Abstract
Lamivudine is the first nucleoside analogue that was shown to have a potent effect on hepatitis B virus (HBV). However, the emergence of mutants resistant or cross-resistant to nucleoside/nucleotide analogues remains a serious problem. Several assays for the detection and quantification of antiviral-resistant mutants have been reported, but it has been difficult to measure the amounts of mutants accurately, especially when the target strain is a minor component of the mixed population. It has been shown that accurate measurement of a minor strain is difficult as long as a matching reaction with a single probe is included in the assay. We developed a new method for the quantification of lamivudine-resistant strains in a mixed-virus population by real-time PCR using minor groove binder probes and peptide nucleic acids, and we achieved a wide and measurable range, from 3 to 10 log10 copies/ml, and high sensitivity, with a discriminative limit of 0.01% of the predominant strain. The clinical significance of measuring substitutions not only of M204 but also of L180 residues of HBV polymerase was demonstrated by this method. This assay increases the versatility of a sensitive method for the quantification of a single-nucleotide mutation in a heterogeneous population.
Chronic liver diseases due to hepatitis B virus (HBV) infection are still serious problems worldwide, and many patients suffer from liver cirrhosis and hepatocellular carcinoma (2, 6, 21). Nucleoside/nucleotide analogues have been introduced in the treatment of chronic hepatitis B, and inhibition of disease progression has been achieved (20). However, since several analogues have been developed and have come into wide use, the risk of the emergence of resistant or cross-resistant mutations is increasing. This is a serious problem, since it may lead to the virological and biochemical breakthroughs that result in the loss of therapeutic effects (18).
Lamivudine (3TC) is the first nucleoside analogue that was demonstrated to have a potent effect on HBV (8, 17). The results of many studies on lamivudine-resistant mutants have been reported, and the impact of alteration from M204 to I204 or V204 (YMDD mutations) was initially investigated (20). Thereafter, the clinical significance of other regions of HBV polymerase was clarified. We reported the significance of dual mutations of the YMDD motif and the LLAQ motif (specifically the L180 residue) for the severity of breakthrough hepatitis (29). Recently, alteration of the L180 residue has been noted as one of the most crucial changes for the emergence of entecavir-resistant mutants.
It is important to detect mutants as early as possible in order to confirm genotypic or phenotypic resistance that may be followed by virological breakthrough and rebound. Several methods using PCR or hybridization for the qualitative detection of antiviral-resistant mutants have been reported.
Some assays have been proposed as sensitive methods for the quantitative measurement of mutant strains (33), and we have reported a quantification assay using type-specific minor groove binder (MGB) probes (40). However, we found that it was difficult to measure the amounts of mutants accurately, especially when the target strain was a minor component of the mixed population.
In this study, we developed a new sensitive assay for the quantification of hepatitis B virus mutants by introducing peptide nucleic acids (PNAs) into real-time PCR with MGB probes. PNAs are DNA mimics that bind to their complementary nucleic acid sequences with high specificity and have PCR-clamping effects (30, 31). We evaluated the efficacy of this assay and demonstrated the clinical significance of estimating the quantitative follow-up of dual mutants.
MATERIALS AND METHODS
Serum samples.
HBs antigen-positive samples were obtained from chronic hepatitis B patients and were stored at −20°C. DNA was extracted and purified from the serum by a spin column method by using a QIAamp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The study was conducted with the approval of our institute's Ethics Committee, and written informed consent was obtained from all patients.
Construction of MGB probes.
Minor groove binder (MGB) probes for the YMDD motif were designed to hybridize with HBV containing reverse transcriptase 204 (rt204) in domain C of the polymerase region in order to be complementary to the wild-type (rt204M) and mutant (rtM204I and rtM204V) strains. For the YIDD motif, two kinds of MGB probes were prepared because of the different kinds of codons for the same amino acid: YIDD1 for codon ATC and YIDD2 for codon ATT. The sequences of primers and MGB probes used were as follows: forward primer, 5′-GGGCTTTCCCCCACTGTT-3′; reverse primer, 5′-AAAGGGACTCAAGATGTTGTACAGACT-3′; YMDD probe, 5′-CTTTCAGTTATATGGATGATGTG-3′; YIDD1 probe, 5′-CTTTCAGTTATATCGATGATGTGG-3′; YIDD2 probe, 5′-CTTTCAGTTATATTGATGATGTGG-3′; YVDD probe, 5′-CTTTCAGTTATGTGGATGATGT-3′.
MGB probes for the LLAQ motif (rt179 to rt182) in domain B were also designed to be complementary to the wild type (rt180L) and a mutant (rtL180M). The sequences of primers and MGB probes used were as follows: forward primer, 5′-CCTATGGGAGTGGGCCTC-3′; reverse primer, 5′-AACAGTGGGGGAAAGCCCT-3′; LLAQ probe, 5′-CTCCTGGCTCAGTTTA-3′; LMAQ probe, 5′-TTCTCATGGCTCAGTTTACTA-3′. Those probes were purchased from Applied Biosystems Japan, Ltd. (Tokyo, Japan).
Construction of PNAs.
PNAs were designed to match the sequence of each MGB probe exactly and were provided by Greiner Bio-One (Tokyo, Japan). The sequences of the PNAs were as follows: PNA-YMDD, 5′-GTTATATGGATGATGTG-3′; PNA-YIDD1, 5′-GTTATATCGATGATGTGG-3′; PNA-YIDD2, 5′-GTTATATTGATGATGTGG-3′; PNA-YVDD, 5′-AGTTATGTGGATGATGT-3′; PNA-LLAQ, 5′-TCCGTTTCTCCTGGCTC-3′; PNA-LLAQ, 5′-CCGTTTCTCATGGCTCA-3′.
Construction of HBV plasmids for the standard.
HBV plasmids were prepared for the construction of a standard for quantification. PCR products that contained both YMDD and LLAQ motifs obtained from HBV-positive patients were cloned, and Escherichia coli XL-1 competent cells were transformed by each of the mutant-specific DNA fragments. Finally, plasmid DNA corresponding to each MGB probe was constructed.
Real-time PCR.
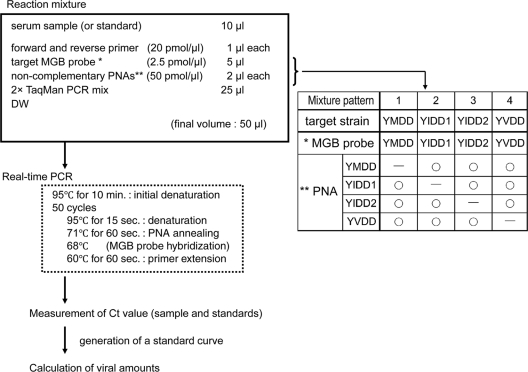
Real-time PCR with the MGB probe and PNAs was performed using an ABI real-time PCR system, model 7300 (Applied Biosystems, Foster City, CA). A sample was measured with an MGB probe that is complementary to a target strain and with PNAs that are complementary to each strain except for the target strain (Fig. 1).
FIG. 1.
Measurement of viral amounts by real-time PCR with the MGB probe and PNAs. The table shows patterns of combination of the MGB probe and noncomplementary PNAs.
The reaction was carried out in a final volume of 50 μl in each well of a plate containing 10 μl of a serum sample, 400 nM forward and reverse primers, 250 nM MGB probe, 2 μM PNAs, and 1× TaqMan Universal PCR master mix (Applied Biosystems, Foster City, CA). Optimal concentrations of MGB probes and PNAs were determined by our own examinations as described below.
The conditions of the PCR were as follows: 10 min at 95°C for the initial denaturation, followed by 50 cycles at 95°C for 15 s for denaturation, 71°C for 1 min for PNA annealing, and 60°C for 1 min for primer extension.
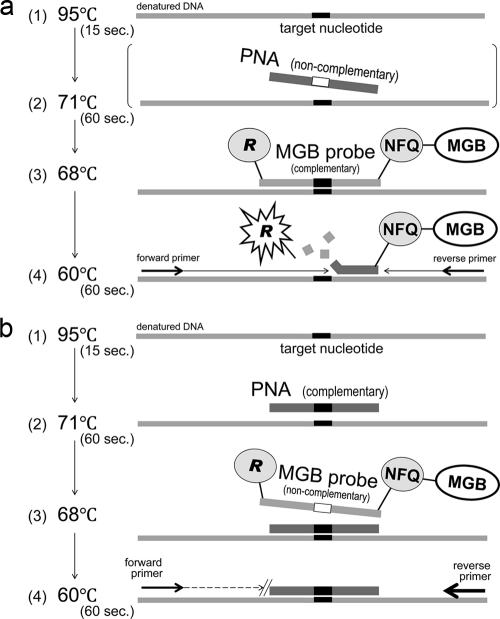
A TaqMan probe has a reporter dye and a nonfluorescent quencher at the 5′ and 3′ ends, respectively. The reporter dye emission is quenched when the probe remains intact. When an MGB probe hybridized with a complementary strain, the fluorescence of a reporter dye became detectable (Fig. 2a).
FIG. 2.
(a) PCR with a complementary MGB probe. During the extension phase with primers, the DNA polymerase cleaves the reporter dye from the probe, and the dye emits its characteristic fluorescence. When a PNA added to the reaction mixture is noncomplementary to the target sequence, it does not affect amplification with a complementary MGB probe. NFQ, nonfluorescent quencher. (b) PCR with a complementary PNA. The mismatch of an MGB probe noncomplementary to the target sequence is blocked by the PNA, which is preferentially bound at a higher melting temperature than the MGB probe.
When PCR was performed with a complementary PNA and an MGB probe noncomplementary to the target nucleotide, the extension and amplification were blocked by the preferentially bound PNA (Fig. 2b).
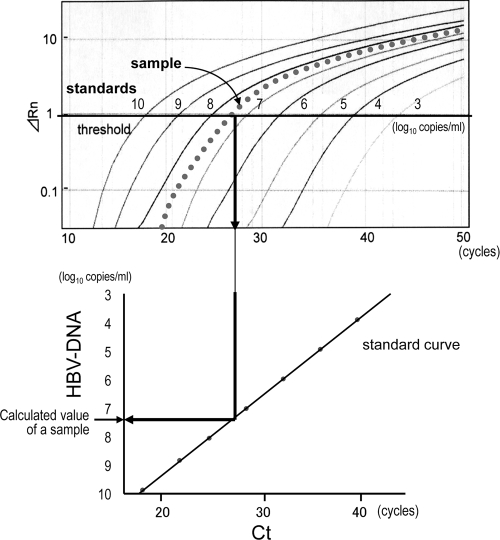
Real-time PCR was performed separately with each MGB probe and with noncomplementary PNAs for all sets of target strains. After the exclusion of background signals by subtracting the signals of early cycles of PCR, a threshold was set in the linearly increasing region of accruals of fluorescence signals. Then the cycle at which the signal reached the threshold (threshold cycle [CT]) was measured. The amount of the target strain was calculated from the CT number and a linear regression curve for standards of each strain (Fig. 3).
FIG. 3.
Calculation of values from the results obtained by real-time PCR. The threshold was set in the linearly increasing region of accruals of fluorescence signals (ΔRn), and the threshold cycle (CT) was measured. The value of the target strain was calculated from the CT number and a standard curve for each strain. The measurement was performed separately with each MGB probe and noncomplementary PNAs for all sets of target strains. Solid lines, 10-fold serially diluted standards; dotted line, a sample of the target strain.
RESULTS
Standard curve for the measurement of a single strand using an MGB probe.
HBV plasmids that had been quantitatively adjusted beforehand were used for 10-fold serial dilutions from 10 log10 copies/ml to 1 log10 copies/ml; from these, the standard curves were generated. A strong inverse correlation between the logarithmic concentration of samples and CT values was obtained, and linearity was confirmed within a wide range from 3 to 10 log10 copies/ml.
Measurement of a target mutant in the mixed strains.
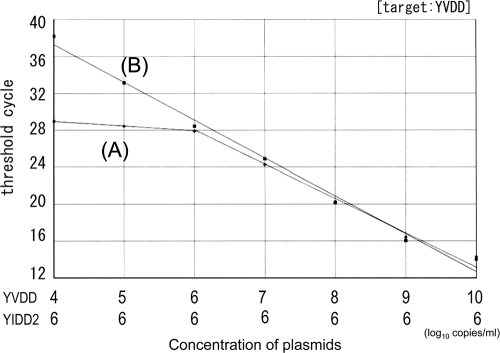
The accuracy of quantification of a single HBV strain was determined in a mixture of multiple strains by real-time PCR with a type-specific MGB probe without PNA. HBV plasmids serially diluted 10-fold within a range of 10 log10 copies/ml to 4 log10 copies/ml were used for quantification under the condition of coexistence of 6 log10 copies/ml of another strain with the same position. The target strain was correctly measured when the concentration of the target strain predominated over that of another strain, and the measurement error was estimated to be less than twice the amount theoretically calculated. However, measurements were overestimated when the concentration of the target strain was lower than that of another strain (Fig. 4).
FIG. 4.
Measurement of a target strain in mixed strains. Samples of a 10-fold serial dilution of the YVDD strain were measured under the condition of coexistence of 6 log10 copies/ml of the YIDD2 strain. Line A, MGB for YVDD; line B, MGB for YVDD with PNAs for YMDD, YIDD1, and YIDD2. Linearity was maintained even at the lower concentration range.
Clamping effects of PNA on PCR.
PNAs were introduced into the assay system to reduce quantitative errors due to mismatching of MGB probes. The clamping effects of PNAs were investigated beforehand in order to determine the concentration at which the noncomplementary PNAs had a negative impact.
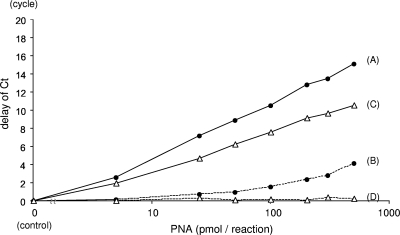
Six log10 copies/ml of HBV plasmids were quantified together with an MGB probe and a PNA that were complementary to the target strain. Delays in the CT with different concentrations of PNA were measured (Fig. 5). The delay correlated with the PNA concentration, and a clamping effect of 1/100 to 1/1,000 was observed when 100 pmol of PNA was used. When real-time PCR was performed with a complementary MGB probe and with all sets of the PNAs that are complementary to the motifs in each strain except for the target strain, a delay in the CT was observed when noncomplementary PNA was present at high concentrations. However, the delays were modest when the amount of PNA was less than 100 pmol. The concentration of PNA for the assay was determined from this result.
FIG. 5.
Clamping effects of PNA on PCR. Delays in the CT with different concentrations of complementary (solid lines) and noncomplementary (dashed lines) PNAs were measured with the MGB probe for the LLAQ strain (filled circles) and the LMAQ strain (open triangles). PNAs used were complementary (A) or noncomplementary (B) to the LLAQ strain and complementary (C) or noncomplementary (D) to the LMAQ strain. A delay in the CT was observed even with noncomplementary PNAs when the concentration was high. The differences in delays between PNAs complementary and noncomplementary to the LLAQ strain are similar to those between PNAs complementary and noncomplementary to the LMAQ strain.
Measurement using both MGB probes and PNA.
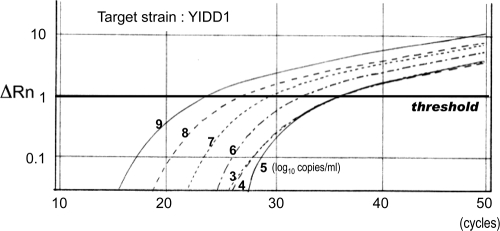
For confirmation of the proper method of determination by this assay, the amount of the HBV strain was measured in a mixture of two different strains of the same position using a new method of real-time PCR with the MGB probe and PNA. At first, 10-fold-diluted samples of target HBV plasmids from 9 log10 copies/ml to 3 log10 copies/ml were measured under the condition of coexistence of 3 log10 copies/ml of another strain. There were no significant differences in the values obtained for the target strain regardless of the presence of the other strain. Next, the amount of a minority strain was determined in the presence of a predominant strain. Tenfold serial dilutions of target HBV plasmids ranging from 9 log10 copies/ml to 3 log10 copies/ml were measured under the condition of coexistence of 9 log10 copies/ml of a different strain. A linear correlation between the CT values and sample concentrations was observed in the range from 9 log10 copies/ml to 5 log10 copies/ml. However, a target strain at a concentration less than 5 log10 copies/ml could not be quantified accurately. Therefore, the detection limit of a nondominant strain was determined to be 0.01% of the predominant strain (Fig. 6).
FIG. 6.
Real-time PCR with the MGB probe and PNAs. Samples of a 10-fold serial dilution of the YIDD1 strain were measured under the condition of coexistence of 9 log10 copies/ml of YMDD strain with an MGB probe complementary to YIDD1 and PNAs complementary to an YMDD motif other than YIDD1 (YMDD, YIDD2, and YVDD). ΔRn, accruals of a fluorescence signal.
A clinical case.
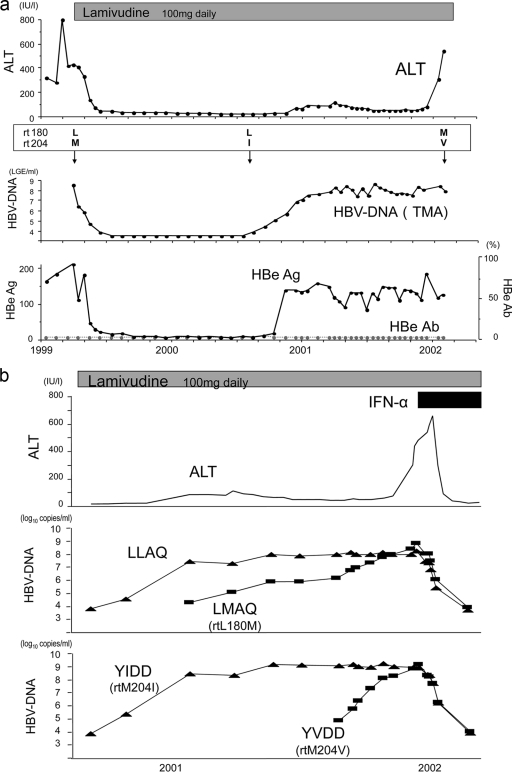
The clinical and virological course of a case of breakthrough hepatitis after the emergence of lamivudine-resistant mutants is described below (Fig. 7a).
FIG. 7.
(a) Course of a clinical case. HBV DNA was measured by a transcription-mediated amplification assay (logarithmic genome equivalent [LGE] per milliliter). L, M, I, and V of rt180 or rt204 represent the amino acids leucine, methionine, isoleucine, and valine, respectively. (b) Changes in viral amounts of the LLAQ and YMDD motifs. ALT levels were stable while the LLAQ/YIDD strain was dominant. When the LMAQ/YVDD strain became dominant over preexisting strains, breakthrough hepatitis occurred.
A 42-year-old male was administered 100 mg of lamivudine once a day (q.d.) in 1999. At first, HBV DNA levels were steadily reduced, and the serum alanine aminotransferase (ALT) level also decreased to the normal range after the start of treatment. About 16 months later, viral breakthrough and mild elevation of the ALT level were observed. Despite the fact that HBV DNA and HBe antigen returned to relatively high levels, the ALT level remained almost within the normal range. Thereafter, the ALT level suddenly increased, despite the fact that HBV DNA remained at the same level. When we measured mutants with resistance to lamivudine by a direct sequencing method, the changes in ALT and HBV DNA levels were thought to be related to the changes in mutant patterns over time. However, we could not predict the time or the severity of breakthrough hepatitis by this limited information, especially when HBV DNA loads did not change.
Precise examination of changes in HBV mutants was performed by real-time PCR with the MGB probe and PNA (Fig. 7b).
ALT levels were stable, whereas total levels of HBV DNA remained high. An LLAQ YIDD strain was dominant in the earlier phase. Next, an LLAQ strain became measurable, and when a YVDD strain emerged and overcame the YIDD strain levels, breakthrough hepatitis occurred. This case is a good example that demonstrates the clinical significance of measuring the amounts of HBV mutants with not only the YMDD motif but also the LLAQ motif by this new assay.
DISCUSSION
Inhibition of disease progression for patients with chronic HBV infection has been achieved by treatment with nucleoside/nucleotide analogues (20). Treatment with these analogues has reduced not only the incidence of hepatic decompensation but also the risk of occurrence of hepatocellular carcinoma (27). The same effect on recurrence has also been reported (4). However, the most serious issue is the emergence of viruses with resistance to nucleoside/nucleotide analogues, because this leads to virological and biochemical breakthroughs. Therapeutic effects were lost for patients who developed the resistant mutations (18).
From a resistance or a cross-resistance perspective, nucleoside/nucleotide analogues are classified into three groups based on their structural characteristics (35, 42): l-nucleoside analogues (lamivudine, telbivudine, clevudine, and emtricitabine), alkyl phosphonates (adefovir and tenofovir), and the d-cyclopentane group (entecavir). Resistance rates after 5 years of treatment have been reported to be 80% for lamivudine, 29% for adefovir, and 1.2% for entecavir. Although the rate of early-period resistance has been reduced as new drugs have been introduced, the long-term results have not yet been confirmed. Moreover, the resistance rate becomes higher for patients with a lamivudine-resistant mutant, as high as 20% within 1 year for adefovir and 51% after 5 years for entecavir.
It is important to detect and quantify these mutant strains, and various methods have been developed for such measurements. The most common method for the detection of mutant viruses is direct sequencing after PCR amplification. However, the detection limit of a minor strain in the heterogeneous virus population is about 20%. Recently, a new method called ultradeep pyrosequencing has been developed (26, 36). This sequencing relies on the detection of DNA polymerase activity by measuring the pyrophosphate (PPi) released by the addition of a deoxyribonucleoside monophosphate (dNMP) to the 3′ end of a primer. The detection limit decreased to 1% by this technique.
There are several methods for hybridization-based genotyping: the line probe assay (LiPA) (12, 22, 37), mixed hybridization-sequencing-PCR (minisequencing) (15), and the oligonucleotide tip assay (13). The sensitivity of the LiPA is 5 to 10%, and that of the minisequencing assay is 2 to 10%. The sensitivity of the high-density DNA chip reported by Tran et al. was 30 to 50%, but it was able to detect 245 mutations, 20 deletions, and 2 insertions at 151 positions (39). Hong et al. reported that mass spectrometry is another method for the detection of mutants. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) had a detection limit of 100 copies/ml and a sensitivity of 1% (10).
Some modified real-time PCR methods have been developed for the quantification of mutant viruses: PCR with molecular beacons (32), PCR with a minor groove binder (MGB), PCR with a locked nucleic acid (LNA)-mediated probe (3, 19, 38), and amplification refractory mutation system PCR (ARMS-PCR) (33).
An MGB can fit snugly into the minor groove, which is the deep narrow space between the two phosphate-sugar backbones of a double-stranded DNA helix.
Afonina et al. reported that a DNA probe with a tripeptide {1,2-dihydro-(3H)-pyrrolo[3,2-e]indole-7-carboxylate} (GDPI3), which is conjugated to the 5′ ends of short oligodeoxynucleotides (ODNs), formed unusually stable hybrids with cDNA and can be used as a PCR primer (1). This MGB probe has a higher melting temperature (Tm) and greater specificity than ordinary DNA probes, especially when a mismatch is in the MGB region of the duplex (7, 16). Zhao et al. reported a real-time PCR method using a TaqMan-MGB probe for the measurement of total amounts of HBV DNA (41), and we have reported the quantification of lamivudine-resistant mutants by the type-specific TaqMan MGB probe assay (40).
Several authors have reported a similar sensitivity of 10% for the detection of minor variants (9, 11, 25). Intraexperimental variability was reported to be 4.9% by Lole and Arankalle (24) and 1.0 to 2.2%, depending on the type of mutant, by our own evaluation (40).
However, our study revealed that the accuracy of quantification was reduced in the case of measurements of minor strains in a mixed population. Therefore, we introduced PNAs to be used in combination with the MGB probes.
PNAs are DNA mimics in which the deoxyribose phosphate backbone of DNA has been replaced by N-(2-aminoethyl)glycine linkages (30). They recognize and bind to their complementary nucleic acid sequences with high thermal stability and specificity (31). PNAs cannot function as primers for DNA polymerase and can be used to block a PCR amplification process in a sequence-specific manner. This PCR clamping allows for direct analysis of single-base mutations by PCR.
Kirishima et al. (14) reported a sensitive method for the detection of a lamivudine-resistant mutant by PNA-mediated PCR clamping with restriction fragment length polymorphism (RFLP) analysis, and Mori et al. performed semiquantitative measurements of mutants using PNA (28).
Under the real-time PCR conditions used in our assay, one reaction cycle started at 95°C for denaturation and finished at 60°C for extension. The Tm of PNA was designed to be 3°C higher than that of an MGB probe. Because of this difference in temperature, PNA is expected to clamp a noncomplementary strain first, and then an MGB probe will hybridize to the target strand. As a result, the assay was able to achieve a significant reduction in mismatches. However, an amplification delay was observed with an excessive amount of noncomplementary PNA. No matter how well an assay is constructed, mismatches cannot be completely avoided as long as the reaction includes a matching process. However, it is thought that the actual measurement can be performed correctly by setting the appropriate concentration of the reaction mixture. As for the limit of detection, we showed that a level of the minority strain equivalent to 0.01% of the predominant strain could be measured by our assay. However, more-robust analysis is required to determine the true analytical sensitivity of the assay by measuring a number of clinical samples.
This requires a precise knowledge of the sequence of the mutants prior to determination by this assay; as such, this assay cannot be construed as a general method for the study of viral quasispecies where mutants and emerging mutants will have an unknown sequence. However, it seems ideally suited for the approach of measuring a known mutant virus, such as HBV mutants resistant to nucleoside/nucleotide analogues.
Although the risk of breakthrough hepatitis is thought to correlate with the duration of infection with mutant viruses (23), not only the duration but also the change in the mutant strains is important for the occurrence of hepatitis. We reported that the patterns of the YMDD and LLAQ motifs often changed from YMDD and LLAQ to YIDD and LLAQ, YIDD and LMAQ, and YVDD and LMAQ, in that order, and that the degree of liver damage increased as the mutations accumulated (29). As demonstrated by the case presented in this report, breakthrough hepatitis seems to have a relationship with the specific types of mutants and the length of time from the emergence of a new mutant. The degree of increase in the viral load after the emergence of dual mutations was smaller than that after the emergence of the first mutation, despite the fact that the level of liver dysfunction was higher with the dual mutations. This result suggested that breakthrough hepatitis correlated closely with changes in viral characteristics. Das et al. (5) reported differences in inhibitory effects on HBV DNA polymerase between different lamivudine-resistant mutants. The inhibitory effects were measured by recombinant HBV DNA polymerase expressed in a baculovirus transfer vector. The fold changes in inhibition of the DNA polymerase were 8.0 for the M204I mutation, 15.2 for the L180M and M204I mutations, and 25.2 for the L180M and M204V mutations, compared to the wild type. The results for our clinical case showed a tendency similar to that reported in the evaluations of Das et al. and similar to the results of a recent molecular modeling study. Sharon and Chu demonstrated that binding energy for mutant HBV polymerases was significantly different between dCTP and 3TC and that 3TC lost binding affinity with the mutants in comparison to the natural substrate dCTP (34). L180 residues did not interact directly with 3TC, but the M180 mutant caused loosening of the hydrophobic pocket formed by the residues A87, F88, P177, L180, and M204. Consequently, dual mutations (L180M plus M204I or L180M plus M204V) were more likely to show a steric clash between I204 or V204 and 3TC.
Despite the high risk of multidrug resistance evoked by lamivudine, it has been widely used, mainly due to its low cost. Substitution of any of the residues T184, S202, and M250, in addition to the M204V and L180M mutations, is necessary for the emergence of an entecavir-resistant mutant. Therefore, it is important to observe the change in the L180 substitution prior to M204V emergence, even for cases where a drug other than lamivudine has been administered.
In conclusion, we have developed a new method for the accurate quantification of lamivudine-resistant strains in a mixed virus population. This is the first report of the combined use of an MGB probe and PNAs for the quantification of viruses using real-time PCR. This methodology should prove immensely useful for better understanding of the dynamics of the emergence of HBV-resistant mutants. Moreover, this assay has the versatility to be applied as a sensitive method for the quantification of a single-nucleotide mutation in a heterogeneous population not only of viruses but also of other nucleic acids.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (20590753).
Footnotes
Published ahead of print on 6 October 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Afonina, I., M. Zivarts, I. Kutyavin, E. Lukhtanov, H. Gamper, and R. B. Meyer. 1997. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 25:2657-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, C. J., H. I. Yang, J. Su, C. L. Jen, S. L. You, S. N. Lu, G. T. Huang, U. H. Iloeje, and R.-H. S. Grp. 2006. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295:65-73. [DOI] [PubMed] [Google Scholar]
- 3.Chieochansin, T., S. Chutinimitkul, S. Payungporn, A. Theamboonlers, P. Tangkijvanich, P. Komolmit, and Y. Poovorawan. 2006. Rapid detection of lamivudine-resistant hepatitis B virus mutations by PCR-based methods. Tohoku J. Exp. Med. 210:67-78. [DOI] [PubMed] [Google Scholar]
- 4.Chuma, M., S. Hige, T. Kamiyama, T. Meguro, A. Nagasaka, K. Nakanishi, Y. Yamamoto, M. Nakanishi, T. Kohara, T. Sho, K. Yamamoto, H. Horimoto, T. Kobayashi, H. Yokoo, M. Matsushita, S. Todo, and M. Asaka. 2009. The influence of hepatitis B DNA level and antiviral therapy on recurrence after initial curative treatment in patients with hepatocellular carcinoma. J. Gastroenterol. 44:991-999. [DOI] [PubMed] [Google Scholar]
- 5.Das, K., X. F. Xiong, H. L. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Franchis, R., G. Meucci, M. Vecchi, M. Tatarella, M. Colombo, E. Delninno, M. G. Rumi, M. F. Donato, and G. Ronchi. 1993. The natural history of asymptomatic hepatitis B surface antigen carriers. Ann. Intern. Med. 118:191-194. [DOI] [PubMed] [Google Scholar]
- 7.de Kok, J. B., E. T. G. Wiegerinck, B. A. J. Giesendorf, and D. W. Swinkels. 2002. Rapid genotyping of single nucleotide polymorphisms using novel minor groove binding DNA oligonucleotides (MGB probes). Hum. Mutat. 19:554-559. [DOI] [PubMed] [Google Scholar]
- 8.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perrillo, H. W. L. Hann, Z. Goodman, L. Crowther, L. D. Condreay, M. Woessner, M. Rubin, and N. A. Brown for the U.S. Lamivudine Investigator Group. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 9.Geng, H. F., B. Hua, H. Wang, Y. H. Cao, Y. Sun, and A. R. Yu. 2006. Dual-probe assay for detection of lamivudine-resistant hepatitis B virus by real-time PCR. J. Virol. Methods 132:25-31. [DOI] [PubMed] [Google Scholar]
- 10.Hong, S. P., N. K. Kim, S. G. Hwang, H. J. Chung, S. Kim, J. H. Han, H. T. Kim, K. S. Rim, M. S. Kang, W. Yoo, and S. O. Kim. 2004. Detection of hepatitis B virus YMDD variants using mass spectrometric analysis of oligonucleotide fragments. J. Hepatol. 40:837-844. [DOI] [PubMed] [Google Scholar]
- 11.Hua, R., Y. Tanaka, K. Fukai, M. Tada, M. Seto, Y. Asaoka, M. Ohta, T. Goto, F. Kanai, N. Kato, H. Yoshida, T. Kawabe, O. Yokosuka, and M. Omata. 2008. Rapid detection of the hepatitis B virus YMDD mutant using TaqMan-minor groove binder probes. Clin. Chim. Acta 395:151-154. [DOI] [PubMed] [Google Scholar]
- 12.Hussain, M., S. Fung, E. Libbrecht, E. Sablon, C. Cursaro, P. Andreone, and A. S. F. Lok. 2006. Sensitive line probe assay that simultaneously detects mutations conveying resistance to lamivudine and adefovir. J. Clin. Microbiol. 44:1094-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang, H., M. Cho, J. Heo, H. Kim, H. Jun, W. Shin, B. Cho, H. Park, and C. Kim. 2004. Oligonucleotide chip for detection of lamivudine-resistant hepatitis B virus. J. Clin. Microbiol. 42:4181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirishima, T., T. Okanoue, Y. Daimon, Y. Itoh, H. Nakamura, A. Morita, T. Toyama, and M. Minami. 2002. Detection of YMDD mutant using a novel sensitive method in chronic liver disease type B patients before and during lamivudine treatment. J. Hepatol. 37:259-265. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, S., K. Shimada, H. Suzuki, K. Tanikawa, and M. Sata. 2000. Development of a new method for detecting a mutation in the gene encoding hepatitis B virus reverse transcriptase active site (YMDD motif). Hepatol. Res. 17:31-42. [Google Scholar]
- 16.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, C. L., R. N. Chien, N. W. Y. Leung, T. T. Chang, R. Guan, D. I. Tai, K. Y. Ng, P. C. Wu, J. C. Dent, J. Barber, S. L. Stephenson, D. F. Gray, and the Asia Hepatitis Lamivudine Study Group. 1998. A one-year trial of lamivudine for chronic hepatitis B. N. Engl. J. Med. 339:61-68. [DOI] [PubMed] [Google Scholar]
- 18.Lai, C. L., J. Dienstag, E. Schiff, N. W. Y. Leung, M. Atkins, C. Hunt, N. Brown, M. Woessner, R. Boehme, and L. Condreay. 2003. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin. Infect. Dis. 36:687-696. [DOI] [PubMed] [Google Scholar]
- 19.Latorra, D., K. Campbell, A. Wolter, and J. M. Hurley. 2003. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum. Mutat. 22:79-85. [DOI] [PubMed] [Google Scholar]
- 20.Liaw, Y. F., J. J. Y. Sung, W. C. Chow, G. Farrell, C. Z. Lee, H. Yuen, T. Tanwandee, Q. M. Tao, K. Shue, O. N. Keene, J. S. Dixon, D. F. Gray, and J. Sabbat, for the Cirrhosis Asian Lamivudine Multicentre Study Group. 2004. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 351:1521-1531. [DOI] [PubMed] [Google Scholar]
- 21.Liaw, Y. F., D. I. Tai, C. M. Chu, and T. J. Chen. 1988. The development of cirrhosis patients with chronic type B hepatitis—a prospective study. Hepatology 8:493-496. [DOI] [PubMed] [Google Scholar]
- 22.Libbrecht, E., J. Doutreloigne, H. Van De Velde, M. F. Yuen, C. L. Lai, F. Shapiro, and E. Sablon. 2007. Evolution of primary and compensatory lamivudine resistance mutations in chronic hepatitis B virus-infected patients during long-term lamivudine treatment, assessed by a line probe assay. J. Clin. Microbiol. 45:3935-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lok, A. S. F., C. L. Lai, N. Leung, G. B. Yao, Z. Y. Cui, E. R. Schiff, J. L. Dienstag, E. J. Heathcote, N. R. Little, D. A. Griffiths, S. D. Gardner, and M. Castiglia. 2003. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology 125:1714-1722. [DOI] [PubMed] [Google Scholar]
- 24.Lole, K. S., and V. A. Arankalle. 2006. Quantitation of hepatitis B virus DNA by real-time PCR using internal amplification control and dual TaqMan MGB probes. J. Virol. Methods 135:83-90. [DOI] [PubMed] [Google Scholar]
- 25.Malmstrom, S., C. Hannoun, and M. Lindh. 2007. Mutation analysis of lamivudine resistant hepatitis B virus strains by TaqMan PCR. J. Virol. Methods 143:147-152. [DOI] [PubMed] [Google Scholar]
- 26.Margeridon-Thermet, S., N. S. Shulman, A. Ahmed, R. Shahriar, T. Liu, C. L. Wang, S. P. Holmes, F. Babrzadeh, B. Gharizadeh, B. Hanczaruk, B. B. Simen, M. Egholm, and R. W. Shafer. 2009. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J. Infect. Dis. 199:1275-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto, A., E. Tanaka, A. Rokuhara, K. Kiyosawa, H. Kumada, M. Omata, K. Okita, N. Hayashi, T. Okanoue, S. Iino, K. Tanikawa, and the Inuyama Hepatitis Study Group. 2005. Efficacy of lamivudine for preventing hepatocellular carcinoma in chronic hepatitis B: a multicenter retrospective study of 2795 patients. Hepatol. Res. 32:173-184. [DOI] [PubMed] [Google Scholar]
- 28.Mori, K., M. Minami, T. Kirishima, K. Kunimoto, M. Okita, M. Nakayama, A. Makiyama, J. Yamaoka, T. Nakajima, K. Yasui, Y. Itoh, and T. Okanoue. 2006. Prediction of breakthrough hepatitis due to lamivudine-resistant hepatitis B virus by a sensitive semiquantitative assay using peptide nucleic acids. Intervirology. 49:274-280. [DOI] [PubMed] [Google Scholar]
- 29.Natsuizaka, M., S. Hige, Y. Ono, K. Ogawa, M. Nakanishi, M. Chuma, S. Yoshida, and M. Asaka. 2005. Long-term follow-up of chronic hepatitis B after the emergence of mutations in the hepatitis B virus polymerase region. J. Viral Hepat. 12:154-159. [DOI] [PubMed] [Google Scholar]
- 30.Ohishi, W., H. Shirakawa, Y. Kawakami, S. Kimura, M. Kamiyasu, S. Tazuma, T. Nakanishi, and K. Chayama. 2004. Identification of rare polymerase variants of hepatitis B virus using a two-stage PCR with peptide nucleic acid clamping. J. Med. Virol. 72:558-565. [DOI] [PubMed] [Google Scholar]
- 31.Ørum, H., P. E. Nielsen, M. Egholm, R. H. Berg, O. Buchardt, and C. Stanley. 1993. Single-base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 21:5332-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pas, S. D., S. Noppornpanth, A. A. van der Eijk, R. A. de Man, and H. G. M. Niesters. 2005. Quantification of the newly detected lamivudine resistant YSDD variants of hepatitis B virus using molecular beacons. J. Clin. Virol. 32:166-172. [DOI] [PubMed] [Google Scholar]
- 33.Punia, P., P. Cane, C. G. Te, and N. Saunders. 2004. Quantitation of hepatitis B lamivudine resistant mutants by real-time amplification refractory mutation system PCR. J. Hepatol. 40:986-992. [DOI] [PubMed] [Google Scholar]
- 34.Sharon, A., and C. K. Chu. 2008. Understanding the molecular basis of HBV drug resistance by molecular modeling. Antivir. Res. 80:339-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, T., A. Bartholomeusz, and S. Locarnini. 2006. HBV drug resistance: mechanisms, detection and interpretation. J. Hepatol. 44:593-606. [DOI] [PubMed] [Google Scholar]
- 36.Solmone, M., D. Vincenti, M. C. F. Prosperi, A. Bruselles, G. Ippolito, and M. R. Capobianchi. 2009. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naïve patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J. Virol. 83:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuyver, L., C. Van Geyt, S. De Gendt, G. Van Reybroeck, F. Zoulim, G. Leroux-Roels, and R. Rossau. 2000. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J. Clin. Microbiol. 38:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, Z., L. F. Zhou, H. Y. Zeng, Z. Chen, and H. H. Zhu. 2007. Multiplex locked nucleic acid probes for analysis of hepatitis B virus mutants using real-time PCR. Genomics 89:151-159. [DOI] [PubMed] [Google Scholar]
- 39.Tran, N., R. Berne, R. Chann, M. Gauthier, D. Martin, M. A. Armand, A. Ollivet, C. G. Teo, S. Ijaz, D. Flichman, M. Brunetto, K. P. Bielawski, C. Pichoud, F. Zoulim, and G. Vernet. 2006. European multicenter evaluation of high-density DNA probe arrays for detection of hepatitis B virus resistance mutations and identification of genotypes. J. Clin. Microbiol. 44:2792-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, S., S. Hige, M. Yoshida, N. Yamashita, S. I. Fujisawa, K. Sato, T. Kitamura, M. Nishimura, M. Chuma, M. Asaka, and H. Chiba. 2008. Quantification of lamivudine-resistant hepatitis B virus mutants by type-specific TaqMan minor groove binder probe assay in patients with chronic hepatitis B. Ann. Clin. Biochem. 45:59-64. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, J. R., Y. J. Bai, Q. H. Zhang, Y. Wan, D. Li, and X. J. Yan. 2005. Detection of hepatitis B virus DNA by real-time PCR using TaqMan-MGB probe technology. World J. Gastroenterol. 11:508-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoulim, F., and S. Locarnini. 2009. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology 137:1593-1608. [DOI] [PubMed] [Google Scholar]