Abstract
Nocardia species identification is difficult due to a complex and rapidly changing taxonomy, the failure of 16S rRNA and cellular fatty acid analysis to discriminate many species, and the unreliability of biochemical testing. Here, Nocardia species identification was achieved through multilocus sequence analysis (MLSA) of gyrase B of the β subunit of DNA topoisomerase (gyrB), 16S rRNA (16S), subunit A of SecA preprotein translocase (secA1), the 65-kDa heat shock protein (hsp65), and RNA polymerase (rpoB) applied to 190 clinical, 36 type, and 11 reference strains. Phylogenetic analysis resolved 30 sequence clusters with high (>85%) bootstrap support. Since most clusters contained a single type strain and the analysis corroborated current knowledge of Nocardia taxonomy, the sequence clusters were equated with species clusters and MLSA was deemed appropriate for species identification. By comparison, single-locus analysis was inadequate because it failed to resolve species clusters, partly due to the presence of foreign alleles in 22.1% of isolates. While MLSA identified the species of the majority (71.3%) of strains, it also identified clusters that may correspond to new species. The correlation of the identities by MLSA with those determined on the basis of microscopic examination, biochemical testing, and fatty acid analysis was 95%; however, MLSA was more discriminatory. Nocardia cyriacigeorgica (21.58%) and N. farcinica (14.74%) were the most frequently encountered species among clinical isolates. In summary, five-locus MLSA is a reliable method of elucidating taxonomic data to inform Nocardia species identification; however, three-locus (gyrB-16S-secA1) or four-locus (gyrB-16S-secA1-hsp65) MLSA was nearly as reliable, correctly identifying 98.5% and 99.5% of isolates, respectively, and would be more feasible for routine use in a clinical reference microbiology laboratory.
As part of the aerobic actinomycetes, Nocardia is a group of filamentous branching bacilli that are characteristically Gram positive and modified acid fast. Although Nocardia species normally exist as soil saprophytes, they have increasingly been isolated as infectious agents in immunosuppressed patients and, in some cases, even healthy individuals. Infections range from pulmonary nocardiosis, characterized by necrotizing pneumonia, to cutaneous nocardiosis and even brain abscess (25).
For nearly a century, since its inception in 1888 by Edmund Nocard, the genus Nocardia comprised only about a dozen species (26), largely because the somewhat biochemically inert nature of the group inhibited characterization (6). However, in 1988, Wallace et al. (38) uncovered latent diversity when they described six antimicrobial susceptibility pattern types among clinical isolates. DNA (e.g., 16S rRNA [16S] gene) sequencing confirmed and further expanded knowledge of the genetic diversity within the genus (6, 22). To date, the National Center for Biotechnology Information (NCBI) lists 86 recognized species (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi). However, the species differ in their abilities to cause human infection and their responses to antimicrobials (2, 6, 21, 25, 27, 33, 38). For this reason, species identification of Nocardia isolates from clinical specimens is relevant to patient treatment and provides important epidemiological information.
Beyond Gram and modified-acid-fast staining, species identification of Nocardia relies heavily on biochemical tests and cellular fatty acid analysis, which are cumbersome, time-consuming, and not definitive. Various molecular identification schemes investigated to date represent promising alternatives (7, 10, 29, 32, 36). However, 16S rRNA gene sequencing, considered to be the “gold standard” for bacterial identification, fails to discriminate many species (7), and the reliability of identification methods on the basis of the DNA sequence of a single housekeeping gene suffers from stochastic genetic variation and horizontal gene transfer and recombination (12).
Recently, multilocus sequence analysis (MLSA) has been suggested as a method to examine prokaryotic taxonomy. From phylogenetic analysis of a concatenated sequence typically consisting of 5 to 7 housekeeping genes, MLSA assigns a species designation on the basis of the assumption that sequence clusters represent species clusters (12). MLSA has been employed to identify the species of a number of genera with very promising results (1, 4, 5, 11, 14, 15, 16, 18, 20, 24, 28, 40). Furthermore, because of its ease of use, accuracy, and discriminatory power, MLSA may soon surpass DNA-DNA hybridization (DDH) as the gold standard for the investigation of prokaryotic taxonomy, species identification, and determination of genetic diversity (34).
The purpose of this study was to develop an MLSA scheme for the species identification of Nocardia clinical isolates. Through phylogenetic analysis of concatenated sequences consisting of partial fragments of gyrase B, the β subunit of a type II DNA topoisomerase (gyrB), 16S, subunit A of the SecA preprotein translocase (secA1), the 65-kDa heat shock protein (hsp65), and RNA polymerase (rpoB) genes, we delineated 30 species clusters. Since most clusters contained a single type strain, the results correlated well with existing knowledge of Nocardia taxonomy and provided a means of species assignment for the clinical isolates on the basis of strain placement within the phylogenetic analysis. Furthermore, the MLSA identifications were consistent with, although more discriminatory than, species assignments based on traditional microscopic evaluation, biochemical testing, and cellular fatty acid analysis. We present MLSA as a practical tool for routine Nocardia species identification in a clinical reference microbiology laboratory.
MATERIALS AND METHODS
Strains.
One hundred ninety clinical isolates of Nocardia were used in the study. The isolates were derived from clinical samples submitted to the Mycology Section of the Ontario Public Health Laboratory from December 2005 through January 2010. Microscopic observation of Gram-positive branching, filamentous bacilli, positive modified (Kinyoun)-acid-fast staining, and the production of aerial hyphae on pyruvate agar confirmed that the isolates were Nocardia (23). Routine species identification of Nocardia isolates was performed by assessing decomposition of adenine, casein, tyrosine, and xanthine; ability to grow at 35°C and 45°C; urease production; and acid production on 19 sugars (adonitol, arabinose, cellobiose, dulcitol, erythritol, galactose, glucose, glycerol, inositol, lactose, maltose, mannitol, melibiose, raffinose, rhamnose, sorbitol, sucrose, trehalose, and xylose). Results were compared to the reactions reported for six medically important Nocardia spp. (6, 23). As well, cellular fatty acid composition was determined by gas chromatography and compared to the compositions of cellular fatty acids in a standard library using the Sherlock Microbial Identification System (MIDI, Inc., Newark, DE). Table 1 lists the 36 type and 11 reference strains used in this study. (To date, “Nocardia rhamnosiphila” is not a validly published taxonomic name.) Type and reference strains were selected on the basis of their known clinical significance and their similarity to 16S sequences of the clinical isolates. All strains were maintained on either brain heart infusion agar (Difco) with 10% sheep's blood or GYM medium (0.4% glucose, 0.4% yeast extract, 1% malt extract, 0.2% CaCO3, 1.2% agar; DSM [German Collection of Microorganisms and Cell Cultures], Braunschweig, Germany) incubated at 37°C.
TABLE 1.
Nocardia species and species clusters, including the type and reference strains, number of clinical isolates, and average genetic diversity of the gyrB-16S-secA1-hsp65-rpoB concatenated sequence within each cluster
| Species or species cluster | Culture collection type and reference strains contained in cluster | No. (%) of clinical isolates | Total no. of isolates | % avg genetic diversitya (CI95b) |
|---|---|---|---|---|
| N. abscessus cluster | DSMc 44432T | 1 (0.53) | 2 | 0.59 |
| N. africana cluster | DSM 44491T, DSM 44500, NRRL B-1519 (N. veterana) | 0 | 3 | 1.07 (0.02, 2.12) |
| N. amamiensis cluster | DSM 45066T | 1 (0.53) | 2 | 1.57 |
| N. anaemiae cluster | DSM 44821T | 1 (0.53) | 2 | 3.02 |
| N. aobensis cluster | DSM 44805T, DSM 44543 (N. paucivorans) | 0 | 2 | 0.04 |
| N. asiatica cluster | DSM 44668T | 6 (3.16) | 7 | 1.10 (0.50, 1.70) |
| N. asteroides cluster | ATCCd 19247T | 2 (1.05) | 3 | 1.10 (0.15, 2.05) |
| N. beijingensis cluster | DSM 44636T | 1 (0.53) | 2 | 1.80 |
| N. brasiliensis cluster | DSM 43758T, DSM 43064 | 8 (4.21) | 10 | 1.39 (0.85, 1.93) |
| N. brevicatena | ATCC 15333T | 0 | 1 | |
| N. caishijiensis | DSM 44831T | 0 | 1 | |
| N. carnea cluster | NRRLe B-1336T, DSM 44558 | 0 | 2 | 1.80 |
| N. cerradoensis cluster | DSM 44546T | 2 (1.05) | 3 | 0.58 (0.01, 1.15) |
| N. cyriacigeorgica cluster | DSM 44484T, DSM 43208 | 41 (21.58) | 43 | 1.62 (1.34, 1.9) |
| N. farcinica cluster | NRRL B-2089T, DSM 43289 | 28 (14.74) | 30 | 0.45 (0.30, 0.60) |
| N. flavorosea | DSM 44480T | 0 | 1 | |
| N. fluminea cluster | DSM 44489T | 1 (0.53) | 2 | 0.91 |
| N. kruczakiae cluster | DSM 44877T | 1 (0.53) | 2 | 0.23 |
| N. neocaledoniensis cluster | DSM 44717T | 1 (0.53) | 2 | 1.38 |
| N. nova cluster | DSM 44481T, DSM 43207 | 17 (8.95) | 19 | 0.28 (0.16, 0.40) |
| N. otitidiscaviarum cluster | DSM 43242T, DSM 44565 | 10 (5.26) | 12 | 0.51 (0.31, 0.71) |
| N. paucivorans | DSM 44386T | 0 | 1 | |
| N. pneumoniae cluster | DSM 44730T | 2 (1.05) | 3 | 1.05 (0.02, 2.08) |
| N. pseudobrasiliensis | DSM 44290T | 0 | 1 | |
| N. rhamnosiphilaf cluster | NRRL B-24637T | 3 (1.58) | 4 | 0.40 (0.10, 0.70) |
| N. sienata cluster | DSM 44766T | 1 (0.53) | 2 | 2.59 |
| N. thailandica cluster | DSM 44808T | 1 (0.53) | 2 | 0.27 |
| N. transvalensis cluster | NRRL B-16037T, DSM 46068 | 0 | 2 | 2.69 |
| N. veterana cluster | NRRL B-24136T | 1 (0.53) | 2 | 0.0 |
| N. vinacea cluster | DSM 44638T | 1 (0.53) | 2 | 0.87 |
| N. wallacei cluster | DSM 45136T | 3 (1.58) | 4 | 1.32 (0.69, 1.95) |
| N. abscessus/N. arthritidis-like | DSM 44557 (N. abscessus) | 18 (9.47) | 19 | 0.36 (0.25, 0.47) |
| N. nova/cerradoensis/N. kruczakiae/N. aobensis-like | 20 (10.53) | 20 | 0.29 (0.21, 0.37) | |
| N. arthritidis/N. gamkensis/N. exalbida cluster | DSM 44731T, DSM 44956T, DSM 44883T | 7 (3.68) | 10 | 1.12 (0.87, 1.37) |
| N. ignorata/N. coubleae cluster | DSM 44496T, DSM 44960T | 1 (0.53) | 3 | 1.02 (0.78, 1.26) |
| Nonclustering strains | 11 (5.79) | 11 | ||
| Total | 47 | 190 | 237 | 6.53 |
Calculated using the K2P substitution model.
CI95, 95% confidence interval.
DSM, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
ATCC, American Type Culture Collection, Manassas, VA.
NRRL, United States Department of Agriculture, Agricultural Research Service, Peoria, IL.
To date, the taxonomic name Nocardia rhamnosiphila is not validly published.
DNA extraction, PCR, and sequencing.
DNA was extracted using PrepMan reagent (Applied Biosystems, Foster City, CA), according to the manufacturer's instructions. Partial 16S, secA1, hsp65, rpoB, and gyrB sequences were amplified using a Phire Hot Start DNA polymerase kit (New England Biolabs, Ipswich, MA) and a Bio-Rad (Hercules, CA) iCycler thermal cycler. The primers and PCR cycling conditions are listed in Table 2. Primers were designed on the basis of sequences found in GenBank. While the majority of secA1, hsp65, gyrB, and rpoB sequences were obtained with the first sets of primers listed in Table 2, a few strains required alternate primers, which are listed subsequently for each gene. Forward and reverse cycle sequencing reactions were performed using the PCR primers and a BigDye Terminator (version 1.1) cycle sequencing kit (Applied Biosystems), with the products subsequently being sequenced on a 3130xl genetic analyzer (Applied Biosystems) according to the manufacturer's instructions.
TABLE 2.
PCR primers and DNA amplification conditions for 16S, gyrB, secA1, hsp65, and rpoB
| Target | Region | Forward primer (5′-3′) |
Reverse primer (5′-3′) |
PCR cycling conditions | Reference or source | ||
|---|---|---|---|---|---|---|---|
| Primer name (position) | Sequence | Primer name (position) | Sequence | ||||
| 16S | bp 31-492 | E8F (bp 11-30) | AGAGTTTGATCCTGGCTCAG | 534r (bp 493-509) | ATTACCGCGGCTGCTGG | 30 s 98°C; 35× 5 s 98°C, 5 s 56°C, 20 s 72°C; 1 min 72°C | 3, this study |
| gyrB | bp 1031-1511 | Noc-gyrB-F (bp 972-992) | CTTCGCCAACACCATCAACAC | Noc-gyrB-R (bp 1563-1582) | TGATGATCGACTGGACCTCG | 30 s 98°C; 35× 5 s 98°C, 5 s 60°C, 20 s 72°C; 1 min 72°C | This study |
| Noc-gyrB-F3 (bp 1011-1030) | CGAGGAGGGCTTCCGCGCGG | Noc-gyrB-R3 (bp 1512-1532) | ATCGACTGGACCTCGTTGTTC | 30 s 98°C; 35× 5 s 98°C, 5 s 60°C, 20 s 72°C; 1 min 72°C | This study | ||
| secA1 | bp 431-875 | secA1-F47 (bp 413-430) | GCGACGCCGAGTGGATGG | secA1-ConR2 (bp 876-896) | TTGGCCTTGATGGCGTTGTTC | 30 s 98°C; 35× 5 s 98°C, 5 s 67°C, 20 s 72°C; 1 min 72°C | This study |
| secA1-F47 (bp 413-430) | GCGACGCCGAGTGGATGG | secA1-ConR (bp 913-933) | GCGGACGATGTAGTCCTTGTC | 30 s 98°C; 35× 5 s 98°C, 5 s 63°C, 20 s 72°C; 1 min 72°C | 10, this study | ||
| hsp65 | bp 165-565 | Noc-hsp65-F (bp 145-164) | ACCAACGATGGTGTGTCCAT | Noc-hsp65-R (bp 566-585) | CTTGTCGAACCGCATACCCT | 30 s 98°C; 35× 5 s 98°C, 5 s 54°C, 20 s 72°C; 1 min 72°C | 37 |
| Noc-hsp65-F2 (bp 109-129) | GTTGTCCTGGAGAAGAAGTGG | Noc-hsp65-R (bp 566-585) | CTTGTCGAACCGCATACCCT | 30 s 98°C; 35× 5 s 98°C, 5 s 54°C, 20 s 72°C; 1 min 72°C | 37, this study | ||
| rpoB | bp 1089-1488 | Noc-rpoB-F2 (bp 897-919) | CCGCTACAAGATCAACAAGAAGC | Noc-rpoB-R4 (bp 1612-1634) | CCCGCGAGGACATCGTCG | 30 s 98°C; 35× 5 s 98°C, 5 s 50°C, 20 s 72°C; 1 min 72°C | This study |
| Noc-rpoB-F2 (bp 897-919) | CCGCTACAAGATCAACAAGAAGC | Noc-rpoB-R2 (bp 1657-1677) | GGCGACGTACTCCATCTCCTC | 30 s 98°C; 35× 5 s 98°C, 5 s 50°C, 20 s 72°C; 1 min 72°C | This study | ||
| Noc-rpoB-F4 (bp 962-979) | CCCGCGAGGACATCGTCG | Noc-rpoB-R4 (bp 1612-1634) | CCCGCGAGGACATCGTCG | 30 s 98°C; 35× 5 s 98°C, 5 s 50°C, 20 s 72°C; 1 min 72°C | This study | ||
| Noc-rpoB-F5 (bp 987-1006) | CGAGTACCTGGTGCGYCTGC | Noc-rpoB-R5 (bp 1588-1607) | TCGACCGGCGAGTTGGCCTG | 30 s 98°C; 35× 5 s 98°C, 5 s 62°C, 20 s 72°C; 1 min 72°C | This study | ||
Cluster analysis.
In BioNumerics (version 6.0.1) software (Applied Maths, Austin, TX), sequences were aligned and trimmed to defined start and end positions to yield fragments of the following sizes: 462 bp for 16S, 482 bp for gyrB, 445 bp for secA1, 401 bp for hsp65, and 400 bp for rpoB. The trimmed sequences were concatenated in the order gyrB-16S-secA1-hsp65-rpoB to generate a 2,190-bp sequence. Unrooted trees obtained using individual gene sequences and concatenated sequences were generated by the neighbor-joining (NJ) algorithm following Kimura 2-parameter (K2P) correction and the maximum-parsimony (MP) algorithm using BioNumerics software. Bootstrap analysis (500 replicates) was used to assess the robustness of the clusters.
RESULTS
MLSA revealed distinct species clusters.
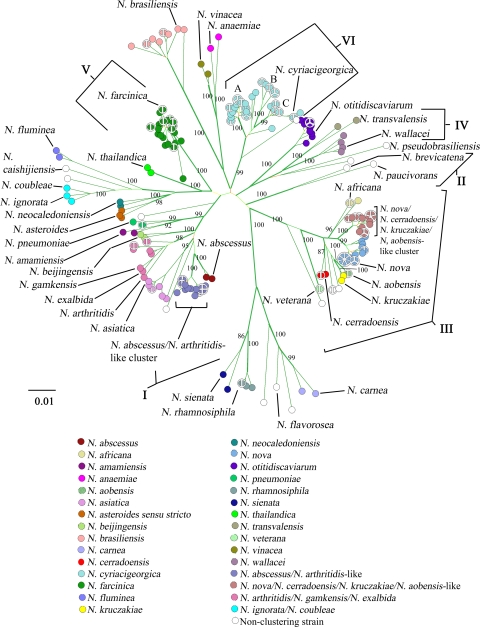
Among all 237 strains, the diversity of the concatenated gyrB-16S-secA1-hsp65-rpoB sequences ranged from 0 to 12.2%, with 0 to 246 nucleotide differences occurring between strains. The NJ tree of the concatenated sequences (Fig. 1) clearly showed 30 sequence clusters with bootstrap values of >85%, many of which contained a single type strain. In keeping with standard MLSA (18, 20, 24, 25, 32, 33, 34), these sequence clusters were considered to represent species clusters, and the isolates within each species cluster were colored coded according to their type strain(s). In total, 93.2% (221/237) of strains were contained within 1 of the 30 species clusters. For 71.3% (169/237) of strains, a species identification was assigned because their MLSA species cluster contained a single type strain. An additional 21.9% (52/237) of strains were contained within 1 of 4 species clusters that lacked a single type strain (Fig. 1; Table 1). On the other hand, 6.8% (16/237) of strains (5 type strains and 11 clinical isolates) failed to cluster with other strains and thus remained uncolored (white).
FIG. 1.
An NJ tree constructed from the 2,190-bp concatenated gyrB-16S-secA1-hsp65-rpoB sequences of 237 strains. Strains were colored to represent species clusters inferred from sequence clusters. Each of the 36 type strains is labeled with its species name. Bootstrap values (500 replicates) for nodes leading to sequence clusters are indicated. Branches indicated in boldface were also recovered in the MP tree. The three subgroups of N. cyriacigeorgica are indicated A, B, and C. Also indicated are the six drug-type patterns (I, II, III, IV, V, VI) previously described (3, 4). Bar, 1% nucleotide substitutions.
MLSA revealed two distinct clades that did not cluster with any of the type strains. The first was a set of 20 strains that showed a high level of concatenated sequence similarity to N. nova DSM 44481T (98.2 to 98.7%), N. cerradoensis DSM 44546T (97.9 to 98.2%), N. kruczakiae DSM 44877T (97.9 to 98.2%), and N. aobensis DSM 44805T (97.9 to 98.2%) and yet clustered separately with a high level of bootstrap support (100%) (Fig. 1). The second was a cluster of 19 strains that were similar to N. abscessus DSM 44432T (97.8 to 98.1%) and N. arthritidis DSM 44731T (97.6 to 97.8%) but again formed a distinct clade with 100% bootstrap support (Fig. 1).
The MLSA scheme also revealed two sets of type strains that fail to form distinct clusters. First, N. coubleae DSM 44960T and N. ignorata DSM 44496T along with one clinical isolate form a distinct clade with 100% bootstrap support. Second, N. arthritidis DSM 44731T, N. gamkensis DSM 44956T, and N. exalbida DSM 44883T together with 7 clinical isolates formed a cluster with 98% bootstrap support.
Moreover, although the N. cyriacigeorgica group was a well-supported species cluster, it did contain three clearly distinct subgroups supported by bootstrap values of 99%, 100%, and 100%.
Table 1 lists the average genetic diversity present within each species cluster. The average genetic diversity varied depending on the species cluster. The N. veterana cluster had the lowest genetic diversity at 0.0%, while the N. anaemiae species cluster had the greatest average genetic diversity at 3.02%.
Three of 11 reference strains failed to cluster according to the species designation reported by their respective culture collections. N. veterana NRRL B-1519 clustered with the N. africana isolates, N. paucivorans DSM 44543 was identical to and therefore clustered with N. aobensis DSM 44805T, and N. abscessus DMS 44557 grouped with the N. abscessus/N. arthritidis-like isolates (Fig. 1; Table 1).
Beyond delineating species clusters, the overall structure of the NJ tree correlated well with the six drug pattern types of the Nocardia asteroides complex described by Wallace et al. (38) and clearly differentiated the well-recognized species N. brasiliensis and N. otitidiscaviarum (6) (Fig. 1).
The concatenated sequences were used to generate a maximum-parsimony tree (data not shown). However, no major differences were observed between the MP tree and the NJ tree. All major branches were supported by both methods, except for a slight rearrangement of N. fluminea isolates with respect to N. coubleae DSM 44960T and N. ignorata DSM 44496T (Fig. 1).
Individual gene sequences failed to resolve species clusters.
Phylogenetic analysis using the NJ algorithm applied to each of the five loci individually (data not shown) also demonstrated sequence diversity within the genus. The range of genetic diversity varied for each of the loci: 0.0 to 17.7% for gyrB, 0.0 to 9.2% for 16S, 0.0 to 18.2% for secA1, 0.0 to 13.0% for hsp65, and 0.0 to 22.3% for rpoB. Despite the observed sequence diversity, these trees failed to discriminate a number of the species clusters resolved using the concatenated sequence. Species clusters that did form generally received weak bootstrap support. Consequently, attempts to identify individual strains on the basis of cluster analysis of individual loci were unsuccessful.
Of the 179 clinical and 11 reference strains assigned to an MLSA species cluster, 148 (77.9%) had alleles at the secA1, gyrB, hsp65, and rpoB loci that were most similar to those of the other strains within its designated MLSA species cluster. However, 20.5% (n = 39) of strains contained a foreign allele at one of the loci; that is, the allele was most similar to that found in the type strain representing another species cluster. This caused aberrant clustering when the single-locus tree was compared to trees constructed using the concatenated sequence, 16S, or any of the other three loci. An additional three strains (1.6%) had foreign alleles at 2 loci. Of the loci, hsp65 was the most promiscuous, with 33 strains (17.4%) exhibiting a foreign allele, compared to secA1 (3 strains, 1.6%), rpoB (6 strains, 3.2%), and gyrB (3 strains, 1.6%). Of note, 22 strains from the N. cyriacigeorgica species cluster had an hsp65 allele that exhibited 97.5 to 98.5% similarity to the hsp65 alleles of N. asiatica DSM 44668T, N. abscessus 44432T, and N. pneumoniae DSM 44730T but only 96.5 to 97.3% similarity to the N. cyriacigeorgica DSM 44484T hsp65 allele. The strains exclusively formed subgroup A of the N. cyriacigeorgica species cluster (Fig. 1). Apart from the hsp65 allele of these 22 N. cyriacigeorgica strains, the majority of the foreign alleles (78.3%) were most similar to alleles of adjacent species clusters on the NJ tree of concatenated sequences.
As mentioned above, two distinct clades failed to cluster with a type strain during MLSA. For the first clade, the rpoB, secA1, and 16S alleles of the N. nova/N. cerradoensis/N. kruczakiae/N. aobensis-like cluster were most similar to those of N. nova DSM 44481T (99.0 to 100%, 98.9 to 99.3%, and 99.6%, respectively), while the hsp65 alleles were most similar to those of N. veterana NRRL B-24136T (97.5 to 98.5%) and the gyrB alleles were most similar to those of N. cerradoensis DSM 44546T (98.1 to 98.5%). For the second clade, when examined individually, the secA1 and hsp65 sequences of the N. abscessus/N. arthritidis-like cluster were most similar to those of N. arthritidis DSM 44731T (96.0 to 96.3% and 99.2 to 99.5%, respectively), while the rpoB and gyrB sequences were most similar to those of N. abscessus DSM 44432T (98.2 to 98.5% and 97.9 to 99.4%, respectively). The 16S sequences were identical to the N. abscessus DSM 44432T 16S sequence and differed by only 1 bp from that of N. arthritidis DSM 44731T.
Identification by MLSA cluster analysis is generally concordant with, although more discriminatory, than identification achieved through microscopic examination, biochemical tests, and cellular fatty acid analysis.
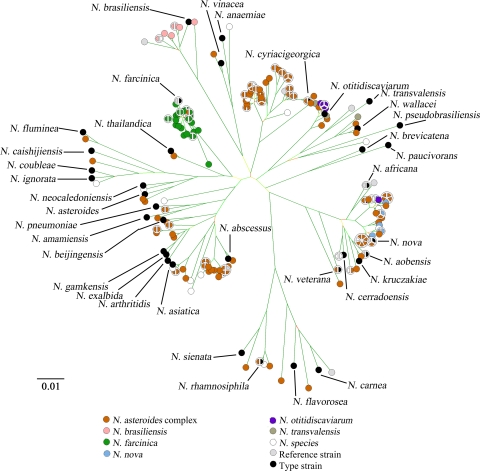
Figure 2 shows the NJ tree constructed using the concatenated gyrB-16S-secA1-hsp65-rpoB sequences color coded to reflect the species designations achieved through traditional morphological and microscopic examination, biochemical testing, and cellular fatty acid analysis. To correlate these results with the MLSA species designations, we considered 139 clinical isolates. (Of the 190 clinical isolates, 30 were excluded from the comparison because they lacked a microscopic/biochemical/cellular fatty acid identification [n = 19], an MLSA species cluster designation [n = 7], or both [n = 4]. An additional 21 isolates were identified as N. asteroides complex by microscopic/biochemical/cellular fatty acid assessment but were assigned to one of the following newly described or rarely encountered species by MLSA: N. amamiensis, N. asiatica, N. beijingensis, N. exalbida/N. gamkensis/N. arthritidis, N. ignorata/N. coubleae, N. fluminea, N. pneumoniae, N. rhamnosiphila, N. sienata, N. thailandica, and N. vinacea. These isolates were also excluded from the comparison because the taxonomic status of these species with respect to the ever-waning N. asteroides complex remains undescribed.) In order to accommodate the evolving taxonomy of Nocardia, all strains designated N. asteroides complex by microscopic/biochemical/cellular fatty acid identification were considered concordant with any MLSA species cluster designation from the following: N. abscessus, N. brevicatena, N. paucivorans, N. nova, N. veterana, N. africana, N. kruczakiae, N. transvalensis, N. wallacei, N. farcinica, and N. cyriacigeorgica (6, 8). Furthermore, the MLSA identification of isolates of the N. abscessus/N. arthritidis-like species cluster and the N. nova/N. cerradoensis/N. kruczakiae/N. aobensis-like species cluster was considered concordant with a microscopic/biochemical/cellular fatty acid identification of N. asteroides complex since the species describing these clusters are part of the N. asteroides complex. The concordance between the microscopic/biochemical/cellular fatty acid identification and the MLSA species cluster designation was 95% (132/139); however, MLSA was far more discriminatory because it identified the species of 84 isolates defined as N. asteroides complex by microscopy, biochemical, and cellular fatty acid testing.
FIG. 2.
An NJ tree constructed from the 2,190-bp concatenated gyrB-16S-secA-hsp65-rpoB sequences of 237 strains. Strains were colored to represent species identification on the basis of microscopic examination, biochemical testing, and cellular fatty acid analysis. Each of the 36 type strains is labeled with its species name. Bar, 1% nucleotide substitutions.
N. cyriacigeorgica and N. farcinica were the most commonly encountered species among clinical isolates.
Table 1 shows the species distribution on the basis of MLSA for the 190 Nocardia clinical isolates received by the Mycology Section at the Ontario Public Health Laboratory from December 2005 through January 2010. N. cyriacigeorgica (n = 41) and N. farcinica (n = 28) were the most commonly encountered species, accounting for 36.32% of strains. Also frequently encountered were isolates of the N. nova/N. cerradoensis/N. kruczakiae/N. aobensis-like species cluster (10.53%), the N. abscessus/N. arthritidis-like species cluster (9.47%), and N. nova (8.95%).
Toward a species identification scheme for Nocardia.
Although five-locus MLSA definitively differentiates many species clusters and facilitates the identification of unknown isolates, the routine amplification of five loci from each and every clinical isolate in a clinical laboratory setting would be too cumbersome. Therefore, we reanalyzed the data in various combinations to produce three- and four-locus NJ trees (data not shown). (Most combinations included the 16S locus because of its predominance in sequence-based identification schemes.) The trees were examined to determine whether they yielded the same species clusters and strain placement as the five-locus tree. As expected, the four-locus trees correctly assigned clinical and reference isolates more frequently than the three-locus trees. The gyrB-16S-secA1-hsp65 and gyrB-16S-secA1 MLSAs yielded the highest correlations (99.5% and 98.5%, respectively) with the five-locus MLSA, while the 16S-secA1-hsp65 MLSA produced the lowest at 88.1%.
DISCUSSION
The aim of this study was to develop a system for the species identification of Nocardia that confidently and powerfully discriminates species, satisfies the most current theories in prokaryotic taxonomy, and yet can be implemented for routine use in a clinical laboratory setting. The MLSA presented here provides such an all-encompassing identification system capable of differentiating currently recognized species and pinpointing anomalous strains potentially representing undescribed species. It is demonstrably superior to identifications based on traditional microscopic examination, biochemical tests, and cellular fatty acid analysis because of its unparalleled ability to comprehensively discriminate a myriad of old and new species recently added to the genus (6, 22).
Of the many approaches to species identification, MLSA represents an ideal method for such a taxonomically diverse and challenging group as the Nocardia. Although DDH has long been held to be the gold standard for prokaryotic species demarcation, MLSA may soon supplant DDH to become the primary measure of bacterial diversity and species identification (34). While the results of MLSA correlate well with DDH taxonomic classifications (5, 28, 30, 31), it is technically easier to perform and allows the rapid concurrent comparison of multiple strains. Compared to single-locus sequence-based identification schemes (e.g., 16S rRNA), it yields a fuller understanding of the genome by sampling multiple alternate regions of the DNA. The use of multiple loci buffers against stochastic genetic variation and horizontal gene transfer with homologous recombination (12, 17, 35). MLSA is less cumbersome than biochemical testing, yet it demonstrates greater discriminatory power (12, 35).
In keeping with the standard MLSA protocol, we chose the following four single-copy housekeeping genes for analysis, in addition to 16S rRNA: secA1, which encodes subunit A of a preprotein translocase; hsp65, which encodes a 65-kDa heat shock protein; rpoB, the gene for RNA polymerase; and gyrB, the gene for the β subunit of a type II DNA topoisomerase. Each of these loci has previously been recommended as a conserved core marker suitable for bacterial systematics (19), and most have been used individually for sequence-based identification of Nocardia species (7, 10, 29, 32, 36).
MLSA using the NJ phylogenetic tree generated from the concatenated gyrB-16S-secA1-hsp65-rpoB sequences of 237 strains demonstrates a tremendous amount of genetic diversity among the Nocardia. As recommended for MLSA analysis, species clusters were delineated on the basis of visual inspection of the tree rather than an arbitrary percent similarity threshold largely because intraspecies genetic diversity levels generally vary between species clusters (4, 12). Indeed, our analysis also showed that the degree of genetic diversity within Nocardia species clusters fluctuated substantially, discouraging the use of a cutoff value. Remarkably, 30 species clusters were identified within a genus that until recently contained only a few species (26). While individual treatment of the loci produced ambiguous and unreliable species identification, concatenation of all five loci resolved the species clusters with very strong (>85%) bootstrap support, providing a high degree of confidence in the species assignments. As a method of determining the species identity of the strains, MLSA was highly successful: 71.3% received a species assignment, while important taxonomic information was revealed about an additional 21.9% of strains which comprised clusters that did not contain a single type strain.
MLSA ideally utilizes a large number of strains that are representative of the diversity within the genus. Large numbers of isolates produce robust species clusters, which in turn increases confidence in species identification (12, 17). In this study, many of the clusters contained <5 isolates. While this is less than ideal, the MLSA scheme presented here did demarcate and cluster the available strains with a high degree of bootstrap support. The analysis provides a solid foundation to which more strains can be added in the future. As more concatenated sequences are added, the compositions of the clusters and their connections can be expected to change to provide a more accurate picture of the genus (12).
Despite the need for more isolates in some species clusters, the results of MLSA correlate well with current knowledge of Nocardia taxonomy and the results of microscopic/biochemical/cellular fatty acid evaluation. Most (26 of 30) of the species clusters contain a single previously described type strain. The six drug-type patterns of the N. asteroides complex plus the well-recognized species N. otitidiscaviarum and N. brasiliensis (6, 38) are clearly delineated. Furthermore, strain-by-strain comparison of the MLSA species assignment with the identifications provided by microscopic examination, biochemical tests, and cellular fatty acid analysis is generally consistent (95%), although MLSA is more discriminatory. For instance, MLSA confirmed and then further refined the species assignments of a large number (n = 84) of N. asteroides complex isolates. The 7 inconsistencies in identification among 139 clinical isolates (5%) and 3 of the 11 (27.3%) reference strains reflect the widely acknowledged difficulties with Nocardia species identification (6). MLSA is a superior identification method because it identified a number of rarely encountered or newly described species among 21 clinical isolates previously identified as N. asteroides complex by traditional microscopic/biochemical/cellular fatty acid analysis. Although DDH has historically been considered the gold standard for prokaryotic species determination, its laborious nature prevented a direct comparison with MLSA in this study. However, the strong concordance with literature reports and the results of microscopic/biochemical/cellular fatty acid species identification methods validates the MLSA system as accurate for Nocardia species identification.
As with MLSA studies involving other bacterial genera (4, 11, 16, 18, 28, 40), 22.1% (42/190) of the Nocardia isolates had foreign alleles discernible at one or more loci, often from a neighboring cluster, indicating that a history of interspecies recombination has distorted the phylogenetic relationships between species clusters. This phenomenon as well as the discovery of two well-resolved species clusters that lack a type strain discourages the practice of comparing a single housekeeping locus allele sequence to the sequences of a set of type strains as a method for species delineation. However, during MLSA, the phylogenetic signals from multiple loci generally buffered the effect of the foreign allele, such that species clusters were still resolved, albeit fuzzy (16), due to the presence of intermediate genotypes and provided a method for identifying novel species clusters.
Studies with other bacterial genera suggest using MLSA to inform the discovery of new species or taxonomic revisions (5, 13, 14, 20, 24, 30, 31). In our study, MLSA did not distinguish either the N. arthritidis, N. gamkensis, and N. exalbida type strains or the N. ignorata and N. coubleae type strains. While sequence analysis of additional genes may demarcate these type strains, failure to do so would prompt an extensive evaluation of the legitimacy of their species status. The N. abscessus/N. arthritidis-like and N. nova/N. cerradoensis/N. kruczakiae/N. aobensis-like clusters appear to be a fusion of alleles from two or three type strains. Additional sequences should either reassign these isolates to type strain-defined species clusters or prompt the characterization of new species. The species or subspecies status of the three subclusters of N. cyriacigeorgica would benefit from further taxonomic investigation, especially considering its importance as a human pathogen (9, 39). Nonclustering clinical isolates may also represent new species, provided that they fail to cluster with additional type strains when they are incorporated into the analysis. In all cases, novel species discovery must be supported by MLSA as well as DDH data, extensive biochemical and phenotypic characterizations, and perhaps ecological distinction (12, 13, 20, 30, 31).
The species distribution recovered among Ontario clinical isolates correlates well with the distributions presented in other reports, which also list N. cyriacigeorgica, N. farcinica, N. nova, and N. abscessus among the most frequently encountered species from clinical specimens (2, 21, 25, 27, 33). Lack of information regarding clinical symptoms precludes any speculation as to whether the various species infect or simply colonize humans.
While MLSA may involve anywhere from 3 to 8 loci, depending on the genus, at some point additional loci fail to enhance species delineation (4, 17). In order to avoid excess labor and cost in a routine laboratory setting, we sought to identify the best combination of 3 or 4 loci that maintain a good resolution of species clusters. Either the gyrB-16S-secA1-hsp65 MLSA or the gyrB-16S-secA1 MLSA is a good option, as each provides the correct species cluster assignment for 99.5% and 98.5% of the isolates, respectively. Frequently encountered species could be confidently assigned on the basis of either of these truncated MLSA methods, with the 5-locus MLSA being reserved for identification of rare species.
In summary, we have developed a MLSA method capable of demarcating various species clusters from among type strains, reference strains, and clinical isolates. This system is superior to biochemical testing for the identification of unknown strains because of its simplicity and ease of use, its discriminatory power, and its ability to concurrently provide taxonomic information on virtually all strains of Nocardia encountered in a clinical setting.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 2.Agterof, M. J., T. van der Bruggen, M. Tersmette, E. J. ter Borg, J. M. M. van den Bosch, and D. H. Biesma. 2007. Nocardiosis: a case series and a mini review of clinical and microbiological features. Neth. J. Med. 65:199-202. [PubMed] [Google Scholar]
- 3.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, C. J., D. M. Aanensen, G. E. Jordan, M. Kilian, W. P. Hanage, and B. G. Spratt. 2009. Assigning strains to bacterial species via the Internet. BMC Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady, C., I. Cleenwerck, S. Venter, M. Vancanneyt, J. Swings, and T. Coutinho. 2008. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 31:447-460. [DOI] [PubMed] [Google Scholar]
- 6.Brown-Elliott, B. A., J. M. Brown, P. S. Conville, and R. J. Wallace, Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin. Microbiol. Rev. 19:259-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conville, P. S., J. M. Brown, A. G. Steigerwalt, B. A. Brown-Elliott, and F. G. Witebsky. 2008. Nocardia wallacei sp. nov. and Nocardia blacklockiae sp. nov., human pathogens and members of the “Nocardia transvalensis complex.” J. Clin. Microbiol. 46:1178-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conville, P. S., and F. G. Witebsky. 2007. Organisms designated as Nocardia asteroides drug pattern type VI are members of the species Nocardia cyriacigeorgica. J. Clin. Microbiol. 45:2257-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conville, P. S., A. M. Zelazny, and F. G. Witebsky. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J. Clin. Microbiol. 44:2760-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deletoile, A., V. Passet, J. Aires, I. Chambaud, M.-J. Butel, T. Smokvina, and S. Brisse. 2010. Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Res. Microbiol. 161:82-90. [DOI] [PubMed] [Google Scholar]
- 12.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrankt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739. [DOI] [PubMed] [Google Scholar]
- 13.Glass, M. B., A. G. Steigerwalt, J. G. Jordan, P. P. Wilkins, and J. E. Gee. 2006. Burkholderia oklahomensis sp. nov., a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int. J. Syst. Evol. Microbiol. 56:2171-2176. [DOI] [PubMed] [Google Scholar]
- 14.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, Y., W. Zheng, X. Rong, and Y. Huang. 2008. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 58:149-159. [DOI] [PubMed] [Google Scholar]
- 16.Hanage, W. P., C. Fraser, and B. G. Spratt. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1917-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanage, W. P., T. Kaijalainen, E. Herva, A. Saukkoriipi, R. Syrjanen, and B. G. Spratt. 2005. Using multilocus sequence data to define the Pneumococcus. J. Bacteriol. 187:6223-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig, W. 2007. Nucleic acid techniques in bacterial systematics and identification. Int. J. Food Microbiol. 120:225-236. [DOI] [PubMed] [Google Scholar]
- 20.Margos, G., S. A. Vollmer, M. Cornet, M. Garnier, V. Fingerle, B. Wilske, A. Bormane, L. Vitorino, M. Collares-Pereira, M. Drancourt, and K. Kurtenbach. 2009. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 75:5410-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez, R., S. Reyes, and R. Menendez. 2008. Pulmonary nocardiosis: risk factors, clinical features, diagnosis and prognosis. Curr. Opin. Pulm. Med. 14:219-227. [DOI] [PubMed] [Google Scholar]
- 22.McNabb, A., G. Geddes, C. Shaw, S. Mithani, and J. Isaac-Renton. 2006. Identification of Nocardia species by partial sequencing of the 16S rRNA gene: the sun sets on Nocardia asteroides. Can. J. Med. Lab. Sci. 68:18-33. [Google Scholar]
- 23.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menna, P., F. G. Barcellos, and M. Hungria. 2009. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD, and dnaK genes. Int. J. Syst. Evol. Microbiol. 59:2934-2950. [DOI] [PubMed] [Google Scholar]
- 25.Minero, M. V., M. Marin, E. Cercenado, P. M. Rabadan, E. Bouza, and P. Munoz. 2009. Nocardiosis at the turn of the century. Medicine 88:250-261. [DOI] [PubMed] [Google Scholar]
- 26.Mishra, S. K., R. E. Gordon, and D. A. Barnett. 1980. Identification of nocardiae and streptomycetes of medical importance. J. Clin. Microbiol. 11:728-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz, J., B. Mirelis, L. M. Aragon, N. Gutierrez, F. Sanchez, M. Espanol, O. Esparcia, M. Gurgui, P. Domingo, and P. Coll. 2007. Clinical and microbiological features of nocardiosis 1997-2003. J. Med. Microbiol. 56:545-550. [DOI] [PubMed] [Google Scholar]
- 28.Rivas, P., M. Martens, P. De Lajudie, and A. Willems. 2009. Multilocus sequence analysis of the genus Bradyrhizobium. Syst. Appl. Microbiol. 32:101-110. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Nava, V., A. Couble, G. Devulder, J.-P. Flandrois, P. Boiron, and F. Laurent. 2006. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J. Clin. Microbiol. 44:536-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rong, X., Y. Guo, and Y. Huang. 2009. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and the taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst. Appl. Microbiol. 32:314-322. [DOI] [PubMed] [Google Scholar]
- 31.Rong, X., and Y. Huang. 2010. Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA-DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. Int. J. Syst. Evol. Microbiol. 60:696-703. [DOI] [PubMed] [Google Scholar]
- 32.Roth, A., S. Andrees, R. M. Kroppenstedt, D. Harmsen, and H. Mauch. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation of division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J. Clin. Microbiol. 41:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saubolle, M. A., and D. Sussland. 2003. Nocardiosis: review of clinical and laboratory experience. J. Clin. Microbiol. 41:4497-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shneyer, V. S. 2007. On the species-specificity of DNA: fifty years later. Biochemistry (Moscow) 72:1377-1384. [DOI] [PubMed] [Google Scholar]
- 35.Staley, J. T. 2006. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1899-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda, K., Y. Kang, K. Yazawa, T. Gonoi, and Y. Mikami. 2010. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J. Med. Microbiol. 59:165-171. [DOI] [PubMed] [Google Scholar]
- 37.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Botteger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace, R. J., Jr., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 32:1776-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witebsky, F. G., P. S. Conville, J. R. Wallace, Jr., and B. A. Brown-Elliott. 2008. Nocardia cyriacigeorgica—an established rather than an emerging pathogen. J. Clin. Microbiol. 46:2469-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zelazny, A. M., J. M. Root, Y. R. Shea, R. E. Colombo, I. C. Shamputa, F. Stock, S. Conlan, S. McNulty, B. A. Brown-Elliott, R. J. Wallace, Jr., K. N. Olivier, S. M. Holland, and E. P. Sampaio. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J. Clin. Microbiol. 47:1985-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]