Abstract
Reference isolates of Mycobacterium neoaurum, Mycobacterium aurum, and the nonvalidated species “Mycobacterium lacticola” were the focus of two recent molecular taxonomic studies. On the basis of this grouping, we identified 46 clinical pigmented, rapidly growing mycobacterial isolates. By 16S rRNA gene sequencing, only two major taxa were identified: M. neoaurum and a previously uncharacterized “M. neoaurum-like” group. The M. neoaurum-like group exhibited only 99.7% identity to M. neoaurum by 16S rRNA gene sequencing and 96.5% identity to M. neoaurum by rpoB sequencing and was named M. bacteremicum. No clinical isolates of M. aurum or M. lacticola were identified. Of isolates with known sources, 4/8 (50%) of M. bacteremicum isolates and 22/34 (65%) of M. neoaurum isolates were recovered from blood, and 35% of these were known to be from patients with catheter-related sepsis. MIC and clinical data on these 46 isolates of M. neoaurum and M. bacteremicum along with a review of 16 previously reported cases of infection with the M. neoaurum-M. lacticola group demonstrated that the isolates were highly susceptible to all drugs tested except clarithromycin, and most clinical cases were successfully treated. The clarithromycin resistance suggested the presence of an inducible erm gene reported in other species of rapidly growing mycobacteria. Sequencing studies are currently required to identify these two species. Strain ATCC 25791 (originally submitted as an example of Mycobacterium aurum) is proposed to be the type strain of M. bacteremicum.
Recently, we reported on the phylogenetic analysis of the Mycobacterium neoaurum-“M. lacticola” group and redescription of reference American Type Culture Collection (ATCC) isolates previously classified as Mycobacterium aurum (18). These pigmented, rapidly growing Mycobacterium spp. have been associated with clinical disease, especially catheter-related sepsis. In a previous study by Simmon and colleagues (18), complete 16S rRNA gene sequence analysis showed four reference strains of M. aurum to have 100% identity to the type strain of the nonvalidated species M. lacticola, which differed by 8 bp from the type strain of M. neoaurum. With the addition of hsp65 and rpoB gene targets, these four isolates remained closely clustered as M. lacticola, with intraspecies variabilities being only 0.7% for hsp65 and 1.5% for rpoB. One reference strain submitted as M. aurum (ATCC 25791) did not match either of these two taxa. All of these reference strains were from environmental sources (18).
Phylogenetic studies need to be performed with clinical isolates to determine their relatedness to these reference strains of M. neoaurum, M. lacticola, and ATCC 25791, which we tentatively named “M. neoaurum-like” (11, 18). We initiated a detailed study for 32 of the previously reported clinical isolates of M. neoaurum-M. lacticola group from the laboratory at the Associated Regional and University Pathologists (ARUP) Institute for Clinical and Experimental Pathology and the University of Texas Health Science Center at Tyler (UTHSCT), along with 14 additional isolates of the M. neoaurum-M. neoaurum-like group which were recovered following the submission of the first study.
As noted in the previous publication, there is a paucity of case reports of M. neoaurum and M. lacticola infections in the literature. The incidence of M. neoaurum-M. lacticola group infections likely has been underestimated, since most clinical laboratories have not been able to identify pigmented, rapidly growing mycobacteria of this species. Biochemical and cell wall analysis and high-performance liquid chromatography (HPLC) are not validated methods for such identification. Hence, we reviewed published cases of this pigmented group to determine how well they met current molecular taxonomic criteria and how they compared to the well-studied isolates in the present study.
MATERIALS AND METHODS
Organisms.
Clinical isolates of pigmented, rapidly growing mycobacteria identified to be M. neoaurum, M. neoaurum-like, or M. lacticola by hsp65 PCR restriction enzyme analysis (PRA) (at UTHSCT) or 16S partial gene sequencing (500 bp) (at ARUP) were the focus of this study (7, 11, 18, 19, 21). Isolates had been stored at −70°C after initial laboratory testing. They were later subcultured and underwent complete 16S rRNA gene sequencing as previously described (18). Isolates whose complete 16S rRNA gene sequence was a 100% match to the complete 16S rRNA gene sequence of the type strain of M. neoaurum (ATCC 25795) or M. lacticola (ATCC 9626) or that exhibited >99.5% identity (difference of 6 bp or less) to the complete 16S rRNA gene sequence of the M. neoaurum type strain (designated M. neoaurum-like) were included in the study.
Forty-six clinical isolates (21 from ARUP, 25 from UTHSCT) were studied. These included 32 isolates from the study of Simmon et al. (18) and 14 additional isolates recovered after the submission of the manuscript for the first study.
Patient demographics.
Patient information, including geographical source, clinical site, age, sex, and associated or underlying disease or risk factor, were compiled for all isolates at submission (Table 1). Both of these studies were conducted under protocols approved by the institutional review boards (IRBs) at both UTHSCT and ARUP (18).
TABLE 1.
Demographic and phylogenetic information for isolates of Mycobacterium neoaurum and the M. neoaurum-like group from the current studya
| Organism and laboratory strain no. | Ageb | Sex | Source | Geog loc. | Underlying disease | Phylogenetic clusterc |
||
|---|---|---|---|---|---|---|---|---|
| 16S rRNA | hsp65 | rpoB | ||||||
| M. neoaurum-like group | ||||||||
| MO-2218 | 2 | M | Central catheter | OH | Bone marrow transplant | C | H | S |
| MO-1171 | 31 | F | Blood | TX | Unknown | C | E | N |
| MO-1875 | 54 | M | Blood | IN | Unknown | C | G | P |
| MO-2886 | 36 | M | Blood | LA | Retinitis/HIV infection | C | E | R |
| MO-2842 | 5 | M | Toe wound | NY | Unknown | C | H | Q |
| AR-19 | 59 | F | Sputum | TN | Unknown | C | G | P |
| AR-04 | 72 | F | Finger | UT | Unknown | C | G | O |
| AR-08 | 38 | M | Sputum | OH | Unknown | C | G | O |
| AR-01 | 39 | M | Unknown | TX | Unknown | C | G | R |
| AR-25 | 38 | M | Unknown | CA | Unknown | C | G | T |
| M. neoaurum | ||||||||
| MO-2355 | 5 | M | Blood | PA | Unknown | B | A | H |
| MO-2441 | 4 | 4 | Blood | CA | Myeloblastic leukemia | B | D | B |
| MO-2117 | 2 | M | Central catheter | NC | Hemophilia | B | A | D |
| MO-2123 | 4 | F | Blood | TX | Leukemia | B | A | A |
| MO-2432 | 61 | M | Blood | MA | Unknown | B | A | D |
| MO-2442 | 61 | F | Central catheter | MA | Pulmonary embolism | B | A | D |
| AS-544 | 67 | F | Central catheter | OH | Unknown | B | A | H |
| MO-1110 | 67 | F | Central catheter | ME | Unknown | B | D | B |
| MO-1302 | 32 | F | Blood | FL | Systemic lupus erythematosus | B | A | C |
| MO-696 | 36 | F | Central catheter | CA | Pulmonary hypertension septicemia | B | A | C |
| MO-2765 | 61 | F | Blood | LA | Unknown | B | A | D |
| MO-2921 | 64 | M | Blood (port also) | PA | End-stage renal disease on hemodialysis | B | A | H |
| MO-2933 | 33 | F | Central catheter | KS | Unknown | B | A | L |
| AR-20 | 24 | F | Central catheter | IL | Unknown | B | A | G |
| MO-2225 | 35 | F | Sputum | IL | Unknown | B | A | H |
| MO-1351 | 86 | F | Sputum | WI | Unknown | B | D | K |
| MO-1934 | 31 | M | Sputum | NY | Unknown | B | A | H |
| MO-891 | 50 | F | Sputum | TX | Unknown | B | C | J |
| MO-2573 | 52 | M | Palm tissue | MA | Unknown | NA | NA | NA |
| MO-2496 | NA | F | Sputum | MA | Unknown | NA | NA | NA |
| MO-1156 | 80 | F | Urine | OH | Unknown | B | A | K |
| AR-21 | 84 | M | Sputum | OR | Unknown | B | B | I |
| AR-22 | 81 | F | Sputum | MA | Unknown | B | D | B |
| AR-18 | 30 | M | Sputum | HI | Unknown | B | A | B |
| AR-11 | 22 | M | Blood | CA | Unknown | B | A | C |
| AR-05 | 56 | M | Blood | MN | Unknown | A | A | H |
| AR-14 | 11 mo | F | Blood | PA | Unknown | B | A | H |
| AR-09 | 4 | F | Blood | CA | Unknown | B | B | H |
| AR-16 | 52 | F | Blood | HI | Unknown | B | A | F |
| AR-15 | 10 | M | Blood | PA | Unknown | B | B | G |
| AR-12 | 10 mo | F | Blood | IL | Unknown | B | A | G |
| AR-10 | 61 | F | Wound | MD | Unknown | B | D | B |
| AR-06 | 17 | M | Wound | PA | Unknown | B | A | D |
| AR-02 | 55 | M | Unknown | FL | Unknown | B | D | B |
| AR-03 | 81 | F | Unknown | NY | Unknown | B | A | D |
| AR-24 | 1 | F | Blood | WV | Unknown | B | D | E |
Sequencing.
Forty-four of 46 (96%) of the clinical isolates were subjected to full sequencing of the 16S rRNA gene and partial sequencing of the hsp65 and the rpoB genes as previously described (18, 23). Two isolates previously identified by hsp65 PRA were not available for multigene sequencing.
Susceptibility testing.
Susceptibilities to 14 antimicrobial agents were determined by broth microdilution using the recommended Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) guidelines for rapidly growing mycobacteria (26). The antimicrobials tested included those recommended by the CLSI: amikacin, cefoxitin, ciprofloxacin, clarithromycin, doxycycline, imipenem, sulfamethoxazole, tobramycin, and linezolid. Additional antimicrobials for which there are no current CLSI recommendations were tested and included gatifloxacin, minocycline, moxifloxacin, tigecycline, and trimethoprim-sulfamethoxazole. The breakpoints for the last group of agents were those recommended by the CLSI in document M100-S18 for aerobic bacteria (4) for all agents except moxifloxacin and gatifloxacin, for which the breakpoints used were 2 and 4 μg/ml, respectively (1 and 2 dilutions higher, respectively, than the breakpoints recommended for bacterial isolates), and tigecycline, whose breakpoint has not been addressed by the CLSI. Because not all isolates were tested simultaneously, some isolates were not tested with all antimicrobials, and the concentrations of antimicrobials tested varied in the MIC panels.
Susceptibility to clarithromycin was read after incubation for 3 and 14 days in order to ascertain isolates that had inducible macrolide resistance (15, 26) (proposals for modification of the document cited in reference (26) have been submitted to the CLSI).
Sequence analysis.
Sequence alignment and phylogenetic trees were constructed using the neighbor-joining method with Kimura's two-parameter distance correction model and 1,000 bootstrap replications in the MEGA (version 4) software package (20).
Nucleotide sequence accession numbers.
The 16S rRNA, hsp65, and rpoB gene sequences of the clinical isolates were deposited in GenBank with accession numbers HM011124 to HM011258. The M. lacticola hsp65 and rpoB gene sequences were deposited in GenBank with accession numbers HMO30495 and HMO30494, respectively.
RESULTS
Organisms.
Forty-four isolates underwent DNA sequencing (18): 34 were identified as M. neoaurum and 10 were identified as M. neoaurum-like. Clinical information on all of the isolates was provided at the time of submission (Table 1). Of 42 isolates of the M. neoaurum and M. neoaurum-like groups from UTHSCT and ARUP with known sources, 26 (62%) were recovered from blood, 5 (12%) were recovered from tissue or wound specimens, 10 (24%) were recovered from sputum, and 1 (2%) was recovered from urine. Within the two groups, 4/8 (50%) of the M. neoaurum-like isolates and 22/34 (65%) of the M. neoaurum isolates with known sources were from blood (Table 1).
Patient demographics.
There were 10 patients under age 6 years, and the remaining 36 patients were adolescents, adults, or of unknown ages. Patient ages and geographical locations along with their underlying diseases or conditions are presented in Table 1.
Underlying diseases or conditions were known for only nine (19%) of the cases (Table 1). Of the 26 isolates recovered from blood cultures, 35% were known to be associated with catheter-related sepsis.
The majority (60%) of patients were from California, Pennsylvania, Massachusetts, Texas, Ohio, New York, and Illinois; but overall, patients were also from 14 other states.
Sequencing. (i) 16S rRNA gene.
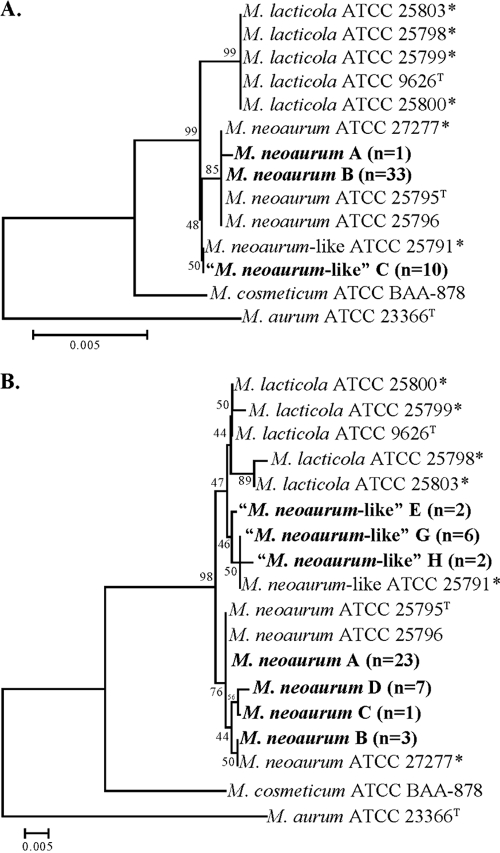
By full 16S rRNA gene sequencing (1,413 bp), two major clusters with three unique sequences were identified among clinical isolates. The first cluster contained 34 clinical isolates whose sequences showed 99.9% to 100% identity to those of M. neoaurum type strain ATCC 25795 (10) and ATCC reference strains ATCC 27277, ATCC 25796, and ATCC 23072 (18) (Fig. 1A). A sole isolate in this cluster differed by a single nucleotide from the M. neoaurum type strain. A second cluster consisted of 10 clinical isolates, which shared 100% identity to ATCC 25791. This group does not match any recognized species. The sequence of these isolates differed by 4 bp from the sequence of the M. neoaurum (2 substitutions and 2 indels) and 4 bp from the sequence of M. lacticola (4 substitutions) type strains (99.7% identity). Compared to the sequence of M. neoaurum, M. lacticola had the same 4-bp substitutions as the ATCC 25791 group plus four additional unique substitutions.
FIG. 1.
Neighbor-joining trees of 16S rRNA (A) and hsp65 (B) genes of M. neoaurum, M. lacticola, and M. neoaurum-like clinical isolates and culture collection strains. Branch support is recorded at the nodes as a percentage of 1,000 bootstrap iterations. M. cosmeticum is present as the most closely related species. M. aurum is included as an outgroup. *, annotated as a different species by the American Type Culture Collection (18).
Of interest, the full rRNA gene sequence of none of the clinical isolates matched the full 16S rRNA gene sequence of the nonvalidated species of M. lacticola (ATCC 9626) (11, 18).
(ii) hsp65 gene.
By hsp65 partial gene sequencing (402 bp), the grouping of the clinical isolates was the same as that seen by 16S rRNA gene sequencing (Fig. 1B). Among the 34 clinical isolates whose 16S rRNA gene sequences had 100% identity to the 16S rRNA gene sequence of the M. neoaurum type strain, all formed a tight cluster of hsp65 sequence variants (sequevars). The largest sequevar cluster consisted of 23 clinical strains whose hsp65 sequences showed 100% identity to the hsp65 sequences of M. neoaurum type strain ATCC 25795 and reference strain ATCC 25796. The other three sequevars included 7 clinical strains, 3 clinical strains, and 1 clinical strain each, respectively. These four sequevars showed up to 0.5% intraspecies variability and differed by a G → T at position 112 and/or a C → T at position 211 compared to the sequence of the M. neoaurum type strain. Among the 10 clinical isolates whose 16S rRNA genes showed 100% sequence identity to the 16S rRNA gene of ATCC 25791, three sequevars of the hsp65 gene were seen. The largest cluster consisted of six clinical isolates whose hsp65 sequences had 99.2% identity (3 bp differences) and 99.8% identity (1 bp difference) to the hsp65 sequences of the M. neoaurum and M. lacticola type strains, respectively. The other two sequevars included two clinical strains each. The sequences of these three sequevars showed up to 0.3% intraspecies variability and differed by 1 or 2 bp. All but one sequevar had the same position 88 C → T and position 115 G → C substitutions seen with M. lacticola compared to the sequence of M. neoaurum, and all sequevars had one additional unique position 367 C → G substitution. Thus, no hsp65 sequences shared 100% identity to the hsp65 sequence of the M. lacticola type strain. It should be noted the hsp65 amino acid sequence was identical for all clinical strains.
(iii) rpoB gene.
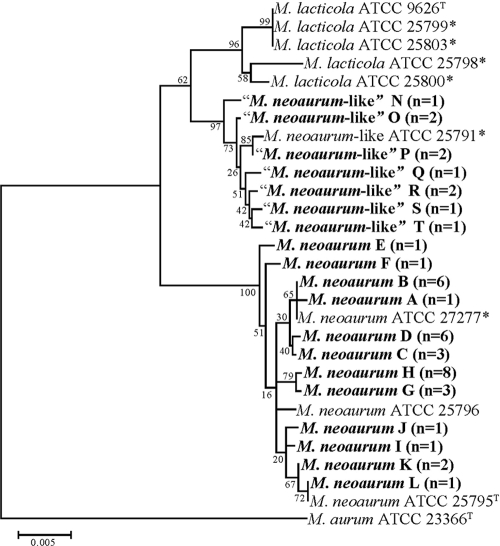
By rpoB partial gene sequencing (621 bp), the grouping of the clinical isolates was the same as that seen with the 16S rRNA gene, but with overall greater sequence heterogeneity. In total, for the two taxa, 19 sequence variants (sequevars) were seen among the clinical isolates by rpoB sequencing and 7 were seen for hsp65.
Among the 34 clinical isolates whose 16S rRNA gene showed 100% sequence identity to that of M. neoaurum type strain ATCC 25795, the same grouping uncovered with the hsp65 gene was detected (Fig. 2 ). Eleven sequevars were identified among the 34 isolates that group with M. neoaurum, with no more than 8 isolates being in any one group (Fig. 2). The rpoB sequence of one sequevar of 6 clinical isolates matched that of ATCC 27277. Five sequevars contained only 1 strain, while the remaining six consisted of 8, 6, 6, 3, 3, and 2 clinical isolates each, respectively. Overall the 11 sequevars showed up to 1.1% intraspecies variability, with a total of 11 positions being variable.
FIG. 2.
Neighbor-joining trees of the rpoB gene of M. neoaurum, M. lacticola, and M. neoaurum-like clinical isolates and culture collection strains. Branch support is recorded at the nodes as a percentage of 1,000 bootstrap iterations. M. aurum is included as an outgroup. *, annotated as a different species by the American Type Culture Collection (18).
Among the 10 clinical isolates that showed 100% 16S rRNA gene sequence identity to ATCC 25791, seven sequevars were identified which contained three groups of two isolates each and four isolates that each had a unique sequence. These seven sequevars showed up to 1.0% intraspecies variability in their rpoB sequences, which varied by 2 to 6 bp from each other. The closest identities observed for any of the 10 isolates to the M. neoaurum and M. lacticola type strains were 96.4 and 97.9%, respectively.
The 19 unique rpoB sequences seen among the clinical isolates resulted in only 3 unique amino acid sequences. Thirty-four clinical isolates shared 100% amino acid homology with the M. neoaurum type strain (Fig. 2). The 10 clinical isolates that showed 100% 16S rRNA gene sequence identity to ATCC 25791 shared 100% amino acid homology with M. lacticola.
The four reference strains that showed 100% identity to M. lacticola ATCC 9626 by 16S rRNA gene sequencing exhibited three rpoB sequevars that showed only 1.5% intraspecies variability and that exhibited a 3.9% difference from the M. neoaurum type strain and a 2.3% difference from M. neoaurum-like reference strain ATCC 25791.
Susceptibility testing.
Susceptibilities by broth microdilution were available for 46 isolates in this study. The majority of the 46 isolates tested in this study were pansusceptible, having MICs indicating susceptibility to amikacin (46/46), cefoxitin (46/46), tobramycin (46/46), ciprofloxacin (46/46), doxycycline (21/21), gatifloxacin (38/38), imipenem (46/46), linezolid (46/46), moxifloxacin (23/23), sulfamethoxazole (20/20), tigecycline (22/22), and trimethoprim-sulfamethoxazole (45/45).
The one exception was for clarithromycin. Clarithromycin susceptibility primarily exhibited a bimodal distribution with 3 days of incubation, with 12/46 (26%) isolates having highly resistant MICs of ≥16 μg/ml and 25/46 (54%) having susceptible MICs of ≤2 μg/ml. A total of 18/46 (39%) isolates had resistant clarithromycin MICs (≥8 μg/ml) at 3 days. With 14 days of incubation, most susceptible isolates remained susceptible or intermediate to clarithromycin, although 25/46 (54%) were resistant. Tables 2 and 3 show the antimicrobial susceptibility results by broth microdilution for the isolates in the current study. Of the 36 isolates of M. neoaurum, 12 (47%) were susceptible to clarithromycin at 3 days of incubation, whereas only 3 isolates (8%) were susceptible to clarithromycin at 14 days of incubation. Likewise, for the 10 isolates in the M. neoaurum-like group, 80% were clarithromycin susceptible at 3 days of incubation, whereas only 30% were susceptible after 14 days of incubation. There was no apparent difference in susceptibility to antimicrobials other than clarithromycin between the two groups of isolates.
TABLE 2.
Comparison of MIC ranges, MIC50s, and MIC90s and percent susceptible to antimicrobials for clinical and reference isolates of M. neoaurum-like group
| Antimicrobial | No. of isolates tested | MIC (μg/ml) |
% susceptible | |||
|---|---|---|---|---|---|---|
| ATCC 25791 | Range | 50% | 90% | |||
| Amikacin | 10 | ≤1 | ≤1-2 | ≤1 | 2 | 100 |
| Cefoxitin | 10 | ≤4 | ≤2-8 | ≤8 | 8 | 100 |
| Ciprofloxacin | 10 | ≤0.12 | ≤0.06-0.5 | ≤0.12 | 0.25 | 100 |
| Clarithromycin, 3 days | 10 | 0.5 | ≤0.06->16 | 0.5 | 8 | 80 |
| Clarithromycin, 14 days | 10 | 4 | 1->64 | 4 | >64 | 30 |
| Doxycycline | 5 | ≤0.12 | ≤0.12-≤0.25 | ≤0.25 | ≤0.25 | 100 |
| Gatifloxacin | 10 | NDa | ≤0.03-0.12 | ≤0.06 | 0.12 | 100 |
| Imipenem | 10 | ≤2 | ≤0.5-1 | ≤0.5 | ≤1 | 100 |
| Linezolid | 10 | ≤1 | ≤1-≤2 | ≤1 | 1 | 100 |
| Minocycline | 10 | ≤1 | ≤0.06-≤0.5 | ≤0.5 | ≤0.5 | 100 |
| Moxifloxacin | 5 | ≤0.25 | 0.03-≤0.25 | ≤0.06 | ≤0.25 | 100 |
| Sulfamethoxazole | 5 | ND | ≤2-32 | 4 | 32 | 100 |
| Trimethoprim-sulfamethoxazole | 10 | ≤0.25/4.8 | ≤0.25/4.8-0.25/4.8 | ≤0.25/4.8 | 0.25/4.8 | 100 |
| Tigecycline | 4 | 0.03 | 0.06-0.12 | ≤0.03 | 0.12 | —b |
| Tobramycin | 10 | 2 | ≤2-4 | 2 | 4 | 100 |
ND, not done.
—, breakpoints for tigecycline have not been addressed by the CLSI.
TABLE 3.
Comparison of MIC ranges, MIC50s, and MIC90s and percent susceptible to antimicrobials for clinical and reference isolates of Mycobacterium neoaurum
| Antimicrobial | No. of isolates tested | MIC (μg/ml) |
% susceptible | |||
|---|---|---|---|---|---|---|
| ATCC 25795T | Range | 50% | 90% | |||
| Amikacin | 36 | ≤1 | ≤1-2 | ≤1 | 2 | 100 |
| Cefoxitin | 36 | 8 | 4-16 | ≤8 | 16 | 100 |
| Ciprofloxacin | 36 | ≤0.12 | ≤0.12-1 | ≤0.12 | ≤0.25 | 100 |
| Clarithromycin, 3 days | 36 | 4 | 0.5->32 | 4 | 32 | 47 |
| Clarithromycin, 14 days | 36 | 8 | 2->64 | 8 | >64 | 8 |
| Doxycycline | 19 | ≤0.12 | ≤0.06-≤0.25 | ≤0.25 | ≤0.25 | 100 |
| Gatifloxacin | 29 | NDa | ≤0.06-0.25 | ≤0.06 | ≤0.12 | 100 |
| Imipenem | 36 | ≤2 | ≤0.5-≤2 | ≤0.5 | 1 | 100 |
| Linezolid | 36 | ≤1 | ≤0.5-2 | ≤1 | 2 | 100 |
| Minocycline | 35 | ≤1 | ≤0.25-≤1 | ≤0.5 | 0.5 | 100 |
| Moxifloxacin | 18 | ≤0.25 | ≤0.06-0.5 | ≤0.12 | ≤0.25 | 100 |
| Sulfamethoxazole | 15 | ND | ≤4-32 | 8 | 16 | 100 |
| Trimethoprim-sulfamethoxazole | 35 | 0.5/9.5 | ≤0.25/4.8-1/19 | 0.25/4.8 | 0.5/9.5 | 100 |
| Tigecycline | 18 | 0.06 | ≤0.03-0.25 | 0.06 | 0.12 | —b |
| Tobramycin | 36 | 2 | ≤1-4 | 2 | 4 | 100 |
ND, not done.
—, breakpoints for tigecycline have not been addressed by the CLSI.
DISCUSSION
The first reported human infection of M. neoaurum was in 1988 in Queensland, Australia, from a patient with a cystadenocarcinoma of the ovary with involvement of abdominal nodes and peritoneal metastases (22). Parenteral nutrition was provided via a Hickman catheter, from which the isolate was recovered. This report is of a study performed prior to the use of molecular identification techniques. The investigators identified this isolate by biochemical testing and thin-layer chromatography and compared the results for the isolate to those for the type strain of M. neoaurum. The results were identical to those for the M. neoaurum type strain, ATCC 27595 (5). Prior to that report, the species (including the type strain) had been isolated only from soil, dust, and water (22). Since the original report, 15 other human cases have been documented in Australia, the United States, Italy, Canada, and China (Table 4) .
TABLE 4.
Published cases of Mycobacterium neoaurum, M. neoaurum-like, and M. lacticola infectionsa
| Yr (reference) | Geographic location | Underlying disease | Ageb | Sex | Catheter removed (type) | Antibiotics given | Length of antibiotic treatment | Resolution of infection | Isolate cultured | Method(s) of ID | Susceptibility pattern |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1988 (5) | Australia | Ovarian adenocarcinoma | 55 | F | No (Hickman) | Gentamicin, cefoxitin | 7 wk | Yes | Yes | TLC, Bios | Susceptible to amikacin, kanamycin, gentamicin, tetracycline, minocycline, doxycycline, SXT, cefoxitin, vancomycin, imipenem, streptomycin, capreomycin; resistant to ethambutol, isoniazid, cycloserine, cefotaxime, amoxicillin |
| 1994 (10) | Australia | ALL (allogeneic bone marrow transplant) | 17 | M | Yes | Ticarcillin-clavulanate, amoxicillin-clavulanate, tobramycin | 3 wk | Yes | Yes | Bios | Susceptible to ticarcillin clavulanate, amikacin, tetracycline, cefoxitin, imipenem, ciprofloxacin, erythromycin, clarithromycin, roxithromycin, azithromycin; resistant to TMP-SMX |
| 1999 (6) | USA | i.v. drug abuse | NA | M | Yes | Piperacillin, gentamicin | 6 wk | Yes | Yesc | HPLC, Bios | NA |
| 1999 (6) | USA | Primary pulmonary | 46 | M | Yes (Hickman) | Vancomycind | 3 days | Yes | Yes | HPLC, Bios | NA |
| hyper tension, | Ceftriaxoned | 3 days | |||||||||
| recurring fevers | Amoxicillin-clavulanated | 10 days | |||||||||
| 2000 (25) | Hong Kong | ALL | 9 | F | Yes (Hickman) | Ceftazidime, amikacin | 3 wk | Yes | Yes | 16S rRNA sequencing | Susceptible to ampicillin, amoxicillin clavulanate, imipenem, meropenem, ofloxacin, ciprofloxacin, amikacin, tetracycline, doxycycline, co-trimoxazole |
| 2000 (13) | USA | Diabetes mellitus hyper tension, renal failure | 54 | M | Yes (peritoneal) | Cefoxitin, ethambutol, rifampin, and clarithromycin; after ID changed to ciprofloxacin, amikacin, and rifampin | 4 wk | Yes | Yes | TLC, HPLC | Susceptible to amikacin, ampicillin-sulbactam, ciprofloxacin, doxycycline, ofloxacin, rifampin; resistant to clarithromycin, ethambutol, ethionamide, isoniazid |
| 2000 (27) | Italy | Recurrent UTI (progressive renal failure) | 62 | F | NA | NA | NA | Yes | Yese | hsp65 PRA, 16S rRNA, HPLC | Susceptible to streptomycin, vancomycin, imipenem, levofloxacin, ofloxacin; intermediate to amoxicillin; resistant to isoniazid, rifampin, ethambutol, ethionamide |
| 2003 (1) | Canada | Diabetes mellitus, renal failure on hemo- dialysis | 40 | F | Removal not clear (graft/line infection) | Ciprofloxacin, doxycycline | ∼ 4 wk | Yes | Yes | 16S rRNA sequencing | Susceptible to ciprofloxacin, doxycycline, rifampin, imipenem; resistant to ethambutol, isoniazid, clarithromycin |
| 2004 (11) | USA | Autologous stem cell transplant | 4 | F | Yes (Hickman) | Azithromycin, rifampin | 5 days | Yes | Yes | hsp65 PCR, 16S rRNA sequencing | NA |
| 2006 (14) | USA | Pulmonary gastro esophageal, reflux disease, corticosteroids | 67 | F | NA | Clarithromycin, amikacin | 6 mo | Yes | Yes | 16S rRNA sequencing | Susceptible to amikacin, kanamycin, tobramycin, cefoxitin, imipenem, doxycycline, ciprofloxacin, gatifloxacin, moxifloxacin, clarithromycin, SXT, linezolid, amoxicillin clavulanate |
| 2007 (12) | Australia | Cutaneous infection, sarcoid | 53 | F | NA | Moxifloxacin, roxithromycin | 4 mo | Yes | No | PCR of tissue | NA |
| 2007 (24) | |||||||||||
| Patient 1 | USA | Neuroblastoma | 32 mo | F | Yes (Hickman) | Meropenemf | 7 days | Yes | Yes | HPLC, Bios | Resistant to clarithromycin |
| Amikacinf | 7 days | ||||||||||
| Clarithromycinf | 7 days | ||||||||||
| Ciprofloxacin | 3 wk | ||||||||||
| Linezolid | 3 wk | ||||||||||
| Patient 2 | USA | Liver transplant | 15 mo | F | NA | NA | Yes | Yes | HPLC, Bios | NA | |
| Patient 3 | USA | Rhabdomyosarcoma | 3 | M | Yes (Broviac) | Amikacin | 12 days | Yes | Yes | HPLC, Bios | Resistant to clarithromycin |
| Clarithromycin | 6 wk | ||||||||||
| Levofloxacin | 6 wk | ||||||||||
| Patient 4 | USA | Colon cancer, short gut | 59 | F | Yes (PICC) | Cefoxitin | 2 wk | Yes | Yes | HPLC, Bios | NA |
| syndrome | Levofloxacin | 4 wk | |||||||||
| Clarithromycin | 4 wk | ||||||||||
| Ethambutol | 4 wk |
ID, identification; AST, antimicrobial susceptibility testing; TLC, thin-layer chromatography; i.v., intravenous; ALL, acute lymphocytic leukemia; M, male; F, female; NA, not available; Bios, biochemical testing, TMP, trimethoprim; SXT, trimethoprim-sulfamethoxazole; UTI, urinary tract infection; PICC, peripherally inserted central catheter.
Ages are in years unless indicated otherwise.
Polymicrobial infection consisting of two strains of Pseudomonas aeruginosa and Comamonas acidovorans with M. neoaurum.
Prior to catheter removal.
Two separate isolates were recovered (2 months apart).
Initial treatment that was changed to ciprofloxacin and linezolid.
A review of the previously published cases of M. neoaurum and M. lacticola shows results similar to those obtained in the current study, in that 11/15 (73%) of the infections were associated with catheter- or line-related sepsis. Of these 11 cases, however, only three isolates were identified using molecular techniques (1, 11, 25). The first case of M. neoaurum infection identified using 16S rRNA sequencing was in 2000 in a 9-year-old girl with acute lymphoblastic leukemia in Hong Kong. Blood cultures performed with blood drawn through a Hickman catheter were positive for a pigmented, rapidly growing mycobacterium that was identified by complete 16S rRNA gene sequencing as M. neoaurum (25). In 2004, the first documented case of M. lacticola, a newly characterized but currently unvalidated species was described in a patient with catheter-related sepsis following autologous stem cell transplantation (11). Identification was based on complete 16S rRNA gene sequencing. The third case was a patient in Canada with diabetic mellitus and renal failure on hemodialysis (1).
Four patients (27%) without catheters also developed infection with M. neoaurum, with three of these isolates being identified by molecular techniques. These cases included an Italian patient with recurrent urinary tract infections and renal failure (27), a patient from the United States with chronic pulmonary disease receiving long-term corticosteroids (14), a pediatric liver transplant patient from the United States (24), and an immunocompetent Australian patient with cutaneous infection of the scalp for whom cultures were negative but PCR of tissue detected M. neoaurum (12).
Recently, a case of meningoencephalitis followed by rapidly progressive dementia in which M. neoaurum was identified as the possible pathogen was reported (9). However, a subsequent investigation of the case suggested that the M. neoaurum DNA recovered in the previously reported case was likely a laboratory contaminant, and this case was excluded from the cases documented in Table 4 (8).
Susceptibility testing was recorded in 9 of 15 (60%) of the previously reported cases of infection due to M. neoaurum. The susceptibility testing results for the previously published cases, shown in Table 4, indicate susceptibility of most isolates to the antimicrobials recommended for treatment of infections caused by rapidly growing mycobacteria, including amikacin (5/5), doxycycline (5/5), cefoxitin (3/3), imipenem (6/6), ciprofloxacin (5/5), tobramycin (1/1), linezolid (1/1), and sulfonamides (3/4). Four of six (67%) of the isolates were resistant to clarithromycin. Isolates of the M. neoaurum-M. neoaurum-like group in the current study were also generally susceptible to the antimicrobials recommended by the CLSI for susceptibility testing of rapidly growing mycobacteria, including amikacin, cefoxitin, ciprofloxacin, doxycycline, imipenem, tobramycin, trimethoprim-sulfamethoxazole, and linezolid; but similar to the previously documented cases, almost half of the isolates showed resistance to clarithromycin (26).
The outcomes of the previous infections presumed to be caused by the M. neoaurum-M. lacticola group were good. In all 15 of the previously described cases, resolution of infection was documented. For the 14 patients who were treated with antimicrobials, various regimens were successful, although all of the regimens contained at least two antimicrobials. As with other serious nontuberculous mycobacterial infections, combination antimicrobial therapy is probably warranted. Since only one of nine patients (11%) maintained a catheter after discovery of the M. neoaurum-M. lacticola group infection, it seems expedient to recommend removal of the catheter when an M. neoaurum-M. lacticola group infection is diagnosed.
The resistance of M. neoaurum and M. neoaurum-like organisms to clarithromycin suggests the presence of an inducible erm gene. These genes have been shown to be the basis for intrinsic macrolide resistance in other mycobacterial species, including M. tuberculosis [erm(37)] (3), M. smegmatis [erm(38)] (15), M. fortuitum [erm(39)] (17), M. mageritense [erm(40)] (15), and most recently, M. abscessus [(erm(41)] (16). M. smegmatis [erm(38)], like M. neoaurum and the M. neoaurum-like group, is a pigmented, rapidly growing mycobacterial species.
As for identification of the majority of the other nontuberculous mycobacterial species, identification by molecular techniques is currently necessary for definitive identification of isolates of the M. neoaurum-M. neoaurum-like-M. lacticola group, as three closely related species or taxonomic groups (M. neoaurum, M. neoaurum-like, and M. lacticola) are present within the group and are associated with mycobacteremia. Moreover, in a previous report, Simmon et al. indicated that multigene target sequencing, including sequencing of rpoB and hsp65 genes, in addition to DNA relatedness studies, may be necessary to differentiate the M. neoaurum-M. lacticola group from other closely related species (18). It is noteworthy that only five of the previously documented cases of infection caused by the M. neoaurum-M. lacticola group were confirmed by 16S rRNA gene sequencing (Table 4), and the percent similarities to other species in the previous studies was not indicated. Partial sequencing was performed in two of the five cases. Two of the cases used a >1,200-bp sequence, and in one case the number of base pairs sequenced was not specified.
Interestingly, the M. neoaurum, M. neoaurum-like group, and M. lacticola all appear to have pathogenicity similar to that of another rapidly growing mycobacterial species, M. mucogenicum, which is also primarily associated with bloodstream and central line-related infections (2, 22).
Thus, by using multilocus gene sequencing, this study confirms that two closely related taxa of pigmented, rapidly growing mycobacteria are associated with mycobacteremia and central catheter line-related sepsis. The study also demonstrates that these taxa are relatively susceptible to all drugs except the macrolides, and therapy is usually successful. No clinical cases of mycobacteremia due to M. aurum or the nonvalidated species M. lacticola were identified. Continued use of sequencing likely will identify other pathogenic pigmented, rapidly growing mycobacteria in the future.
Mycobacterium bacteremicum.
Mycobacterium bacteremicum (pertaining to the organism's association with bloodstream infections) is an acid-fast bacillus that grows aerobically within 7 days on standard mycobacterial media. It is scotochromogenic (yellow). It is susceptible (100%) to amikacin, cefoxitin, imipenem, trimethoprim-sulfamethoxazole, doxycycline, minocycline, tigecycline, linezolid, ciprofloxacin, moxifloxacin, and gatifloxacin. It shows variable susceptibility to clarithromycin. It is an established cause of central venous catheter-related infections, and as its name suggests, it is most commonly recovered from blood. It has also been recovered from posttraumatic wound infections. The proposed type strain is ATCC 25791, which was originally submitted to the ATCC as an example of M. aurum. By complete 16S rRNA gene sequencing, ATCC 25791T has a unique sequence that is the most closely related to the sequences of M. neoaurum (4 bp, or 99.7%) and M. lacticola (4 bp, or 99.7%) (GenBank accession number FJ172308) (21). By partial sequencing of the hsp65 gene, three closely related sequence variants that showed 0.3% intraspecies variability were identified and were most closely related to M. neoaurum (99.2% identity) (GenBank accession number FJ172314). By rpoB partial gene sequencing, six closely related sequence variants that showed 0.8% intraspecies variability were identified. They were most closely related to M. neoaurum (3.6% difference) and M. lacticola (2.3%) (GenBank accession number FJ172329).
Acknowledgments
We acknowledge the assistance of the Mycobacteria/Nocardia Laboratory staff at the University of Texas Health Science Center. We also thank Leslie Hall, Nancy Wengenack, and staff at the Mayo Clinic for sequencing select isolates; and we thank Kevin Nash of the Saban Research Institute of Children's Hospital, Los Angeles, CA, and of the Department of Pathology, University of Southern California, for discussions about the potential presence of the erm gene. We express appreciation to Joanne Woodring for preparation of the manuscript. A special thank you goes to Christine Turenne for her helpful discussions and critical review of the manuscript.
Funding for this study was provided by institutional funds of the University of Texas Health Science Center and by the ARUP Institute for Clinical and Experimental Pathology.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Becker, M. L., A. A. Suchak, J. N. Wolfe, R. Zarychanski, A. Kabani, and L. E. Nicolle. 2003. Mycobacterium neoaurum bacteremia in a hemodialysis patient. Can. J. Infect. Dis. 14:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmented rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buriankova, K., F. Doucet-Populaire, O. Dorson, A. Gondran, J. C. Ghnassia, J. Weiser, and J. L. Pernodet. 2004. Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 48:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Davison, M. B., J. G. McCormack, Z. M. Blacklock, D. J. Dawson, M. H. Tilse, and F. B. Crimmins. 1988. Bacteremia caused by Mycobacterium neoaurum. J. Clin. Microbiol. 26:762-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George, S. L., and L. S. Schlesinger. 1999. Mycobacterium neoaurum an unusual cause of infection of vascular catheters: case report and review. Clin. Infect. Dis. 28:682-683. [DOI] [PubMed] [Google Scholar]
- 7.Hall, L., K. A. Doerr, S. L. Wohlfiel, and G. D. Roberts. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han, X. Y. 2005. Mycobacterium neoaurum contamination. Emerg. Infect. Dis. 11:1316-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heckman, G. A., C. Hawkins, A. Morris, L. L. Burrows, and C. Bergeron. 2004. Rapidly progressive dementia due to Mycobacterium neoaurum meningoencephalitis. Emerg. Infect. Dis. 10:924-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland, D. J., S. C. Chen, W. W. Chew, and G. L. Gilbert. 1994. Mycobacterium neoaurum infection of a Hickman catheter in an immunosuppressed patient. Clin. Infect. Dis. 18:1002-1003. [DOI] [PubMed] [Google Scholar]
- 11.Kiska, D. L., C. Y. Turenne, A. S. Dubansky, and J. B. Domachowske. 2004. First case report of catheter-related bacteremia due to “Mycobacterium lacticola.” J. Clin. Microbiol. 42:2855-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin, L. K., R. Lawrence, S. Kossard, and D. F. Murrell. 2007. Cutaneous Mycobacterium neoaurum infection causing scarring alopecia in an immunocompetent host. Br. J. Dermatol. 157:204-206. [DOI] [PubMed] [Google Scholar]
- 13.McNally, C. F., and J. E. Mangino. 2000. Mycobacterium neoaurum: a case report and review of the literature. Infect. Dis. Clin. Pract. 9:273-275. [Google Scholar]
- 14.Morimoto, Y., E. D. Chan, L. Heifets, and J. M. Routes. 2007. Pulmonary infection with Mycobacterium neoaurum identified by 16S ribosomal DNA sequencing. J. Infect. 54:e227-e231. [DOI] [PubMed] [Google Scholar]
- 15.Nash, K. A., N. Andini, Y. Zhang, B. A. Brown-Elliott, and R. J. Wallace, Jr. 2006. Intrinsic macrolide resistance in rapidly growing mycobacteria. Antimicrob. Agents Chemother. 50:3476-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nash, K. A., B. A. Brown-Elliott, and R. J. Wallace Jr. 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53:1367-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash, K. A., Y. Zhang, B. A. Brown-Elliott, and R. J. Wallace Jr. 2005. Molecular basis of intrinsic macrolide resistance in clinical isolates of Mycobacterium fortuitum. J. Antimicrob. Chemother. 55:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmon, K. E., Y. Y. Low, B. A. Brown-Elliott, R. J. Wallace, Jr., and C. A. Petti. 2009. Phylogenetic analysis of Mycobacterium aurum and Mycobacterium neoaurum with re-description of M. aurum culture collection strains. Int. J. Syst. Evol. Microbiol. 59:1371-1375. [DOI] [PubMed] [Google Scholar]
- 19.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria J. Clin. Microbiol. 33:149-153. (Erratum, 33: 1686.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 21.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukamura, M. 1972. A new species of rapidly growing scotochromogenic mycobacteria. Mycobacterium neoaurum. Med. Biol. (Tokyo) 85:229-233. [Google Scholar]
- 23.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Washer, L. L., J. Riddell IV, J. Rider, and C. E. Chenoweth. 2007. Mycobacterium neoaurum bloodstream infection: report of 4 cases and review of the literature. Clin. Infect. Dis. 45:e10-e13. [DOI] [PubMed] [Google Scholar]
- 25.Woo, P. C., H. W. Tsoi, K. W. Leung, P. N. Lum, A. S. Leung, C. H. Ma, K. M. Kam, and K. Y. Yuen. 2000. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J. Clin. Microbiol. 38:3515-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods, G. L., B. A. Brown-Elliott, E. P. Desmond, G. S. Hall, L. Heifets, G. E. Pfyffer, M. R. Ridderhof, R. J. Wallace, Jr., N. G. Warren, and F. G. Witebsky. 2003. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; approved standard. M24-A. NCCLS, Wayne, PA. [PubMed]
- 27.Zanetti, S., R. Faedda, G. Fadda, I. Dupré, P. Molicotti, S. Ortu, G. Delogu, M. Sanguinetti, F. Ardito, and L. A. Sechi. 2001. Isolation and identification of Mycobacterium neoaurum from a patient with urinary infection. New Microbiol. 24:189-192. [PubMed] [Google Scholar]