Abstract
We aimed to study the prevalence and clinical implications of hepatitis B virus (HBV) subgenotypes in Chinese patients. A total of 4,300 patients, mainly from northern China, were enrolled, including 182 patients with acute hepatitis B and 4,118 patients with chronic HBV infection who had been exposed to nucleoside or nucleotide analogs. HBV genotypes/subgenotypes were determined by direct sequencing of the HBV S/Pol region. The prevalence rates were 0.40% for HBV/B1, 14.30% for HBV/B2, 0.25% for HBV/B3, 0.35% for HBV/B4, 1.05% for HBV/C1, 81.72% for HBV/C2, 0.93% for HBV/C3, 0.16% for HBV/C4, and 0.84% for HBV/D. In chronic HBV infection, patients with HBV/B2 were younger and had lower ΗBeAg positive rates than patients with HBV/C2. The incidence of lamivudine-resistant mutations was significantly higher in HBV/C2 compared to HBV/B2 (27.9% versus 19.8%; P < 0.01), and the significant difference was observed only for rtM204I and not rtM204V. In addition, compensatory mutations were more frequently detected in HBV/C2. The incidence of adefovir-resistant mutations was similar between the two subsets, but HBV/C2 inclined to show rtA181V (3.6% for C2 versus 0.9% for B2; P < 0.01), while HBV/B2 inclined to show rtN236T (4.5% for versus 2.5% for C2; P < 0.01). The ratios of HBV/B2 to HBV/C2 infection were 1.7 (110/65), 5.7 (2,653/463), 7.5 (520/69), 8.0 (48/6), and 15.3 (183/12) for acute hepatitis B, chronic hepatitis B, liver cirrhosis, acute-on-chronic liver failure, and hepatocellular carcinoma, respectively. In conclusion, HBV/C2 and HBV/B2, two prevalent subgenotypes, differ in lamivudine- and adefovir-resistance-associated mutational patterns. HBV/C2-infected patients are more likely to have disease progression than HBV/B2-infected ones.
Hepatitis B virus (HBV) infection remains a serious health problem that currently affects about 350 million people worldwide and 93 million in China (12, 20). HBV infection is associated with a wide spectrum of clinical presentations, including asymptomatic subclinical infection, acute hepatitis B (AHB), chronic hepatitis (CHB), liver cirrhosis (LC), acute-on-chronic liver failure (ACLF), and hepatocellular carcinoma (HCC). Factors associated with disease progression remain largely unknown. Viral factors have been implicated in the pathogenesis and clinic outcome of HBV infection (23).
HBV exhibits a mutation rate of around 2 × 104 base substitutions/site/year, which is an approximately 100 times higher than that of other DNA viruses (5). HBV is at least classified into eight genotypes, based on nucleotide sequence divergence among strains of >8%. In Asia, HBV genotypes B and C are the predominant genotypes, and HBV genotype C is associated with more severe liver disease, delayed HBeAg seroconversion, and a high risk of HCC (7, 8, 10, 17, 18, 24). Within each HBV genotype, subgenotypes have been identified based on a 4 to 8% difference in the complete nucleotide sequence. HBV/B, HBV/C, and HBV/D have been individually classified into five subgenotypes each, namely, HBV/B1 through HBV/B5, HBV/C1 through HBV/C5, and HBV/D1 through HBV/D5 (22). In China, the majority of genotype B HBV isolates belongs to subgenotype B2, while C1 and C2 were predominant subgenotypes in northern and southern parts of China, respectively (30, 31, 32). HBV subgenotypes may have differences in terms of prevalence of HBeAg and HCC development (25, 39, 40). Patients infected by HBV/C1 were reported to have more severe clinical features than those infected by HBV/C2 (42). A Chinese study showed that HBV/C2 infection was more prone to cause chronic infection than HBV/B2 infection (41). However, the clinical implications of HBV subgenotypes remain controversial; e.g., there was a report showing no correlation between HBV/C subgenotypes and disease progression (29).
Nucleoside and nucleotide analogs (NA) are commonly used clinically for suppressing viral replication to halt the progression of liver diseases caused by chronic HBV infection. However, viral resistance is the main drawback of long-term antiviral therapy. To date, data on the association of HBV subgenotype with drug resistance are very limited and uncertain. A study in Germany suggested that the rate of resistance to lamivudine (LAM) was higher in patients with HBV genotype A infection than in patients with genotype D infection (43), an Indian study of 76 patients reported that genotype D is more likely than genotype A to have a sustained virologic response after LAM therapy (28), and an Italian study on 27 patients showed that biochemical and virological responses to LAM do not vary between genotypes A and D (4). Two investigations from southeast Asian regions suggested that patients infected with HBV genotype C had a poorer virologic response to LAM treatment and a higher rate of posttreatment relapse than patients infected with genotype B (9, 16), but another investigation from the United States suggested that patients infected with HBV genotype B appear to have earlier biochemical resistance to LAM than those infected with HBV genotype C (15). In most of the studies, the sample size was relatively small, and this may bring uncertainty to the results. There is still a paucity of data presenting the association of HBV genotypes/subgenotypes with viral resistance to adefovir (ADV) and entecavir (ETV).
In this study, we aimed to investigate the prevalence and clinical implications of HBV subgenotypes in a large cohort of Chinese patients with HBV infection.
MATERIALS AND METHODS
Patients.
A total of 4,300 patients with AHB (n = 182), CHB (n = 3,247), LC (n = 613), ACLF (n = 55), or HCC (n = 203) who visited Beijing 302 Hospital from July 2007 to June 2009 were enrolled in the study. Patients were mainly from different regions of northern China. The diagnostic criteria were based on the Management Scheme of Diagnostic and Therapy Criteria of Viral Hepatitis and on the Diagnostic and Treatment Guidelines for Liver Failure, which were issued by the Chinese Society of Infectious Diseases and Parasitology and by the Chinese Society of Hepatology and have been described in detail in our previous studies (14, 21, 24, 34, 44). All patients were positive for HBV DNA (≥100 IU/ml or 500 copies/ml) at sampling. All subjects except AHB patients were exposed to different regimens of nucleoside/nucleotide analogs. The study was approved by the Ethics Committee of Beijing 302 Hospital.
Serological markers and quantitation of HBV DNA.
Serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), prothrombin activity (PTA), and other biochemical parameters were measured by standard procedures. HBeAg/anti-HBe, HBsAg/anti-HBs, and anti-HBc were detected by enzyme-linked immunosorbent assay (Kewei Diagnostic Ltd., Beijing, China) or chemiluminescent assay (Abbott Laboratories, Chicago, IL). The HBV DNA level was determined with a real-time PCR kit (Fosun Pharmaceutical Co., Shanghai, China) with a lower limit of detection of 100 IU/ml.
HBV genotype/subgenotype classification.
HBV genotype/subgenotype assignment was based on phylogenetic analysis of the 1,225-bp-long S/Pol gene sequence (nucleotides [nt] 54 to 1278) as described previously (21). The sense and antisense primers for the first-round PCR were 5′-AGTCAGGAAGACAGCCTACTCC-3′ (nt 3146 to 3167) and 5′-AGGTGAAGCGAAGTGCACAC-3′ (nt 1577 to 1596), respectively. The sense and antisense primers for second-round PCR were 5′-TTCCTGCTGGTGGCTCCAGTTC-3′ (nt 54 to 75) and 5′-TTCCGCAGTATGGATCGGCAG-3′ (nt 1258 to 1278), respectively. Direct sequencing was performed using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). Phylogenetic and molecular evolutionary analyses were performed in MEGA version 4 (25a). Phylogenetic trees were constructed using neighbor-joining (NJ) analysis with bootstrap test confirmation performed on 1,000 resampling standard reference sequences acquired from the online Hepatitis Virus Database (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi).
Analysis of genotypic drug resistance.
Drug-resistance-associated mutations in the reverse transcriptase (RT) region of the HBV genome were analyzed as previously described (38). Substitutions at positions rt80, rt173, rt180, rt181, rt184, rt202, rt204, rt236, and rt250 were taken as resistance-associated mutations for analysis.
Statistical analysis.
Continuous variables were expressed as means ± standard deviations or medians. Differences in continuous data were examined by Student's t test, analysis of variance, or the nonparametric Wilcoxon signed-rank test where appropriate, while differences in categorical data were examined by the chi-square test or Fisher's exact test. Logistic regression was used to evaluate P values in multivariate analysis. Statistical analysis was carried out with SPSS 16.0 software. A P value of <0.05 was considered statistically significant.
RESULTS
Classification of HBV genotypes/subgenotypes.
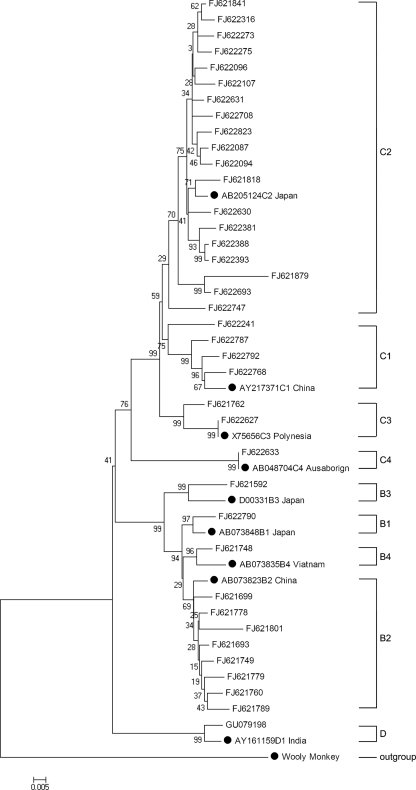
Among a total of 4,300 patients, the subgenotype distribution was as follows: 17 (0.40%) for B1, 615 (14.30%) for B2, 11 (0.25%) for B3, 15 (0.35%) for B4, 45 (1.05%) for C1, 3,514 (81.72%) for C2, 40 (0.93%) for C3, 7 (0.16%) for C4, and 36 (0.84%) for genotype D. Within genotype D, 20 samples were D1, 3 were D2, 4 were D3, 7 were D4, and 2 failed to be classified into subgenotypes. No other genotypes (A, E, F, G, or H) were detected in the patients enrolled in this study. Thus, HBV/C2 was the most predominant subgenotype, followed by HBV/B2 in northern China. A phylogenetic tree based on the 38 representative analyzed HBV genetic sequences with GenBank accession numbers is presented in Fig. 1.
FIG. 1.
Neighbor-joining phylogenetic tree based on the 38 representative analyzed HBV genetic sequences with GenBank accession numbers. Standard reference sequences are marked by circles.
HBV subgenotypes in relation to clinical features.
In patients with AHB, no obvious differences in age, gender, HBV DNA level, TBIL, PTA, ALT, AST, or HBeAg-positive rate were observed between HBV/C2- and HBV/B2-infected patients (Table 1). In patients with chronic HBV infection, however, those with HBV/B2 were younger and had lower HBeAg-positive rates than patients with HBV/C2. There was no significant difference between the two subsets in the other observed parameters (Table 2).
TABLE 1.
HBV subgenotypes in relation to clinical features in patients with acute hepatitis B (n = 182)
| HBV subgenotype | No. of cases | % | Mean age (yr) ± SD | No. male | Mean HBV DNA (log IU/ml) ± SD | Mean TBIL (μmol/liter) ± SD | Mean PTA (%) ± SD | Mean ALT (IU/liter) ± SD | Mean AST (IU/liter) ± SD | % HBeAg+ |
|---|---|---|---|---|---|---|---|---|---|---|
| B1 | 2 | 1.10 | 40 ± 3 | 2 | 2.7 | 96.0 ± 46.7 | 117.1 ± 35.5 | 1,049 ± 969 | 203 ± 93 | 50 |
| B2 | 65 | 35.71 | 36 ± 12 | 54 | 4.0 ± 1.6 | 126.1 ± 96.5 | 95.2 ± 21.7 | 1,698 ± 1,000 | 885 ± 1188 | 24.6 |
| B3 | 1 | 0.55 | 30 | 1 | 3.7 | 24.4 | 30 | 1,388 | 327 | 0 |
| B4 | 1 | 0.55 | 25 | 1 | 4.4 | 107 | 97.4 | 3,181 | 2,787 | 0 |
| C1 | 2 | 1.10 | 34 ± 6 | 2 | 3.8 ± 1.6 | 67.1 ± 56.8 | 94.8 ± 105.6 | 2,393 ± 1,626 | 847 ± 44 | 50 |
| C2 | 110 | 60.44 | 37 ± 13 | 88 | 4.2 ± 1.4 | 135.1 ± 107.5 | 95.4 ± 233.1 | 1,661 ± 1,058 | 758 ± 680 | 37.2 |
| C3 | 1 | 0.55 | 18 | 1 | 6.7 | 233 | 30 | 890 | 890 | 0 |
| C4 | 0 | |||||||||
| D | 0 | |||||||||
| Pa | 0.56 | 0.45 | 0.44 | 0.47 | 0.67 | 0.74 | 0.46 | 0.09 |
The P values were obtained from comparison of parameters between HBV subgenotypes B2 and C2 and evaluated by multivariate analysis.
TABLE 2.
HBV subgenotypes in relation to clinical features in patients with chronic HBV infection (n = 4,118)
| HBV subgenotype | No. of cases | % | Mean age (yr) ± SD | No. male | Mean HBV DNA (log IU/ml) ± SD | Mean TBIL (μmol/liter) ± SD | Mean PTA (%) ± SD | Mean ALT (IU/liter) ± SD | Mean AST (IU/liter) ± SD | % HBeAg+ |
|---|---|---|---|---|---|---|---|---|---|---|
| B1 | 15 | 0.36 | 31 ± 12 | 15 | 4.3 ± 1.7 | 17.0 ± 5.7 | 88.0 ± 11.7 | 56 ± 49 | 38 ± 29 | 33.3 |
| B2 | 550 | 13.35 | 34 ± 13 | 475 | 5.2 ± 1.7 | 19.7 ± 34.6 | 83.2 ± 23.3 | 92 ± 176 | 63 ± 103 | 60.5 |
| B3 | 10 | 0.24 | 40 ± 14 | 7 | 4.5 ± 1.3 | 17.4 ± 9.0 | 88.0 ± 15.4 | 5 ± 42 | 59 ± 57 | 40 |
| B4 | 14 | 0.34 | 33 ± 15 | 10 | 5.0 ± 2.0 | 30.0 ± 52.9 | 92.4 ± 13.4 | 160 ± 261 | 86 ± 119 | 64.3 |
| C1 | 43 | 1.04 | 38 ± 13 | 36 | 5.4 ± 1.7 | 30.8 ± 55.1 | 76.1 ± 16.9 | 84 ± 143 | 80 ± 153 | 69.8 |
| C2 | 3,404 | 82.66 | 40 ± 13 | 2,854 | 5.1 ± 1.7 | 25.6 ± 53.3 | 79.5 ± 21.5 | 84 ± 144 | 67 ± 94 | 64.3 |
| C3 | 39 | 0.95 | 37 ± 12 | 32 | 5.3 ± 1.5 | 15.9 ± 9.1 | 82.2 ± 16.5 | 74 ± 78 | 61 ± 59 | 79.5 |
| C4 | 7 | 0.17 | 29 ± 12 | 6 | 5.0 ± 1.7 | 8.3 ± 2.8 | 88.3 ± 10.4 | 46 ± 41 | 45 ± 44 | 71.4 |
| D | 36 | 0.87 | 35 ± 14 | 28 | 5.2 ± 1.9 | 16.7 ± 10.9 | 80.0 ± 14.6 | 100 ± 168 | 69 ± 114 | 58.3 |
| Pa | <0.01 | 0.2 | 0.64 | 0.68 | 0.75 | 0.33 | 0.18 | 0.01 |
The P values were obtained from comparison of parameters between HBV subgenotypes B2 and C2 and evaluated by multivariate analysis.
Comparison of drug-resistant mutational patterns between HBV/B2 and HBV/C2.
There was no significant difference between HBV-B2- and HBV/C2-infected patients in the proportion and duration of treatment with individual NAs (Table 3). Comparison of drug-resistant mutational patterns between HBV/B2- and HBV/C2-infected patients showed that the two subsets had differences in LAM- and ADV-resistant mutational patterns. With respect to LAM-resistance-associated mutations, HBV/C2-infected patients had a significantly higher incidence of total primary resistance mutations (sum of rtM204I, rtM204V, and rtM204I/V), rtM204I, and compensatory mutations rtL80I, rtV173L, and rtL180M. In addition, concomitance of rtM204I with the compensatory mutation(s) (with any of L80I, V173L, and L180M) was more frequently detected in HBV/C2- than in HBV/B2-infected patients. The proportion of the concomitant mutation(s) in total M204I-positive samples was 43.6% for HBV/B2 and 67.2% for HBV/C2 (P < 0.01) (Table 4). The incidences of adefovir-resistant mutations were similar in the two subsets. However, HBV/C2-infected patients had a higher rtA181V incidence but a lower rtN236T incidence than HBV/B2-infected patients. No obvious difference in occurrence of ETV-resistant mutations was observed between HBV/C2- and HBV/B2-infected patients (Table 5).
TABLE 3.
Comparison of nucleoside/nucleotide analog administration between HBV/B2- and HBV/C2-infected patientsa
| HBV subgenotype (n) | Lamivudine treatment |
Adefovir treatment |
Entecavir treatment |
Telbivudine treatment |
||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of cases | Mean duration (mo) ± SD | No. (%) of cases | Mean duration (mo) ± SD | No. (%) of cases | Mean duration (mo) ± SD | No. (%) of cases | Mean duration (mo) ± SD | |
| B2 (550) | 368 (66.8) | 22.6 ± 19.6 | 377 (68.4) | 18.0 ± 13.6 | 59 (10.8) | 16.0 ± 9.6 | 52 (9.5) | 10.8 ± 6.7 |
| C2 (3,404) | 2,223 (65.3) | 24.9 ± 20.7 | 2220 (65.2) | 18.7 ± 14.4 | 428 (12.6) | 16.0 ± 11.5 | 322 (9.5) | 12.4 ± 10.2 |
| P | 0.464 | 0.237 | 0.127 | 0.591 | 0.222 | 0.601 | 0.997 | 0.538 |
Including monotherapy and sequential and combined therapies in various administration schedules. The duration that the patient was exposed to the individual drug was independently counted.
TABLE 4.
Comparison of incidences of lamivudine-resistance-associated mutations between patients chronically infected with HBV/B2 (n = 550) and HBV/C2 (n = 3,404)
| HBV subgenotype | Mutation incidence (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total | M204I | M204V | M204I + V | L80I | V173L | L180M | M204I + CMa | |
| B2 | 19.8 | 10.0 | 8.7 | 1.1 | 5.3 | 0 | 10.2 | 8.6 |
| C2 | 27.9 | 15.5 | 10.3 | 2.1 | 8.2 | 3.8 | 19.0 | 18.7 |
| P | <0.01 | <0.01 | 0.26 | 0.09 | 0.02 | <0.01 | <0.01 | <0.01 |
CM, compensatory mutation. The proportions of M204I + CM (with any of L80I, V173L, and L180M) in all M204I-positive samples are 43.6% for HBV/B2 and 67.2% for HBV/C2 (P < 0.01).
TABLE 5.
Comparison of incidences of adefovir and entecavir resistance mutations between patients chronically infected with HBV/B2 (n = 550) and HBV/C2 (n = 3,404)
| HBV subgenotype | Mutation incidence (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Adefovir resistance mutations |
Entecavir resistance mutationsa |
|||||||
| Total | A181V | N236T | A181V + N236T | Total | T184A/I/L/S | S202C/G | M250I/L/V | |
| B2 | 6.4 | 0.9 | 4.5 | 0.9 | 1.1 | 0.2 | 0.2 | 0.7 |
| C2 | 7.1 | 3.6 | 2.5 | 0.9 | 1.7 | 0.7 | 0.2 | 0.7 |
| P | 0.56 | <0.01 | <0.01 | 0.99 | 0.29 | 0.13 | 0.81 | 0.89 |
All merge in conjunction with lamivudine resistance mutation (rtM204V and/or rtM204I).
HBV subgenotypes in relation to disease category.
Table 6 summarizes the HBV subgenotypes in relation to the different disease categories. The ratios of HBV/B2 to HBV/C2 infection were 1.7 (110/65), 5.7 (2,653/463), 7.5 (520/69), 8.0 (48/6), and 15.3 (183/12) for AHB, CHB, LC, ACLF, and HCC, respectively.
TABLE 6.
HBV subgenotypes in relation to disease progression
| HBV subgenotype | No. of patients witha: |
||||
|---|---|---|---|---|---|
| AHB (n = 182) | CHB (n = 3,247) | LC (n = 613) | ACLF (n = 55) | HCC (n = 203) | |
| B1 | 2 | 13 | 2 | 0 | 0 |
| B2 | 65 | 463 | 69 | 6 | 12 |
| B3 | 1 | 8 | 1 | 0 | 1 |
| B4 | 1 | 12 | 1 | 1 | 0 |
| C1 | 2 | 34 | 5 | 0 | 4 |
| C2 | 110 | 2,653 | 520 | 48 | 183 |
| C3 | 1 | 26 | 10 | 0 | 3 |
| C4 | 0 | 7 | 0 | 0 | 0 |
| D | 0 | 31 | 5 | 0 | 0 |
| C2 vs B2 | 1.7 | 5.7 | 7.5 | 8.0 | 15.3 |
AHB, acute hepatitis B; CHB, chronic hepatitis B; LC, liver cirrhosis; ACLF, acute-on-chronic liver failure; HCC, hepatocellular carcinoma.
DISCUSSION
Previous studies have shown that that HBV genotype C predominates in northern China, genotype B predominates in southern China, genotype D is prevalent in Xinjiang, genotype A is rare, and genotypes E, F, G, and H are not found in China. Recently, investigations of HBV subgenotypes have attracted much attention. A study of 211 Chinese patients in Guangdong Province in southern China showed that HBV subgenotypes B2, C1, and C2 were the most prevalent HBV variants and that the three subgenotypes had different mutational incidences in the basal core promoter (BCP) and precore regions (33). Another study investigated 304 HBV-infected patients and showed that C2 is the most prevalent subgenotype in northeast China (30). However, the clinical implications of HBV subgenotype are far from well understood. Further studies are needed to provide knowledge about HBV genotypes/subgenotypes with respect to viral latency, HBeAg seroconversion, pathogenesis of liver disease, immune escape, treatment response, and resistance to antiviral drug therapy.
In this study, we systematically analyzed the association between disease categories and HBV subgenotypes. Interestingly, the ratio of HBV subgenotypes C2 and B2 rose in the order of CHB, LC, ACLF, and HCC. In addition, HBV/C2-infected patients were older and had a higher HBeAg-positive rate than HBV/B2-infected patients. These results suggested that HBV/C2 is associated with disease progression, whereas HBV/B2 is associated with resolution. It is suggested that HBV/B2 infection is more likely to induce optimal immune responses than HBV/C2 infection due to intrinsic virologic features. Consistent with previous studies (30, 33), we found that HBV/B2 had a significantly lower incidence of BCP mutations A1762T/G1764A but a higher incidence of precore mutation G1896A than HBV/C2 (data not shown). As BCP and precore mutations may influence HBV replication and/or HBeAg expression, whether the BCP/PC mutations may account for the different outcomes for patients with HBV/B2 and HBV/C2 warrants further clarification.
Currently, PCR-restriction fragment length polymorphism (RFLP) and direct PCR sequencing are the most popular methods for HBV subgenotyping. Although direct PCR sequencing is more time-consuming, it may offer abundant information and is taken as a “gold standard.” However, the sensitivity of direct PCR sequencing is usually lower, and it may not detect samples with a low viral load, which is often the case. We developed a highly sensitive nested PCR assay allowing us to analyze samples with quite low viral loads (≥20 IU/ml), as we had done in a study of severe acute respiratory syndrome (SARS) coronavirus (35, 36, 37). The 1,225-bp-long S/Pol gene fragment amplified in our study encompasses the complete S gene and partial PreS and Pol gene fragments for phylogenetic tree analysis of HBV subgenotypes. Similar methods have been used by other investigators (11, 13, 22). To verify the results, we sequenced 113 complete HBV genomes from enrolled samples (GenBank accession numbers FJ386574 through FJ386689, except FJ386599, FJ386646, and FJ386647) and compared the classification of subgenotypes determined on the basis of S/Pol gene fragments with those determined on the basis of complete HBV genomes. The results showed a 99.1% (112/113) concordance between the two assays. Only one sequence assigned to HBV/C1 based on the 1,225-bp fragment was classified as HBV/C2 based on the complete HBV genome (FJ386664). It was verified to be a recombinant strain because it was classified as HBV/C2 based on the precore/core region. As the 1,225-bp amplicon encompasses the complete reverse transcriptase (RT) region of the HBV polymerase gene, all well-recognized drug resistance mutations could be analyzed simultaneously.
rtM204I and rtM204V are classical LAM resistance mutations and often coexist with compensatory mutations (rtL80I, rtV173L, and rtL180M) (1). rtN236T and rtA181V are two well-recognized ADV resistance mutations (2, 3). Substitutions in rtT184, rtS202, or rtM250 in conjunction with LAM resistance mutations result in ETV resistance (26, 27). Unlike in a rigorously designed clinical trial, the patients with chronic HBV infection enrolled in this study were from routine clinical practice with different treatment regimens and durations. The duration of treatment may influence the incidence of resistant HBV strains, although the influence was relatively minor in the large-population samples of our study. Nevertheless, HBV/B2 and HBV/C2 exhibited different LAM- and ADV-resistant mutational patterns, suggesting that HBV subgenotypes might have an impact on drug resistance. To our knowledge, an association of ADV resistance with HBV subgenotypes B2 and C2 has not been documented so far. Compared to rtA181V, rtN236T confers a greater reduction of ADV sensitivity in vitro (3). On the other hand, rtL80I is reported to be associated with a poor response to ADV (19), and the coselection of rtL180M in patients with rtM204I decreased serum ALT normalization significantly after ADV therapy (6). Further study is required to clarify the impact of the difference in drug-resistant mutational patterns on the antiviral response to ADV treatment in clinical practice.
In conclusion, the present study verifies that HBV/C2 is the most predominant subgenotype, followed by HBV/B2 in northern China. HBV/C2 and HBV/B2 differ in LAM- and ADV-resistance-associated mutational patterns, and HBV/C2-infected patients are more likely to have disease progression than HBV/B2-infected ones. Our results provide new insight into features of HBV subgenotypes which may have important clinical implications for management of HBV infection in China.
Acknowledgments
This work was supported by the Beijing Natural Science Foundation (7091006), the National Key Basic Research Developing Project (2007CB512803), and the National 11th Five-Year Special Grand Project for Infectious Diseases (2008ZX10002-005-6, 2008ZX10002-011, and 2009ZX10004-314).
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, and L. D. Condreay. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Angus, P., R. Vaughan, S. Xiong, H. Yang, W. Delaney, C. Gibbs, C. Brosgart, D. Colledge, R. Edwards, A. Ayres, A. Bartholomeusz, and S. Locarnini. 2003. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology 125:292-297. [DOI] [PubMed] [Google Scholar]
- 3.Borroto-Esoda, K., M. D. Miller, and S. Arterburn. 2007. Pooled analysis of amino acid changes in the HBV polymerase in patients from four major adefovir dipivoxil clinical trials. J. Hepatol. 47:492-498. [DOI] [PubMed] [Google Scholar]
- 4.Buti, M., M. Cotrina, A. Valdes, R. Jardi, F. Rodriguez-Frias, and R. Esteban. 2002. Is hepatitis B virus subtype testing useful in predicting virological response and resistance to lamivudine? J. Hepatol. 36:445-446. [DOI] [PubMed] [Google Scholar]
- 5.Buti, M., F. Rodriguez-Frias, R. Jardi, and R. Esteban. 2005. Hepatitis B virus genome variability and disease progression: the impact of pre-core mutants and HBV genotypes. J. Clin. Virol. 34(Suppl. 1):S79-S82. [DOI] [PubMed] [Google Scholar]
- 6.Cha, C. K., H. C. Kwon, J. Y. Cheong, S. W. Cho, S. P. Hong, S. O. Kim, and W. D. Yoo. 2009. Association of lamivudine-resistant mutational patterns with the antiviral effect of adefovir in patients with chronic hepatitis B. J. Med. Virol. 81:417-424. [DOI] [PubMed] [Google Scholar]
- 7.Chan, H. L., S. W. Tsang, M. L. Wong, C. H. Tse, N. W. Leung, F. K. Chan, and J. J. Sung. 2002. Genotype B hepatitis B virus is associated with severe icteric flare-up of chronic hepatitis B virus infection in Hong Kong. Am. J. Gastroenterol. 97:2629-2633. [DOI] [PubMed] [Google Scholar]
- 8.Chan, H. L., M. L. Wong, A. Y. Hui, L. C. Hung, F. K. Chan, and J. J. Sung. 2003. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J. Clin. Microbiol. 41:1277-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, R. N., C. T. Yeh, S. L. Tsai, C. M. Chu, and Y. F. Liaw. 2003. Determinants for sustained HBeAg response to lamivudine therapy. Hepatology 38:1267-1273. [DOI] [PubMed] [Google Scholar]
- 10.Chu, C. J., M. Hussain, and A. S. Lok. 2002. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 122:1756-1762. [DOI] [PubMed] [Google Scholar]
- 11.Di Lello, F. A., Y. L. F. G. Pineiro, G. Munoz, and R. H. Campos. 2009. Diversity of hepatitis B and C viruses in Chile. J. Med. Virol. 81:1887-1894. [DOI] [PubMed] [Google Scholar]
- 12.European Association for the Study of the Liver. 2009. EASL clinical practice guidelines: management of chronic hepatitis B. J. Hepatol. 50:227-242. [DOI] [PubMed] [Google Scholar]
- 13.Fang, Z. L., C. A. Sabin, B. Q. Dong, S. C. Wei, Q. Y. Chen, K. X. Fang, J. Y. Yang, X. Y. Wang, and T. J. Harrison. 2009. The association of HBV core promoter double mutations (A1762T and G1764A) with viral load differs between HBeAg positive and anti-HBe positive individuals: a longitudinal analysis. J. Hepatol. 50:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, J., D. Xu, Z. Liu, M. Shi, P. Zhao, B. Fu, Z. Zhang, H. Yang, H. Zhang, C. Zhou, J. Yao, L. Jin, H. Wang, Y. Yang, Y. X. Fu, and F. S. Wang. 2007. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 132:2328-2339. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh, T. H., T. C. Tseng, C. J. Liu, M. Y. Lai, P. J. Chen, H. L. Hsieh, D. S. Chen, and J. H. Kao. 2009. Hepatitis B virus genotype B has an earlier emergence of lamivudine resistance than genotype C. Antivir. Ther. 14:1157-1163. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, J., Y. Ueno, Y. Wakui, H. Niitsuma, K. Fukushima, Y. Yamagiwa, M. Shiina, Y. Kondo, E. Kakazu, K. Tamai, N. Obara, T. Iwasaki, and T. Shimosegawa. Four-year study of lamivudine and adefovir combination therapy in lamivudine-resistant hepatitis B patients: influence of hepatitis B virus genotype and resistance mutation pattern. J. Viral Hepat., in press. [DOI] [PubMed]
- 17.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2003. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124:327-334. [DOI] [PubMed] [Google Scholar]
- 18.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 19.Lee, Y. S., Y. H. Chung, J. A. Kim, S. E. Kim, J. W. Shin, K. M. Kim, Y. S. Lim, N. H. Park, H. C. Lee, and D. J. Suh. 2009. Hepatitis B virus with the rtL80V/I mutation is associated with a poor response to adefovir dipivoxil therapy. Liver Int. 29:552-556. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., C. M. Wang, J. Cheng, Z. L. Liang, Y. W. Zhong, X. Q. Ren, Z. H. Xu, F. Zoulim, and D. P. Xu. 2009. Hepatitis B virus in tenofovir-naive Chinese patients with chronic hepatitis B contains no mutation of rtA194T conferring a reduced tenofovir susceptibility. Chin. Med. J. 122:1585-1586. [PubMed] [Google Scholar]
- 21.Liu, Y., Y. Zhong, Z. Zou, Z. Xu, B. Li, X. Ren, S. Bai, L. Wang, X. Li, J. Dai, Y. Wang, P. Mao, and D. Xu. 2010. Features and clinical implications of hepatitis B virus genotypes and mutations in basal core promoter/precore region in 507 Chinese patients with acute and chronic hepatitis B. J. Clin. Virol. 47:243-247. [DOI] [PubMed] [Google Scholar]
- 22.Lusida, M. I., V. E. Nugrahaputra, Soetjipto, R. Handajani, M. Nagano-Fujii, M. Sasayama, T. Utsumi, and H. Hotta. 2008. Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia. J. Clin. Microbiol. 46:2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehermann, B., and M. Nascimbeni. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 5:215-229. [DOI] [PubMed] [Google Scholar]
- 24.Ren, X., Z. Xu, Y. Liu, X. Li, S. Bai, N. Ding, Y. Zhong, L. Wang, P. Mao, F. Zoulim, and D. Xu. Hepatitis B virus genotype and basal core promoter/precore mutations are associated with hepatitis B-related acute-on-chronic liver failure without pre-existing liver cirrhosis. J. Viral Hepat., in press. [DOI] [PMC free article] [PubMed]
- 25.Sugauchi, F., E. Orito, T. Ichida, H. Kato, H. Sakugawa, S. Kakumu, T. Ishida, A. Chutaputti, C. L. Lai, R. G. Gish, R. Ueda, Y. Miyakawa, and M. Mizokami. 2003. Epidemiologic and virologic characteristics of hepatitis B virus genotype B having the recombination with genotype C. Gastroenterology 124:925-932. [DOI] [PubMed] [Google Scholar]
- 25a.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 26.Tenney, D. J., S. M. Levine, R. E. Rose, A. W. Walsh, S. P. Weinheimer, L. Discotto, M. Plym, K. Pokornowski, C. F. Yu, P. Angus, A. Ayres, A. Bartholomeusz, W. Sievert, G. Thompson, N. Warner, S. Locarnini, and R. J. Colonno. 2004. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob. Agents Chemother. 48:3498-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenney, D. J., R. E. Rose, C. J. Baldick, S. M. Levine, K. A. Pokornowski, A. W. Walsh, J. Fang, C. F. Yu, S. Zhang, C. E. Mazzucco, B. Eggers, M. Hsu, M. J. Plym, P. Poundstone, J. Yang, and R. J. Colonno. 2007. Two-year assessment of entecavir resistance in lamivudine-refractory hepatitis B virus patients reveals different clinical outcomes depending on the resistance substitutions present. Antimicrob. Agents Chemother. 51:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thakur, V., S. K. Sarin, S. Rehman, R. C. Guptan, S. N. Kazim, and S. Kumar. 2005. Role of HBV genotype in predicting response to lamivudine therapy in patients with chronic hepatitis B. Indian J. Gastroenterol. 24:12-15. [PubMed] [Google Scholar]
- 29.Tseng, T. C., C. J. Liu, P. J. Chen, M. Y. Lai, C. L. Lin, J. H. Kao, and D. S. Chen. 2007. Subgenotypes of hepatitis B virus genotype C do not correlate with disease progression of chronic hepatitis B in Taiwan. Liver Int. 27:983-988. [DOI] [PubMed] [Google Scholar]
- 30.Wang, H. Y., D. Li, W. Liu, X. Jin, B. Du, Y. P. Li, H. X. Gu, and S. Y. Zhang. 2010. Hepatitis B virus subgenotype C2 is the most prevalent subgenotype in northeast China. Clin. Microbiol. Infect. 16:477-481. [DOI] [PubMed] [Google Scholar]
- 31.Wang, Z., J. Hou, G. Zeng, S. Wen, Y. Tanaka, J. Cheng, F. Kurbanov, L. Wang, J. Jiang, N. V. Naoumov, M. Mizokami, and Y. Qi. 2007. Distribution and characteristics of hepatitis B virus genotype C subgenotypes in China. J. Viral Hepat. 14:426-434. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Z., Y. Huang, S. Wen, B. Zhou, and J. Hou. 2007. Hepatitis B virus genotypes and subgenotypes in China. Hepatol. Res. 37:S36-S41. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Z., Y. Tanaka, Y. Huang, F. Kurbanov, J. Chen, G. Zeng, B. Zhou, M. Mizokami, and J. Hou. 2007. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J. Clin. Microbiol. 45:1491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, D., J. Fu, L. Jin, H. Zhang, C. Zhou, Z. Zou, J. M. Zhao, B. Zhang, M. Shi, X. Ding, Z. Tang, Y. X. Fu, and F. S. Wang. 2006. Circulating and liver resident CD4+CD25+ regulatory T cells actively influence the antiviral immune response and disease progression in patients with hepatitis B. J. Immunol. 177:739-747. [DOI] [PubMed] [Google Scholar]
- 35.Xu, D., Z. Zhang, F. Chu, Y. Li, L. Jin, L. Zhang, G. F. Gao, and F. S. Wang. 2004. Genetic variation of SARS coronavirus in Beijing Hospital. Emerg. Infect. Dis. 10:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, D., Z. Zhang, L. Jin, F. Chu, Y. Mao, H. Wang, M. Liu, M. Wang, L. Zhang, G. F. Gao, and F. S. Wang. 2005. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur. J. Clin. Microbiol. Infect. Dis. 24:165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, D., Z. Zhang, and F. S. Wang. 2004. SARS-associated coronavirus quasispecies in individual patients. N. Engl. J. Med. 350:1366-1367. [DOI] [PubMed] [Google Scholar]
- 38.Xu, Z., Y. Liu, T. Xu, L. Chen, L. Si, Y. Wang, X. Ren, Y. Zhong, J. Zhao, and D. Xu. 2010. Acute hepatitis B infection associated with drug-resistant hepatitis B virus. J. Clin. Virol. 48:270-274. [DOI] [PubMed] [Google Scholar]
- 39.Yin, J., H. Zhang, C. Li, C. Gao, Y. He, Y. Zhai, P. Zhang, L. Xu, X. Tan, J. Chen, S. Cheng, S. Schaefer, and G. Cao. 2008. Role of hepatitis B virus genotype mixture, subgenotypes C2 and B2 on hepatocellular carcinoma: compared with chronic hepatitis B and asymptomatic carrier state in the same area. Carcinogenesis 29:1685-1691. [DOI] [PubMed] [Google Scholar]
- 40.Yuan, J., B. Zhou, Y. Tanaka, F. Kurbanov, E. Orito, Z. Gong, L. Xu, J. Lu, X. Jiang, W. Lai, and M. Mizokami. 2007. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J. Clin. Virol. 39:87-93. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, H. W., J. H. Yin, Y. T. Li, C. Z. Li, H. Ren, C. Y. Gu, H. Y. Wu, X. S. Liang, P. Zhang, J. F. Zhao, X. J. Tan, W. Lu, S. Schaefer, and G. W. Cao. 2008. Risk factors for acute hepatitis B and its progression to chronic hepatitis in Shanghai, China. Gut 57:1713-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, L., C. H. Tse, V. W. Wong, A. M. Chim, K. S. Leung, and H. L. Chan. 2008. A complete genomic analysis of hepatitis B virus genotypes and mutations in HBeAg-negative chronic hepatitis B in China. J. Viral Hepat. 15:449-458. [DOI] [PubMed] [Google Scholar]
- 43.Zollner, B., J. Petersen, E. Puchhammer-Stockl, J. Kletzmayr, M. Sterneck, L. Fischer, M. Schroter, R. Laufs, and H. H. Feucht. 2004. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology 39:42-50. [DOI] [PubMed] [Google Scholar]
- 44.Zou, Z., D. Xu, B. Li, S. Xin, Z. Zhang, L. Huang, J. Fu, Y. Yang, L. Jin, J. M. Zhao, M. Shi, G. Zhou, Y. Sun, and F. S. Wang. 2009. Compartmentalization and its implication for peripheral immunologically-competent cells to the liver in patients with HBV-related acute-on-chronic liver failure. Hepatol. Res. 39:1198-1207. [DOI] [PubMed] [Google Scholar]