasiformis S. B. Saksena, the only species of the genus, is a filamentous fungus reported in soil, driftwood, and grains (1, 12, 34), characterized by flask-shaped sporangia, short sporangiophores, oval sporangiospores, and dark rhizoids. It is a thermotolerant fungus that grows between 25°C and 44°C (13, 24, 44) and is able to cause severe human infections in both immunocompromised and immunocompetent hosts. Mucormycosis caused by S. vasiformis most often occurs after traumatic implantation of the fungus but can also be due to inhalation of spores (18), spider bites, insect stings, and the use of indwelling catheters (11, 22, 28, 31). Clinical cases seems to be more common in tropical and subtropical climates than elsewhere and have been reported from Australia (16, 19, 22, 40, 48), India (6, 7, 11, 33), the United States (1, 8, 31, 35, 45), Thailand (44), Tunisia (28), the Middle East (2, 25), and Central and South America (9, 47).
Recent molecular studies, based mostly on internal transcribed spacer (ITS) sequences, which have proven to be a good phylogenetic marker in the Mucorales (5), have demonstrated unexpectedly high genetic diversity within the most relevant clinical species of this order (3, 4, 21). In the case of Saksenaea vasiformis, a few studies have also demonstrated relatively high intraspecific genetic diversity (9, 28), suggesting that more than one phylogenetic species may be present within this morphospecies.
To identify possible cryptic species in S. vasiformis, we performed a polyphasic study, based on sequence analysis of three loci, and evaluated different morphological and physiological characteristics for a diverse panel of strains.
MATERIALS AND METHODS
Fungal strains.
A total of 11 strains from different reference culture collections were included in the study (Table 1). They were provided by the American Type Culture Collection (ATCC) Manassas, VA; the Centraalbureau voor Schimmelcultures (CBS) Utrecht, Netherlands; the National Reference Center for Mycoses and Antifungal Agents (NRCMA), Institut Pasteur, Paris, France; the Facultad de Medicina de Reus (FMR), Reus, Spain; the Fungus Testing Laboratory at the University of Texas Health Science Center (UTHSC), San Antonio, TX; and the ARS (NRRL) Culture Collection, Peoria, IL. The strains were cultured on potato dextrose agar (PDA; Pronadisa, Madrid, Spain) and were incubated at 37°C ± 1°C for 2 to 5 days.
TABLE 1.
List of Saksenaea isolates included in the study
| Isolatea | Source | GenBank accession no. |
||
|---|---|---|---|---|
| ITS | D1/D2 domains of 28S rRNA gene | EF-1α | ||
| ATCC 28740 | Craniofacial tissue and brain, Mississippi | FR687322 | HM776674 | HM776685 |
| ATCC 60625 | Human wrist lesion at arterial catheter site, Louisiana | FR687323 | HM776675 | HM776686 |
| CBS 133.90 | Forest soil, Brazil | FR687324 | HM776676 | HM776687 |
| CNRMAF/9-83 | Skin lesion, France | FR687325 | HM776677 | HM776688 |
| FMR 10131 | Cutaneous lesion, Tarragona, Spain | FR687326 | HM776678 | HM776689 |
| NRRL 2443T | Soil, India | FR687327 | HM776679 | HM776690 |
| UTHSC 08-3606 | Bovine fetus, Texas | FR687328 | HM776680 | HM776691 |
| UTHSC 09-528 | Human tissue, United States | FR687329 | HM776681 | HM776692 |
| UTHSC 08-379 | Eye, Utah | FR687330 | HM776682 | HM776693 |
| UTHSC 06-576 | Blood, Middle East | FR687331 | HM776683 | HM776694 |
| UTHSC R-2974 | Human tissue, Texas | FR687332 | HM776684 | HM776695 |
ATCC, American Type Culture Collection, Manassas, VA; CBS, Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; CNRMA, Centre National de Référence Mycologie et Antifongiques, Paris, France; FMR, Facultat de Medicina i Ciències de la Salut, Reus, Spain; NRRL, ARS Culture Collection (also known as the NRRL Collection), Peoria, IL; UTHSC, Fungus Testing Laboratory, University of Texas Health Science Center, San Antonio, TX; T, type strain.
DNA extraction, amplification, and sequencing.
For sequencing of the ITS region, DNA was extracted and purified directly from fungal colonies by following a slightly modified Fast DNA kit protocol (Bio 101, Vista, CA) consisting of a homogenization step repeated three times with a FastPrep FP120 instrument (Thermo Savant, Holbrook, NY). DNA was quantified with GeneQuant pro (Amersham Pharmacia Biotech, Cambridge, England). The ITS was amplified with the primer pair ITS5 and ITS4. The PCR mixture (25 μl) included 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 2.5 mM MgCl2 (10× Perkin-Elmer buffer II plus MgCl2 solution [Roche Molecular Systems, Branchburg, NJ]), 100 μM each deoxynucleoside triphosphate (dNTP) (Promega, Madison, WI), 1 μM each primer, and 1.5 U of AmpliTaq DNA polymerase (Roche). The amplification program for the three DNA fragments included an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing for 1 min at 56°C, and extension for 1 min at 72°C. A final extension step at 72°C for 7 min was included at the end of the amplification. The products were purified with an Illustra GFX PCR DNA and gel band purification kit (General Electric Healthcare, Buckinghamshire, United Kingdom) and were stored at −20°C until they were used in sequencing. PCR products were sequenced by using the same primers employed for amplification and following the Taq Dye Deoxy Terminator cycle sequencing kit protocol (Applied Biosystems, Gouda, Netherlands). Reactions were run on a 310 DNA sequencer (Applied Biosystems). Consensus sequences were obtained using the Autoassembler program (Perkin-Elmer Applied Biosystems) and Seqman software (Lasergene, Madison, WI).
The D1 and D2 domains of the 28S rRNA gene (D1/D2) and the elongation factor 1α (EF-1α) locus were sequenced at the NRCMA. The mycelium was grown as described previously (17). Genomic DNA was extracted for approximately 100 mg of mycelium. The mycelium was homogenized using green ceramic MagNA lyser beads (diameter, 1.4 mm; Roche, Mannheim, Germany) for three cycles of 30 s at 5,000 rpm. The tubes were then centrifuged, and 450 μl of supernatant was recovered. Three microliters of RNase A (100 mg/ml) was added, and the tubes were incubated for 10 min at 65°C. DNA was then extracted by using a DNeasy plant minikit (Qiagen, Germany) according to the manufacturer's instructions.
D1/D2 was amplified with the primer pair NL-1 and NL-4 (26), and a small region of EF-1α was amplified with primers MEF-11 and MEF-41 (32). PCR amplification of D1/D2 and EF-1α and the corresponding sequence analysis were carried out as described previously (17).
Phylogenetic analyses.
The sequences were aligned using the Clustal X (version 1.8) computer program, and alignments were corrected using the Gblocks tool (version 0.91b) (10), followed by manual adjustments with a text editor. Neighbor-joining (NJ) phylogenetic trees were constructed using the MEGA package, version 4.1 (43). The distance matrix was based on the maximum composite likelihood method with the pairwise deletion option.
For the phylogenetic inferences using maximum-likelihood (ML) analysis, trees were inferred with PhyML (version 3.0) (20), and we used heuristic searches, with starting trees obtained by random addition with 100 replicates and nearest-neighbor interchange (NNI) branch swapping. For bootstrap ML analysis, we performed 1,000 replicates (NJ starting tree with NNI branch swapping). Bayesian analyses were carried out using MrBayes, version 3.1 (23). Bayesian analyses were performed by running 1,000,000 generations in four chains, saving the current tree every 100 generations. The last 18,000 trees were used to construct a 50% majority-rule consensus tree. The best-fit model of nucleotide substitution for the combined data set selected by Modeltest (36) was the general time-reversible (GTR) model (27, 37). Apophysomyces elegans (CBS 476.78) was chosen as the outgroup.
Morphological study.
The strains were subcultured on PDA, Czapek agar (CZA; Difco, Becton Dickinson, France), and malt extract agar (MEA; 10 g of malt extract, 20 g of agar, and 1,000 ml of distilled water) and were incubated at 37°C. The microscopic features were determined in wet mounts on water and on lactic acid, which were examined under a light microscope. The strains were identified using the traditional schemes based on morphological characteristics (13).
Physiological study.
Growth rates at 4, 15, 24, 30, 35, 37, 42, and 50 ± 1°C were determined on PDA, MEA, and CZA for all strains. The petri dishes were inoculated in the center and were incubated in darkness, and the colony diameters (in millimeters) were measured daily.
Carbon source assimilation profiles were determined with the commercial API 50 CH kit (bioMérieux, Marcy-l'Etoile, France) by following protocols described previously (39). The strains were cultured for 6 days on CZA at 37°C. A final spore concentration of 5 × 105 CFU/ml was prepared in 20 ml of yeast nitrogen base (7.7 g/liter; Difco, Becton Dickinson, France) containing 0.5 g/liter chloramphenicol (Sigma-Aldrich Corp., St. Louis, MO) and 0.1% Bacto agar (Difco, Becton Dickinson, France). Each well of the strips was inoculated with 300 μl of medium. The inoculated API 50 CH strips were incubated for 48 to 72 h at 37 ± 1°C in darkness. After incubation, the strips were read visually, and growth or lack of growth was noted. Weak growth was considered a positive result. To determine nitrogen source assimilation, we used the same inoculum described above, except that the yeast nitrogen base broth was replaced with a carbon nitrogen base broth (Difco, Becton Dickinson, France), and testing was performed in sterile, disposable, multiwell microplates. The medium with the nitrogen sources was dispensed into the wells in 150-μl volumes with a multichannel pipette, and each well was inoculated with 50 μl of the conidial suspension. The microplates were incubated at 37°C in darkness for 48 to 72 h. We also evaluated the growth of the strains on NaCl (at 2%, 5%, 7%, and 10%), 2% MgCl2, and 0.1% cycloheximide (14, 49). All tests were performed in duplicate. The production of urease was determined after incubation on Christensen's urea agar slants at 37 ± 1°C for 8 days (30).
Mating tests.
The 11 Saksenaea strains were grown on CZA plates at 37 ± 1°C in the dark, after which they were paired in all combinations, including self-crosses, on CZA. Each strain was streaked onto one-half of a CZA plate opposite the streak of another strain, allowing for a central zone of contact when the strains grew. Plates were incubated at 37 ± 1°C and were examined macroscopically each week for as long as 4 months for the presence of zygospores. All tests were performed in duplicate.
Antifungal susceptibility testing.
All strains were subcultured on CZA for 7 to 20 days at 30°C or 37°C. Sporangiospores were then collected in water, and the suspension was adjusted to 2 × 104 CFU5/ml per well. Pure active powders, of known potency, of amphotericin B (Sigma-Aldrich, Saint Quentin Fallavier, France), voriconazole (Pfizer Central Research, Sandwich, United Kingdom), itraconazole (Janssen-Cilag, Issy-les-Moulineaux, France), posaconazole (Schering-Plough Research Institute, Kenilworth, NJ), terbinafine (Novartis Pharma AG, Basel, Switzerland), caspofungin (Merck & Co., Inc., Rahway, NJ), micafungin (Astellas Pharma, Osaka, Japan), and anidulafungin (Pfizer) were used. Antifungal susceptibility testing was performed by a broth microdilution technique according to the guidelines of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antibiotic Susceptibility Testing for the testing of conidium-forming molds (41), with some modifications (17).
Nucleotide sequence accession numbers.
All the sequences obtained in this study have been deposited in the GenBank database and assigned the accession numbers listed in Table 1.
MycoBank accession numbers.
The descriptions and illustrations of Saksenaea oblongispora and Saksenaea erythrospora are available online (http://www.MycoBank.org/) under MycoBank accession numbers MB 518626 and MB 518627, respectively.
RESULTS
Phylogeny.
With the primers used, we were able to amplify and sequence 641 to 800 bp, 648 to 738 bp, and 486 to 532 bp of the ITS, D1/D2, and EF-1α loci, respectively. The sequences of the three loci were analyzed phylogenetically as separate (data not shown) and combined data sets.
The topologies of the combined data sets by the three methods (NJ, ML, Bayesian inference) were very similar to those observed in the particular trees of the different genes analyzed. Most nodes in the combined analysis showed increased clade support (≥70% bootstrap support for the NJ and ML methods; ≥0.95 for Bayesian inference). Analysis of the combined partitions supports the recognition of three well-supported clades (Fig. 1), each of which represents a different phylogenetic species. Clade 1 comprises eight strains from different countries and sources, mainly from human and animal clinical samples, including the type strain of S. vasiformis (NRRL 2443). This clade shows high intraspecific variability, with three well-supported subclades, two of which contain only one strain each (UTHSC 08-379 and ATCC 28740, respectively). The third subclade contains six strains, including the type strain. The genetic relationships of the members of the clade with the type strain range between 84% and 94% identity, suggesting that this clade represents a complex of species. Clade 2 includes two clinical strains from the United States, and clade 3 consists of only one strain from Brazil.
FIG. 1.
Maximum-likelihood (ML) tree obtained from the combined DNA sequence data from three loci (ITS, D1/D2, EF-1α). Numbers on the branches are bootstrap neighbor-joining (NJ)/ML values above 75%, followed by Bayesian posterior probabilities above 0.95. Support values of <75% (NJ/ML) and <0.95 (Bayesian posterior probabilities) are indicated by minus signs. T, type strain.
Of note, two additional clinical isolates, not shown in Fig. 1, were included in the study. Phylogenetic analysis showed that they belonged to clade 1 (CNRMAF/6-50, recovered from muscle lesions of a French patient) and clade 2 (CNRMAF/4-81, from a patient from French Guiana) (9) (data not shown). These isolates were not viable, however, preventing any further study.
Physiology.
The carbon assimilation patterns of all the strains were positive in 12 tests (glycerol, d-adonitol, d-glucose, d-fructose, d-mannose, d-mannitol, d-sorbitol, N-acetylglucosamine, d-maltose, d-trehalose, amidon, and glycogen). Thirty-two carbon sources were not assimilated by any strain (erythritol, d-arabinose, l-arabinose, d-ribose, d-xylose, l-xylose, methyl-β-d-xylopyranoside, d-galactose, l-sorbose, l-rhamnose, dulcitol, inositol, methyl-d-mannopyranoside, methyl-d-glucopyranoside, amygdalin, arbutin, esculin, salicin, d-cellobiose, d-lactose, d-melibiose, d-saccharose, inulin, d-melezitose, d-raffinose, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, and potassium 5-keto-gluconate). The assimilation profiles of four carbon sources (xylitol, d-arabitol, l-arabitol, and potassium gluconate) were strain dependent. In contrast, variability in the assimilation of nitrogen sources and the tolerance of NaCl, MgCl2, and cycloheximide was null among the species. All the strains were positive for 10 nitrogen sources (creatine, creatinine, l-lysine, nitrate, l-tryptophan, l-proline, l-leucine, l-ornithine, l-cysteine, and arginine). Cadaverine and nitrite were not assimilated by any of the strains. All strains were able to grow in 2% NaCl, 5% NaCl, and 2% MgCl2 but failed to grow in 7% NaCl, 10% NaCl, and 0.1% cycloheximide.
None of the clades obtained in the phylogenetic analysis showed distinctive physiological profiles.
Morphology.
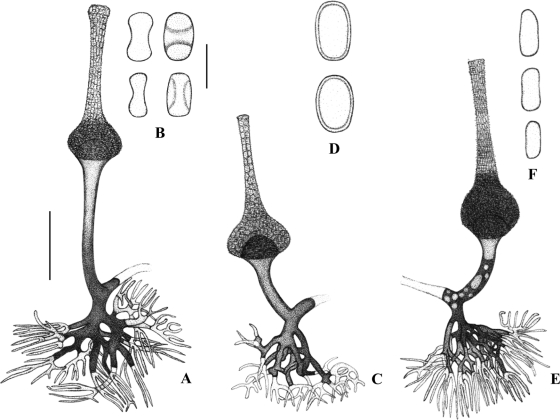
In general, all the strains examined displayed the typical features of the genus Saksenaea. However, a more detailed microscopic observation of these fungi showed important and consistent differences among them, mainly in the morphology of the sporangiophores and sporangiospores, which correlated with the different clades observed in the phylogenetic analysis. The strains that made up clade 1, including the type strain of S. vasiformis, showed the longest sporangiospores (5 to 7 by 2 to 3 μm), which were mainly cylindrical, with rounded ends. The sporangiospores of the strains in clade 2 were mostly ellipsoid, biconcave in a lateral view, and smaller (5 to 5.5 by 2.5 to 3 μm), while the sporangiospores of the clade 3 strain were mainly oblong, measuring 5 to 6.5 by 3 to 4.5 μm (Fig. 2). In addition, the strains of clade 2 showed clearly longer sporangiophores and sporangia than the members of the other clades (Fig. 3).
FIG. 2.
Morphology of the sporangiophores and sporangiospores of Saksenaea spp. (A, B, and C) S. vasiformis NRRL 2443. (A) Sporangiophore; (B) sporangiospores; (C) detail of the upper part of the sporangium. (D and E) Sporangiospores of S. erythrospora UTHSC 08-3606 (D) and S. oblongispora CBS 133.90 (E). Bars, 10 μm in panels A and B and 5 μm in panels C, D, and E.
FIG. 3.
(A and B) Saksenaea erythrospora. (A) Sporangiophore; (B) sporangiospores, frontal and side views. (C and D) Saksenaea oblongispora. (C) Sporangiophore; (D) sporangiospores, frontal and side views. (E and F) Saksenaea vasiformis. (E) Sporangiophore; (F) sporangiospores. Bars, 50 μm for drawings A, C, and E; 5 μm for drawings B, D, F.
Mating test.
After 4 months of incubation, no zygospore formation was observed for any of the mating combinations performed.
Antifungal susceptibility testing.
MICs were determined for the nine strains that produced enough sporangiospores (all except strains ATCC 60625 and ATCC 28740 from clade 1) and are given below as geometric means (ranges), in micrograms per milliliter. High MICs of amphotericin B (4.1 [1 to 8]) and voriconazole (4.7 [2 to 8]) and high minimum effective concentrations (MECs) of echinocandins (4.0 [4 to 8]) were observed for all the strains tested. Itraconazole B (0.2 [0.06 to 1]), posaconazole (0.1 [0.06 to 0.25]), and terbinafine (0.1 [0.03 to 1]) had lower MICs. There was no apparent difference between the species studied, but the small sample size prevents statistical analysis.
TAXONOMY
Based on the morphological differences described, which correlated with the molecular data, we conclude that clades 2 and 3 represent two species of Saksenaea different from S. vasiformis, which we propose here as new species.
Saksenaea oblongispora Alvarez, Stchigel, Cano, Garcia-Hermoso, et Guarro sp. nov.
MycoBank MB 518626. See Fig. 2E and 3C and D.
Coloniae in CZA rapide crescentes, albae, ex mycelium aerium sparsum compositum. Sporangiophora erecta, plerumque simplicia, 80 ad 100 μm longa, 6 ad 10 μm lata, supra massam rhizoidalem saepe bifurcatam positis. Sporangia ampulliformes, 70 ad 110 μm longa. Sporangiosporae plerumque oblongae, 5 ad 6.5 per 3 ad 4.5 μm. Zygospora ignota.
Holotypus, CBS H-20471. Typus lectus in terra, Pará, Capitâo Poço, Brazil, 1990 (cultura viva FMR 6500, CBS 133.90).
Etymology: the epithet refers to the oblong shape of the sporangiospores.
Colonies filling the petri dish (diameter, 9 cm) after 4 days of incubation at 37°C on CZA, whitish, with scarce aerial mycelium; hyphae branched, at first hyaline soon becoming light brown, smooth-walled, 3 to 6 μm in diameter; reverse concolorous. Sporangiophores erect, generally arising singly, hyaline, unbranched, 80 to 100 μm long, 6 to 10 μm wide, with a profusely dichotomously branched rhizoidal complex. Sporangia terminal, multispored, hyaline, flask-shaped, asperulate at low magnification, ornamented with many irregular, bacilliform protuberances under scanning electron microscopy (SEM), 70 to 110 μm long; with a long (60- to 90-μm) neck; apex of the neck closed with a mucilaginous plug, which is gradually dissolved when mature. Sporangiospores mainly oblong, 5 to 6.5 by 3 to 4.5 μm. Zygospores not observed.
Colonies on PDA and MEA showed features similar to those on CZA, but they were more floccose and white, with less sporulation. The optimum growth temperature was 25°C, and the minimum temperature of growth was 15°C. The fungus did not grow at 42°C.
Saksenaea erythrospora Alvarez, Cano, Stchigel, Garcia-Hermoso, et Guarro sp. nov.
MycoBank MB 518627. See Fig. 2D and 3A and B.
Coloniae in CZA rapide crescentes, albae, ex mycelium aerium sparsum compositum. Sporangiophora erecta, plerumque simplicia, 100 ad 150 μm longa, 7 ad 11 μm lata, supra massam rhizoidalem saepe bifurcatam positis. Sporan-gia ampulliformes, 100 ad 220 μm longa. Sporangiosporae plerumque ellipsoidae, maturitate membrana circa sporangiospore collapsa, 5 ad 5.5 per 2.5 ad 3 μm. Zygospora ignota.
Holotypus, CBS H-20472, ex fetu bovis, Texas, 2009 (cultura viva FMR 10672, UTHSC 08-3606).
Etymology: the epithet refers the shape of the sporangiospores, resembling the biconcave form of erythrocytes.
Colonies filling the petri dish after 4 days of incubation at 37°C on CZA, whitish, with scarce aerial mycelium, branched, hyaline, smooth-walled, 3 to 6 μm in diameter; reverse concolorous. Sporangiophores erect, generally arising singly, at first hyaline soon becoming light brown, generally straight, unbranched, 100 to 150 μm long, 7 to 11 μm wide. Sporangia terminal, multispored, flask-shaped, asperulate, ornamented with many irregular, small bacilliform protuberances under SEM, 100 to 220 μm long; with a long (80- to 200-μm) neck; apex of the neck closed with a mucilaginous plug. Sporangiospores mostly ellipsoid, biconcave in a lateral view, smooth-walled, their membranes collapsing at maturity, 5 to 5.5 by 2.5 to 3 μm. Zygospores not observed.
Colonies on PDA and MEA showed features similar to those on CZA, but they were more floccose and white, with less sporulation. The optimum growth temperature was 25°C, and the minimum was 15°C. The fungus grew at 42°C and did not grow at 50°C.
Based on our morphological and physiological studies, the type species of Saksenaea is redefined as follows.
Saksenaea vasiformis S. B. Saksena (Saksena, 1953).
See Fig. 2A to C and 3E and F.
Colonies filling the petri dish after 4 days at 37°C on CZA, whitish, with scarce aerial mycelium; reverse concolorous. Sporangiophores generally arising singly, emerging from aerial hyphae, straight, mainly unbranched, at first hyaline soon becoming light brown, 65 to 100 μm long, 6 to 10 μm wide. Sporangia produced terminally, flask-shaped and at low magnification asperulate, ornamented with small and irregular bacilliform protuberances under SEM, up to 110 μm long; with a long (60 to 90 μm) neck; apex of the neck closed with a mucilaginous plug, which gradually becomes dissolved when mature. Sporangiospores mainly cylindrical, with rounded ends, hyaline, 5 to 7 × 2 to 3 μm.
Colonial features similar to those described on CZA were observed on PDA and MEA, except for reduced mycelium production in the former. The optimum growth temperature was 25°C to 37°C, and the minimum was 15°C. The fungus grew at 42°C and did not grow at 50°C.
DISCUSSION
Despite its reproducibility in severe human infections, Saksenaea remains a poorly studied mucoralean genus, mainly due to the lack of sporulation on the mycological culture media routinely used in clinical laboratories and the scarce number of strains preserved in culture collections. Here we demonstrated high genetic and phenotypic diversity among the 11 strains studied based on a polyphasic approach. The information provided by the three loci evaluated was similar, and they proved to be useful markers for species-level delimitation in Saksenaea. The genus was determined to be genetically heterogeneous, comprising several species. Apart from the type species, at least other two species were characterized phenotypically. The shape and size of the sporangiospores, the length of the sporangiophore and sporangia, and the maximum temperature of growth were the most useful characteristics for their recognition.
Carbon assimilation profiles can be useful for the differentiation of human pathogenic mucoralean fungi at the genus level. In the case of Saksenaea, the test for assimilation of d-cellobiose was negative, while this test was positive for the other genera tested in previous studies (4, 39). However, none of the physiological tests evaluated in this study were discriminatory at the species level.
Antifungal susceptibility profiles also were not discriminatory. However, they matched previously published results (42) for the order Mucorales, except for amphotericin B, which has higher MICs for Saksenaea than Mucor spp.
Up to now, approximately 40 cases of zygomycete infections, mostly cutaneous infections, have been attributed to Saksenaea (6, 40, 46, 47), although for the reasons indicated above, it is likely that the actual number of clinical cases has been underestimated. To avoid difficulties in the detection and identification of Saksenaea in clinical samples, several authors have emphasized the need for special culture techniques, such as the use of floating agar blocks on water, or the use of Borelli′s lactrimel agar (15, 29) to induce sporulation. However, our use of these techniques yielded poor results (data not shown). In contrast, the use of Czapek agar, a culture medium traditionally used for the phenotypic characterization of Aspergillus and Penicillium species, produced good in vitro sporulation of the Saksenaea strains tested in this study. This medium also worked well for the sporulation of another mucoralean genus, Apophysomyces, which also sporulates poorly in routinely used mycological media, such as Sabouraud agar or PDA (4).
In addition to the two new species described here, the phylogenetic analysis also demonstrated the existence of more cryptic species within the first clade. However, since these strains could not be distinguished by using phenotypic characteristics, we prefer to refer to clade 1 as Saksenaea vasiformis complex. Although our study included strains of diverse origins, the number we could obtain was small, due to the fact that most of the clinical isolates previously recovered were not kept in culture collections and/or were no longer viable. It is likely that when more strains become available, additional new species within the genus Saksenaea will be identified.
The study performed here and the DNA sequences generated should be useful for further characterization of clinical isolates and/or identification of causative species from tissue biopsy specimens. They may also allow the identification of other species within the genus or the S. vasiformis complex.
Acknowledgments
This work was supported by the Spanish Ministerio de Ciencia y Tecnología grant CGL 2009-08698/BOS, with cofunding from Fondo Europeo de Desarrollo Regional (FEDER).
We thank Catalina Nuñez for technical assistance. The NRCMA gratefully acknowledges the technical help of Laure Diancourt and Coralie Tran at the sequencing facility (PF-8, Genotyping of Pathogens and Public Health, Institut Pasteur).
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Ajello, L., D. F. Dean, and R. S. Irwin. 1976. The zygomycete Saksenaea vasiformis as a pathogen of humans with a critical review of the etiology of zygomycosis. Mycologia 68:52-62. [PubMed] [Google Scholar]
- 2.Al-Hedaithy, M. 1998. Cutaneous zygomycosis due to Saksenaea vasiformis: case report and literature review. Ann. Saudi Med. 18:428-431. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez, E., D. A. Sutton, J. Cano, A. W. Fothergill, A. Stchigel, M. G. Rinaldi, and J. Guarro. 2009. Spectrum of zygomycete species identified in clinically significant specimens in the United States. J. Clin. Microbiol. 47:1650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez, E., A. Stchigel, J. Cano, D. A. Sutton, A. W. Fothergill, J. Chander, V. Salas, M. G. Rinaldi, and J. Guarro. 2010. Molecular phylogenetic diversity of the emerging mucoralean fungus Apophysomyces: proposal of three new species. Rev. Iberoam. Micol. 27:80-89. [DOI] [PubMed] [Google Scholar]
- 5.Balajee, S. A., A. M. Borman, M. E. Brandt, J. Cano, M. Cuenca-Estrella, E. Dannaoui, J. Guarro, G. Haase, C. C. Kibbler, W. Meyer, K. O'Donnell, C. A. Petti, J. L. Rodriguez-Tudela, D. Sutton, A. Velegraki, and B. L. Wickes. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales in the clinical mycology laboratory: where are we and where should we go from here? J. Clin. Microbiol. 47:877-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baradkar, V. P., and S. Kumar. 2009. Cutaneous zygomycosis due to Saksenaea vasiformis in an immunocompetent host. Indian J. Dermatol. 54:382-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baradkar, V. P., M. Mathur, S. Taklikar, M. Rathi, and S. Kumar. 2008. Fatal rhino-orbito-cerebral infection caused by Saksenaea vasiformis in an immunocompetent individual: first case report from India. Indian J. Med. Microbiol. 26:385-387. [DOI] [PubMed] [Google Scholar]
- 8.Bearer, E. A., P. R. Nelson, M. Y. Chowers, and C. E. Davis. 1994. Cutaneous zygomycosis caused by Saksenaea vasiformis in a diabetic patient. J. Clin. Microbiol. 32:1823-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchet, D., E. Dannaoui, A. Fior, F. Huber, P. Couppié, N. Salhab, D. Hoinard, and C. Aznar. 2008. Saksenaea vasiformis infection, French Guiana. Emerg. Infect. Dis. 14:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti, A., P. Kumar, A. A. Padhye, L. Chatha, S. K. Singh, A. Das, J. D. Wig, and R. N. Kataria. 1997. Primary cutaneous zygomycosis due to Saksenaea vasiformis and Apophysomyces elegans. Clin. Infect. Dis. 24:580-583. [DOI] [PubMed] [Google Scholar]
- 12.Chien, C. Y., D. J. Bhat, and W. B. Kendrick. 1992. Mycological observations on Saksenaea vasiformis (Saksenaeaceae, Mucorales). Trans. Mycol. Soc. Jpn. 33:443-448. [Google Scholar]
- 13.de Hoog, G. S., J. Guarro, J. Gené, and M. J. Figueras. 2000. Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands.
- 14.de Hoog, G. S., F. D. Marvin-Sikkema, G. A. Lahpoor, J. C. Gottschall, R. A. Prins, and E. Guého. 1994. Ecology and physiology of the emerging opportunistic fungi Pseudallescheria boydii and Scedosporium prolificans. Mycoses 37:71-78. [DOI] [PubMed] [Google Scholar]
- 15.Ellis, J. J., and L. Ajello. 1982. An unusual source for Apophysomyces elegans and a method for stimulating sporulation of Saksenaea vasiformis. Mycologia 74:144-145. [Google Scholar]
- 16.Ellis, D. H., and G. W. Kaminski. 1985. Laboratory identification of Saksenaea vasiformis: a rare cause of zygomycosis in Australia. Sabouraudia 23:137-140. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Hermoso, D., D. Hoinard, J. C. Gantier, F. Grenouillet, F. Dromer, and E. Dannaoui. 2009. Molecular and phenotypic evaluation of Lichtheimia corymbifera (formerly Absidia corymbifera) complex isolates associated with human mucormycosis: rehabilitation of L. ramosa. J. Clin. Microbiol. 47:3862-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Martínez, J., F. Lopez-Medrano, A. Alhambra, and A. del Palacio. 2008. Rhinocerebral zygomycosis caused by Saksenaea vasiformis in a diabetic patient. Mycoses 51:549-553. [DOI] [PubMed] [Google Scholar]
- 19.Gonis, G., and M. Starr. 1997. Fatal rhinocerebral mucormycosis caused by Saksenaea vasiformis in an immunocompromised child. Pediatr. Infect. Dis. J. 16:714-716. [DOI] [PubMed] [Google Scholar]
- 20.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, K., S. Discher, and K. Voigt. 2007. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol. Res. 111:1169-1183. [DOI] [PubMed] [Google Scholar]
- 22.Holland, J. 1997. Emerging zygomycosis of humans: Saksenaea vasiformis and Apophysomyces elegans. Curr. Top. Med. Mycol. 8:27-34. [PubMed] [Google Scholar]
- 23.Huelsenbeck, J. P., and F. Rodquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman, L., A. A. Padhye, and S. Parker. 1988. Rhinocerebral zygomycosis caused by Saksenaea vasiformis. J. Med. Vet. Mycol. 26:237-241. [PubMed] [Google Scholar]
- 25.Koren, G., I. Polacheck, and H. Kaplan. 1986. Invasive mucormycosis in a non-immunocompromised patient. J. Infect. 12:165-167. [DOI] [PubMed] [Google Scholar]
- 26.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanave, C., G. Preparata, C. Saccone, and G. Serio. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86-93. [DOI] [PubMed] [Google Scholar]
- 28.Lechevalier, P., D. Garcia-Hermoso, A. Carol, S. Bonacorsi, L. Ferkdadji, F. Fitoussi, O. Lortholary, A. Bourrillon, A. Faye, E. Dannaoui, and F. Angoulvant. 2008. Molecular diagnosis of Saksenaea vasiformis cutaneous infection after scorpion sting in an immunocompetent adolescent. J. Clin. Microbiol. 46:3169-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lye, G. R., G. Wood, and G. Nimmo. 1996. Subcutaneous zygomycosis due to Saksenaea vasiformis: rapid isolate identification using a modified sporulation technique. Pathology 28:364-365. [DOI] [PubMed] [Google Scholar]
- 30.Marimon, R., J. Gené, J. Cano, L. Trilles, M. Dos Santos Lazéra, and J. Guarro. 2006. Molecular phylogeny of Sporothrix schenckii. J. Clin. Microbiol. 44:3251-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberle, A. D., and R. L. Penn. 1983. Nosocomial invasive Saksenaea vasiformis infection. Am. J. Clin. Pathol. 80:885-888. [DOI] [PubMed] [Google Scholar]
- 32.O'Donnell, K., F. Lutzoni, T. J. Ward, and G. L. Benny. 2001. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia 93:286-296. [Google Scholar]
- 33.Padhye, A. A., G. Koshi, V. Anandi, J. Ponniah, V. Sitaram, M. Jacob, R. Mathai, L. Ajello, and F. W. Chandler. 1988. First case of subcutaneous zygomycosis caused by Saksenaea vasiformis in India. Diagn. Microbiol. Infect. Dis. 9:69-77. [DOI] [PubMed] [Google Scholar]
- 34.Pillai, C. M., and R. Ahmed. 1993. Saksenaea vasiformis, an ecologically important fungus on rice from India. Curr. Sci. 65:291. [Google Scholar]
- 35.Pierce, P. F., M. B. Wood, G. D. Roberts, R. H. Fitzgerald, Jr., C. Robertson, and R. S. Edson. 1987. Saksenaea vasiformis osteomyelitis. J. Clin. Microbiol. 25:933-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez, F., J. L. Oliver, A. Marin, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 38.Saksena, S. B. 1953. A new genus of the Mucorales. Mycologia 45:426-436. [Google Scholar]
- 39.Schwarz, P., O. Lortholary, F. Dromer, and E. Dannaoui. 2007. Carbon assimilation profiles as a tool for identification of zygomycetes. J. Clin. Microbiol. 45:1433-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewardson, A. J., N. E. Holmes, D. H. Ellis, and B. P. Howden. 2009. Cutaneous zygomycosis caused by Saksenaea vasiformis following water-related wound in a 24-year-old immunocompetent woman. Mycoses 52:547-549. [DOI] [PubMed] [Google Scholar]
- 41.Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST), J. L. Rodriguez-Tudela, M. C. Arendrup, S. Arikan, F. Barchiesi, J. Bille, E. Chryssanthou, M. Cuenca-Estrella, E. Dannaoui, D. W. Denning, J. P. Donnelly, W. Fegeler, C. Lass-Flörl, C. Moore, M. Richardson, P. Gaustad, A. Schmalreck, A. Velegraki, and P. Verweij. 2008. EUCAST technical note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Clin. Microbiol. Infect. 14:982-984. [DOI] [PubMed] [Google Scholar]
- 42.Sun, Q. N., L. K. Najvar, R. Bocanegra, D. Loebenberg, and J. R. Graybill. 2002. In vivo activity of posaconazole against Mucor spp. in an immunosuppressed-mouse model. Antimicrob. Agents Chemother. 46:2310-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 44.Tanphaichitr, V. S., A. Chaiprasert, V. Suvatte, and P. Thasnakorn. 1990. Subcutaneous mucormycosis caused by Saksenaea vasiformis in a thalassaemic child: first case report in Thailand. Mycoses 33:303-309. [DOI] [PubMed] [Google Scholar]
- 45.Torell, J., B. H. Cooper, and N. P. Helgeson. 1981. Disseminated Saksenaea vasiformis infection. Am. J. Clin. Pathol. 76:116-121. [DOI] [PubMed] [Google Scholar]
- 46.Trotter, D. J., G. Gonis, E. Cottrill, and C. Coombs. 2008. Disseminated Saksenaea vasiformis in an immunocompetent host. Med. J. Aust. 189:519-520. [DOI] [PubMed] [Google Scholar]
- 47.Vega, W., M. Orellana, L. Zaror, J. Gené, and J. Guarro. 2006. Saksenaea vasiformis infections: case report and literature review. Mycopathologia 162:289-294. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, P. A. 2008. Zygomycosis due to Saksenaea vasiformis caused by a magpie peck. Med. J. Aust. 189:521-522. [DOI] [PubMed] [Google Scholar]
- 49.Yarrow, D. 1998. Methods for the isolation, maintenance and identification of yeasts, p. 95-97. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, Netherlands.