Abstract
Candida bracarensis is a recently described Candida species which is phenotypically similar to Candida glabrata. A case of C. bracarensis bloodstream infection in a bone marrow transplant patient is described and confirms this organism as an opportunistic human pathogen. The organism can be distinguished from C. glabrata by its white color on CHROMagar and by DNA sequence analysis using D1/D2 and internal transcribed spacer (ITS) primers.
CASE REPORT
A 50-year-old male underwent a matched related-donor bone marrow transplant after a diagnosis of chronic lymphocytic leukemia made 7 years previously. The patient's posttransplant course was complicated by significant graft-versus-host disease (GVHD) of the skin, liver, and bowel. Four months after transplant, the GVHD necessitated admission to hospital and he was treated with cyclosporine and corticosteroids. In hospital, he had multiple infectious complications, including herpes simplex virus mucositis and cytomegalovirus viremia, treated with antiviral therapy. Seven weeks after admission to hospital, the patient developed respiratory failure and sepsis, which necessitated transfer to the intensive care unit (ICU). Klebsiella pneumoniae was cultured from both the urine and the blood, and Staphylococcus aureus was cultured from bronchoalveolar lavage fluid. He was treated with appropriate antibiotic therapy.
Three weeks after admission to the ICU, two blood cultures became positive, and yeast cells were seen on the Gram stain. After 24 h of growth, small white colonies were seen on blood and chocolate agar media. A wet prep showed budding yeast cells, and a germ tube test was negative. On Saboraud's agar, colonies appeared white and creamy. On cornmeal agar, hyphae or pseudohyphae were not observed microscopically. Both isolates produced white colonies on BBL CHROMagar (BD diagnostics, Maryland). Biochemically, the isolates were positive for the rapid trehalose assay (Remel, Lenexa, KS) and positive for assimilation of lysine and glucose (Table 1). Identification using the API 20C AUX system (bioMérieux, Inc., NC) showed a low-percentage identity match with C. glabrata (<50%). Thus, molecular analysis was applied to further characterize these strains.
TABLE 1.
Characteristics of the two clinical isolates
| Isolate | Assay result |
Assimilation at 48 h |
|||||
|---|---|---|---|---|---|---|---|
| C. glabrata-specific PCR | CHROMagar | Rapid trehalose | l-lysine | d-glucose | Glycerol | d-trehalose | |
| Clinical isolate 1 | − | White | + | + | + | − | ± |
| Clinical isolate 2 | − | White | + | + | + | − | ± |
| C. bracarensis type strain | − | White | + | + | + | − | + |
| C. nivariensis type strain | − | White | − | + | + | + | + |
| C. glabrata type strain | + | Pink | + | − | + | − | + |
| C. albicans type strain | − | Green | − | + | + | − | − |
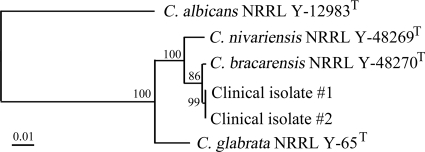
Both isolates were negative as determined by a C. glabrata-specific PCR, with gel electrophoresis endpoint detection using primers targeting the internal transcribed spacer (ITS) region and PCR conditions described previously (8) (Table 1). The ITS1-5.8S-ITS2 region and the D1/D2 region of 26S ribosomal DNA (nucleotides 63 to 642) were amplified and sequenced. PCRs were conducted using the Phire Hot Start DNA polymerase kit (New England Biolabs, Massachusetts) according to the manufacturer's instructions and using the following amplification conditions: 98°C for 30 s, followed by 35 cycles of 98°C for 5 s, annealing temperature (see below) for 5 s, and 72°C for 20 s, followed by a final extension of 72°C for 1 min. The ITS region was amplified using the primers ITS-1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′) (12) and an annealing temperature of 56°C, while amplification of D1/D2 required the primers NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL-4 (5′-GGTTCCGTGTTTCAAGACGG-3′) (9) and an annealing temperature of 60°C. PCR amplicons were sequenced using the ABI 3130xl genetic analyzer (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Sequence alignment and cluster analysis of the ITS and D1/D2 regions of the clinical isolates along with the type strains C. nivariensis NRRL Y-48269T, C. bracarensis NRRL Y-48270T, C. glabrata NRRL Y-65T, and C. albicans NRRL Y-12983T were performed by the neighbor-joining (NJ) method using the BioNumerics v.6.0 software program (Applied Maths, Inc., Austin, TX). The ITS and D1/D2 sequences of the clinical isolates were identical to each other and showed 98.7% and 99.7% similarities, respectively, to the type strain of C. bracarensis. Furthermore, NJ phylogenetic analysis of both D1/D2 (Fig. 1) and ITS (data not shown) demonstrated the clinical isolates clustering more closely with C. bracarensis than any of the other type strains. Based on the results from phenotypic and molecular analysis (Table 1 and Fig. 1), we identified the isolates as C. bracarensis.
FIG. 1.
Neighbor-joining tree based on nucleotide sequences of the D1/D2 region of 26S rRNA of two clinical isolates and type strains of C. bracarensis, C. nivariensis, C. glabrata, and C. albicans. Bootstrap replication percentages (1,000 replicates) are indicated at the nodes. Bar, 1% nucleotide sequence divergence.
Antifungal drug susceptibilities of the two isolates were tested using the Yeast YO9 panel (Trek Diagnostic, Cleveland, OH). Both isolates were susceptible to all the drugs tested except for itraconazole, to which a category of “susceptible dose dependent” was assigned (Table 2). The patient was treated with a 2-week course of caspofungin. Subsequent blood cultures performed while the patient was on caspofungin therapy were negative. However, he ultimately succumbed to his underlying disease (GVHD) 32 days after the positive Candida culture. His death was felt not to be related to the candidemia.
TABLE 2.
Antifungal drug susceptibility profiles of C. bracarensisa
| Isolate (reference) | Source | MIC (μg/ml) of drug |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5-FC | AMB | FLC | ITC | VRC | PSC | CASP | MICA | ANID | ||
| Clinical isolate 1 (this study) | Blood (Canada) | ≤0.06 | 0.25 | 8 | 0.25 | 0.12 | 0.5 | 0.06 | ≤0.008 | 0.03 |
| Clinical isolate 2 (this study) | Blood (Canada) | ≤0.06 | 0.5 | 8 | 0.25 | 0.12 | 0.5 | 0.06 | ≤0.008 | 0.03 |
| C. bracarensis TSa (3) | Vagina (Portugal) | 0.03 | 0.023 | 8 | 0.5 | 0.12 | 0.5 | 0.25 | ||
| Cagl-78 (3) | Stool (USA) | 0.12 | 1 | 8 | 0.25 | 0.06 | 0.5 | 0.25 | ||
| Cagl-112 (3) | Abscess (USA) | 0.12 | 0.094 | 4 | 0.25 | 0.06 | 0.25 | 0.25 | ||
| Cagl-121 (3) | Throat (USA) | 0.25 | 0.19 | 256 | >16 | 8 | >8 | 0.12 | ||
| C. bracarensis no. 1 (7) | Sputum (USA) | 8 | 16 | 0.03 | 0.015 | 0.06 | ||||
| C. bracarensis no. 2 (7) | Blood (USA) | 1 | 2 | 0.03 | 0.015 | 0.06 | ||||
C. bracarensis TS, C. bracarensis type strain NRRL Y-48270T (CBS10154); AMB, amphotericin B; FLC, fluconazole; ITC, itraconazole; VRC, voriconazole; PSC, posaconazole; CASP, caspofungin; MIC, micafungin; ANID, anidulafungin.
Candida species are the third-most-common cause of nosocomial bloodstream infections in ICU patients (13). Although C. albicans remains the predominant cause of candidemia, recent epidemiological data show that C. glabrata has emerged as the second-most-common cause of candidemia in the United States (11). Recently, two new species phenotypically similar to C. glabrata have been described, C. nivariensis (1) and C. bracarensis (5). While C. nivariensis has been described as a new multidrug-resistant pathogen (4), little is known about C. bracarensis. To date, only seven human clinical isolates have been described in the literature. One was isolated from a vaginal exudate in a Portuguese hospital and the other from a blood culture in a United Kingdom hospital (5). Subsequently, three more isolates were identified by retesting 137 C. glabrata clinical isolates using a C. bracarensis-specific probe in a tertiary hospital in the United States (3). Of these, one was from a pelvic abscess of a patient with perforated diverticulitis and two were from the throat and stool of oncology patients without evidence of infection. More recently, 2 more isolates were identified from a large global collection of C. glabrata isolates (1,598 isolates), of which 1 was isolated from sputum culture and the other from a blood culture (7).
Similar to other Candida species, C. bracarensis has been recovered from multiple body sites, especially mucosal surfaces (Table 2), and is clearly associated with infection and colonization (3, 5, 7). Our patient presented with multiple risk factors for developing invasive candidiasis. He was immunocompromised by therapeutic immunosuppression (cyclosporine and methylprednisolone) and by his underlying disease condition (leukemia, bone marrow transplant, and GVHD). GVHD of the skin, liver, and bowel may result in mucosal damage leading to increased fungal translocation of C. bracarensis into the bloodstream (6). Our patient was in the ICU for 20 days prior to the positive culture results. Other relevant risk factors included central venous catheterization and broad-spectrum antibiotic therapy.
C. bracarensis and C. glabrata are morphologically and biochemically indistinguishable except for the color change on CHROMagar and lysine assimilation (Table 1). C. bracarensis and C. nivariensis both produced white colonies on CHROMagar, but C. bracarensis was positive for the rapid trehalose test whereas C. nivariensis was negative (Table 1). Unlike findings in other studies, our two isolates were not initially identified as C. glabrata by the API 20C system, suggesting that C. bracarensis can be either misidentified as C. glabrata or left unidentified by the biochemical analysis. Based on data from ourselves and others (1-3, 5, 7), C. bracarensis can be presumptively identified based on the production of white colonies on CHROMagar agar, microscopic features of small budding yeast cells without hyphae or pseudohyphae, and a positive rapid trehalose test. Identification can be further confirmed by DNA sequence analysis targeting the ITS and/or D1/D2 regions. Therefore, CHROMagar should be included in the workup if small budding yeast cells are seen from a positive blood culture.
The current prevalence of this organism is based on retesting of C. glabrata isolates (3, 7). This approach may possibly underestimate the current prevalence of C. bracarensis, since our two C. bracarensis isolates were initially not identified as C. glabrata. Accurate identification of Candida species can have important implications for the treatment of invasive candidiasis (10). This species is an opportunistic fungal pathogen causing invasive candidiasis in immunocompromised patients. Including the case reported here, three of the patients from whom C. bracarensis was isolated were patients with hematological malignancies (3). Moreover, it will be important to determine the antifungal drug susceptibilities of future isolates of this organism. Two of the eight C. bracarensis isolates tested for antifungal drug susceptibilities were drug resistant, of which one was resistant to amphotericin B and one was resistant to all the azoles (Table 2). The true incidence of this organism in cases of invasive candidiasis has yet to be determined.
Acknowledgments
We thank the staff of the Medical Mycology section at the Public Health Laboratory, Ontario Agency for Health Protection and Promotion, for help with culture identification and collection.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Alcoba-Florez, J., S. Mendez-Alvarez, J. Cano, J. Guarro, E. Perez-Roth, and M. del Pilar Arevalo. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, J. A., N. Chase, R. Lee, C. P. Kurtzman, and W. G. Merz. 2008. Production of white colonies on CHROMagar Candida medium by members of the Candida glabrata clade and other species with overlapping phenotypic traits. J. Clin. Microbiol. 46:3498-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, J. A., N. Chase, S. S. Magill, C. P. Kurtzman, M. J. Fiandaca, and W. G. Merz. 2008. Candida bracarensis detected among isolates of Candida glabrata by peptide nucleic acid fluorescence in situ hybridization: susceptibility data and documentation of presumed infection. J. Clin. Microbiol. 46:443-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., R. Petch, C. J. Linton, M. D. Palmer, P. D. Bridge, and E. M. Johnson. 2008. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J. Clin. Microbiol. 46:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Correia, A., P. Sampaio, S. James, and C. Pais. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 56:313-317. [DOI] [PubMed] [Google Scholar]
- 6.Guiot, H. F., W. E. Fibbe, and J. W. van 't Wout. 1994. Risk factors for fungal infection in patients with malignant hematologic disorders: implications for empirical therapy and prophylaxis. Clin. Infect. Dis. 18:525-532. [DOI] [PubMed] [Google Scholar]
- 7.Lockhart, S. R., S. A. Messer, M. Gherna, J. A. Bishop, W. G. Merz, M. A. Pfaller, and D. J. Diekema. 2009. Identification of Candida nivariensis and Candida bracarensis in a large global collection of Candida glabrata isolates: comparison to the literature. J. Clin. Microbiol. 47:1216-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donnell, K. 1993. Fusarium and its near relatives, p. 225-233. In D. R. Reynolds and J. W. Tayor (ed.), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, United Kingdom.
- 10.Pappas, P. G., C. A. Kauffman, D. Andes, D. K. Benjamin, Jr., T. F. Calandra, J. E. Edwards, Jr., S. G. Filler, J. F. Fisher, B. J. Kullberg, L. Ostrosky-Zeichner, A. C. Reboli, J. H. Rex, T. J. Walsh, and J. D. Sobel. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White, T. J., T. Burns, S. Lee, and J. W. Tayor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY.
- 13.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]