Abstract
We studied the accuracy of various susceptibility testing methods, including the 2009, 2010, and updated 2010 CLSI recommendations, to identify Klebsiella pneumoniae isolates with reduced susceptibility to carbapenems associated with different mechanisms of resistance. Forty-three wild-type (WT) strains, 42 extended-spectrum β-lactamase (ESBL) producers, 18 ESBL producers with outer membrane porin protein loss (ESBL/Omp strains), and 42 blaKPC-possessing K. pneumoniae (KPC-Kp) isolates were evaluated. Imipenem (IPM), meropenem (MEM), ertapenem (ERT), and doripenem (DOR) were tested by broth microdilution (BMD), Etest, and disk diffusion (DD), and the modified Hodge test (MHT) was performed using IPM and MEM disks. Results were interpreted according to original as well as recently updated interpretative criteria. MHT was positive for all 42 KPC-Kp isolates and 10 of 18 ESBL/Omp strains and therefore had poor specificity in differentiating between KPC-Kp and ESBL/Omp isolates. Based on the updated CLSI standards, phenotypic susceptibility testing by BMD and DD differentiated most carbapenem-susceptible from carbapenem-nonsusceptible K. pneumoniae isolates without the need for MHT, while the Etest method characterized many KPC-Kp isolates as susceptible, and breakpoints may need to be lowered for this method. However, both the original and updated CLSI criteria do not adequately differentiate between isolates in the KPC-Kp group, which are unlikely to respond to carbapenem therapy, and those in the ESBL/Omp group, which are likely to respond to carbapenem therapy if MICs are within pharmacokinetic/pharmacodynamic targets. Further studies are required to determine if there is a clinical need to differentiate between KPC-Kp and ESBL/Omp groups.
In the last decade, the frequent occurrence of infections due to resistant Gram-negative organisms, particularly Klebsiella pneumoniae, producing a variety of extended-spectrum β-lactamases (ESBLs) has resulted in physicians frequently resorting to the use of carbapenems (35). As a result, an increasing number of Enterobacteriaceae with resistance to carbapenems has been observed (19). So far, the most common mechanism of carbapenem resistance detected among members of the Enterobacteriaceae in the United States is the production of class A K. pneumoniae carbapenemases (KPCs) (8, 9, 33). Although these enzymes have been identified in many species of Enterobacteriaceae, Pseudomonas spp., and Acinetobacter spp., K. pneumoniae remains the most common organism carrying these worrisome resistance genes (33, 41). However, carbapenem resistance in K. pneumoniae may also be due to production of other carbapenemases (e.g., VIM- and IPM-type enzymes and OXA-48) (3, 40) or to changes in outer membrane porin (Omp) proteins, including OmpK35, OmpK36, and OmpK37, often combined with production of an ESBL, AmpC, or both (11, 20, 25, 27).
Determination of carbapenem resistance among members of the Enterobacteriaceae is still performed routinely by traditional phenotypic testing. However, detection of blaKPC-possessing K. pneumoniae (KPC-Kp) isolates presents a significant challenge for clinical laboratories (1, 28). These isolates are difficult to detect because the majority of them do not manifest high-level resistance to carbapenems (e.g., MICs of 1 to 8 μg/ml for imipenem [IPM] and meropenem [MEM]), based on susceptibility breakpoints currently in use (2, 15). A phenotypic test using boronic acid recently demonstrated an excellent ability in detecting KPC-Kp isolates (10, 34, 42). However, this method is not commercially available and requires an additional day before results are available.
Since clinical failures have been reported for patients infected with KPC-Kp isolates that appeared to be susceptible to imipenem or meropenem by routine susceptibility testing (43), detection and accurate reporting of these strains are major concerns. To address this problem, the Clinical and Laboratory Standards Institute (CLSI) issued recommendations in 2009, reaffirmed in January 2010, to improve the detection of carbapenemases among Enterobacteriaceae (5, 6). These recommendations include performing a modified Hodge test (MHT) on isolates resistant to at least one extended-spectrum cephalosporin and with carbapenem MICs or zone diameters at the upper end of the susceptible range (MICs of 2 to 4 μg/ml for imipenem or meropenem or 2 μg/ml for ertapenem [ERT] or meropenem or ertapenem zone diameters of 16 to 21 or 19 to 21 mm, respectively), as they may produce carbapenemases such as KPC (Table 1) (5, 6). If the MHT is negative, carbapenem MICs are to be reported as indicating susceptibility; if the MHT is positive, carbapenem MICs are to be reported without an interpretation and with the following comment. “This isolate demonstrates carbapenemase production. The clinical efficacy of the carbapenems has not been established for treating infections caused by Enterobacteriaceae that test carbapenem susceptible but demonstrate carbapenemase production in vitro.” Performing the MHT also delays the availability of results for a day. Interpretative MIC breakpoints for doripenem (DOR) for use against Enterobacteriaceae were set by the U.S. Food and Drug Administration (FDA) at ≤0.5 μg/ml for susceptibility and ≥1 μg/ml for nonsusceptibility. The CLSI subsequently issued an update in June 2010, lowering the susceptibility breakpoints to ≤1 μg/ml for imipenem and meropenem and ≤0.25 μg/ml for ertapenem and adding a susceptibility breakpoint of ≤1 μg/ml for doripenem (Table 2) (7).
TABLE 1.
Original 2009 and 2010 CLSI and FDA criteria for interpretation of susceptibility testing of carbapenems and for detection of carbapenemase production among Enterobacteriaceaea
| Agent | MIC breakpoint (μg/ml) |
Disk diffusion breakpoint (mm) |
||||||
|---|---|---|---|---|---|---|---|---|
| S | MHTb | I | R | S | MHTb | I | R | |
| IPM | ≤1 | 2-4 | 8 | ≥16 | ≥16 | NA | 14-15 | ≤13 |
| MEM | ≤1 | 2-4 | 8 | ≥16 | ≥22 | 16-21 | 14-15 | ≤13 |
| ERT | ≤1 | 2 | 4 | ≥8 | ≥22 | 19-21 | 16-18 | ≤15 |
| DORc | ≤0.5 | NA | NA | NA | ≥23 | NA | NA | NA |
See references 5 and 6. S, susceptible; I, intermediate; R, resistant; MHT, modified Hodge test; NA, not applicable.
MHT is required to detect carbapenemase production. If MHT is positive, report MICs with no interpretation and with the following comment. “This isolate demonstrates carbapenemase production. The clinical efficacy of the carbapenems has not been established for treating infections caused by Enterobacteriaceae that test carbapenem susceptible but demonstrate carbapenemase production in vitro.” If MHT is negative, report carbapenems as susceptible (5, 6).
Food and Drug Administration criteria are given for DOR.
TABLE 2.
Updated CLSI criteria for interpretation of susceptibility testing of carbapenemsa
| Agent | MIC breakpoint (μg/ml) |
Disk diffusion breakpoint (mm) |
||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| IPM | ≤1 | 2 | ≥4 | ≥23 | 20-22 | ≤19 |
| MEM | ≤1 | 2 | ≥4 | ≥23 | 20-22 | ≤19 |
| ERT | ≤0.25 | 0.5 | ≥1 | ≥23 | 20-22 | ≤19 |
| DOR | ≤1 | 2 | ≥4 | ≥23 | 20-22 | ≤19 |
See reference 7. S, susceptible; I, intermediate; R, resistant.
In the present work, we investigate the accuracy of various standard phenotypic methods for testing four commercially available carbapenems, IPM, MEM, ERT, and DOR, to determine the ability of the 2009 and updated 2010 CLSI criteria to detect KPC-Kp isolates.
MATERIALS AND METHODS
Clinical isolates.
One hundred forty-five clinical isolates of K. pneumoniae were chosen for the study. The collection included 43 wild-type (WT) isolates that were isolated at University Hospitals Case Medical Center (Cleveland, OH) during 2009 and 42 well-characterized KPC-Kp isolates collected in 2006 and 2007 in the eastern United States (12, 13, 15, 16). The remaining 60 K. pneumoniae isolates showed an ESBL phenotype. Twenty of these 60 ESBL-producing isolates were part of a previous group of isolates collected from different countries in 1996 and 1997 (35). The remaining 40 ESBL producers were collected at the University of Pittsburgh Medical Center (Pittsburgh, PA) in 2005 and 2006 and at the Cleveland Clinic Foundation (Cleveland, OH) in 2007 and were characterized genetically in the present work.
Molecular characterization.
The last group of 40 ESBL-producing K. pneumoniae isolates was analyzed by PCR amplification and DNA sequencing for blaKPC, blaTEM, blaSHV, blaCTX-M, blaPER, blaAmpC, blaVIM, and blaIPM genes, as previously reported (15). If present, blaAmpC genes were additionally characterized as previously described (15).
For selected blaKPC-negative isolates in the overall collection with raised carbapenem MICs compared to the WT group, PCR and DNA sequencing of three Omp protein genes (ompK35, ompK36, and ompK37) and nine carbapenemase genes (blaSPM, blaSME, blaOXA-48, blaGIM, blaSIM, blaIMI, blaNMC, and blaGES) were performed as previously described (14, 15, 40). DNA sequences were analyzed using Lasergene 7.2 (DNASTAR, Madison, WI). The amino acid sequences were deduced using the ExPASy Proteomics Server (http://ca.expasy.org) and compared with those previously described (GenBank accession numbers are as follows: for OmpK35, AJ303057; for OmpK36, AJ344089; and for OmpK37, AJ011502) (www.lahey.org/Studies/).
Phenotypic characterization.
All isolates were initially characterized by use of a MicroScan system (Siemens, Healthcare Diagnostics) using Gram-negative Combo NBPC34 trays, which include IPM, MEM, ERT, and screening for ESBL production with cefotaxime and ceftazidime alone and combined with clavulanate. All isolates were tested against IPM, MEM, ERT, and DOR by reference CLSI broth microdilution (BMD; Trek Diagnostics), disk diffusion (DD) (6), and Etest (bioMérieux) on cation-adjusted Mueller-Hinton (MH) agar or broth. MHT was performed and interpreted according to the current CLSI guidelines, using IPM and MEM disks, for all 145 isolates (6).
For selected K. pneumoniae isolates with raised carbapenem MICs, a disk enzymatic assay was performed to detect carbapenemase production. Briefly, a crude extract of β-lactamases was obtained as previously reported (36). Ten microliters of crude extract was placed on a disk of IPM (10 μg), with 10 μl of sterile water placed on another IPM disk as a control. Following 24 h of incubation at room temperature, the two IPM disks were placed on an MH agar plate inoculated with a 0.5 McFarland inoculum of Escherichia coli ATCC 25922 and incubated at 35°C overnight. After incubation, the inhibition zone diameter was measured and interpreted as suspicious for carbapenemase production if a ≥2-mm decrease in the zone of inhibition of IPM plus crude β-lactamase extract was detected compared to that of IPM alone.
Data analysis.
All phenotypic results were interpreted according to FDA and updated CLSI criteria for DOR and original and updated CLSI criteria for the other carbapenems, including results of MHT when required (Tables 1 and 2) (6, 7). To assess the ability to detect carbapenemase-producing K. pneumoniae isolates, sensitivity and specificity were calculated for each carbapenem tested by different methods. Data were also analyzed to determine the methods that best distinguish carbapenem-susceptible groups (WT strains and ESBL producers) from carbapenem-nonsusceptible groups (ESBL producers with changes in Omp proteins and KPC-Kp isolates).
RESULTS
Molecular characterization.
The 40 previously uncharacterized K. pneumoniae isolates were confirmed as blaESBL-containing isolates. Different families of ESBL genes were detected, including blaTEM, blaSHV, and blaCTX-M, whereas blaKPC, blaVIM, and blaIPM were not found among these isolates; only one isolate possessed a blaAmpC gene (blaCMY-2-like) (Table 3). Of the 60 confirmed ESBL-producing carbapenemase-negative isolates included in the present study (40 characterized above and 20 previously characterized isolates), 18 were nonsusceptible to ERT (1 intermediate and 17 resistant) but susceptible to IPM or MEM on initial MicroScan screening. This subgroup of isolates was categorized as ESBL producers with Omp changes (ESBL/Omp strains). Ten of the ESBL/Omp isolates were analyzed by PCR and DNA sequencing for the ompK35, -36, and -37 genes. Disrupted porin coding sequences in ompK36 and -37 genes were found in all 10 strains, and disrupted sequences in the ompK35 gene were found in 8 strains. On the basis of the above molecular characterization, we categorized the 145 K. pneumoniae isolates studied into four groups: WT (n = 43), ESBL producers (n = 42), ESBL/Omp strains (n = 18), and KPC-Kp isolates (n = 42) (Table 3).
TABLE 3.
Characteristics of K. pneumoniae isolates used in this study
| K. pneumoniae group | No. of isolates | Area (yr) of detection | Notea | Reference or source |
|---|---|---|---|---|
| WT | 43 | Cleveland, OH (2009) | Isolates were resistant to ampicillin only; molecular characterization was not performed | This study |
| ESBL producers | 20 | Intercontinental distribution (1996-1997) | The following blaESBL genes were detected: blaTEM-10 (n = 3), blaTEM-63 (n = 1), blaSHV-2 (n = 5), blaSHV-5 (n = 12), blaCTX-M-2 (n = 1), and blaPER-1-like (n = 2) | 35 |
| 22 | Pittsburgh, PA (2005-2006), Cleveland, OH (2007) | The following blaESBL genes were detected: blaSHV-2 (n = 2), blaSHV-5 (n = 3), blaSHV-5-like (n = 9), blaSHV-7 (n = 2), blaSHV-12 (n = 2), blaTEM-10-like (n = 3), and blaCTX-M-2-like (n = 3) | This study | |
| ESBL producers with OmpK loss | 18 | Pittsburgh, PA (2005-2006), Cleveland, OH (2007) | The following blaESBL or blaAmpC genes were detected: blaSHV-2 (n = 1), blaSHV-5 (n = 6), blaSHV-5-like (n = 5), blaSHV-12 (n = 5), blaSHV-75 (n = 1), blaCTX-M-2-like (n = 4), and blaCMY-2-like (n = 1); 10 isolates did not possess complete ompK36 and ompK37 genes, and 8 isolates did not possess a complete ompK35 gene | This study |
| KPC producers | 42 | Eastern USA (2006-2007) | The following bla genes were detected: blaKPC-2 (n = 25), blaKPC-3 (n = 17), blaSHV-5 (n = 1), blaSHV-11 (n = 40), blaSHV-12 (n = 21), blaSHV-14 (n = 1), blaSHV-26 (n = 1), blaSHV-68 (n = 1), and blaTEM-1 (n = 38) | 12-16 |
Some isolates possess more than one blaESBL gene.
Phenotypic characterization.
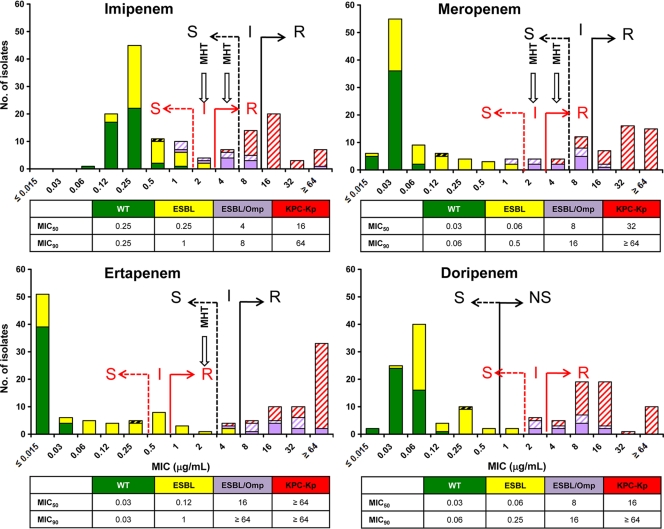
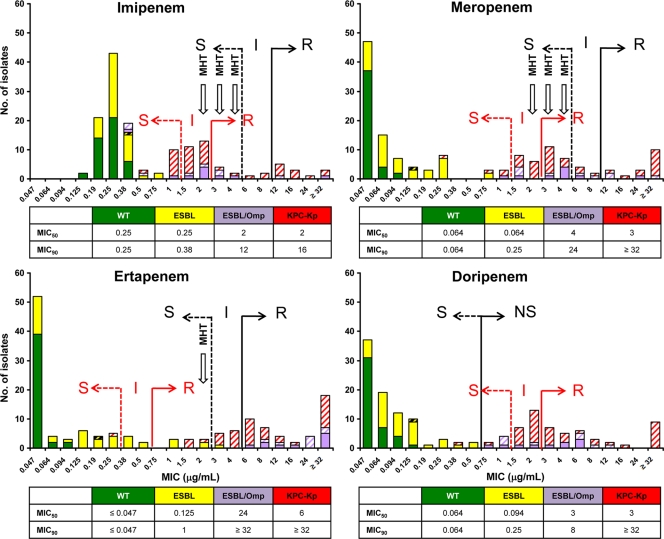
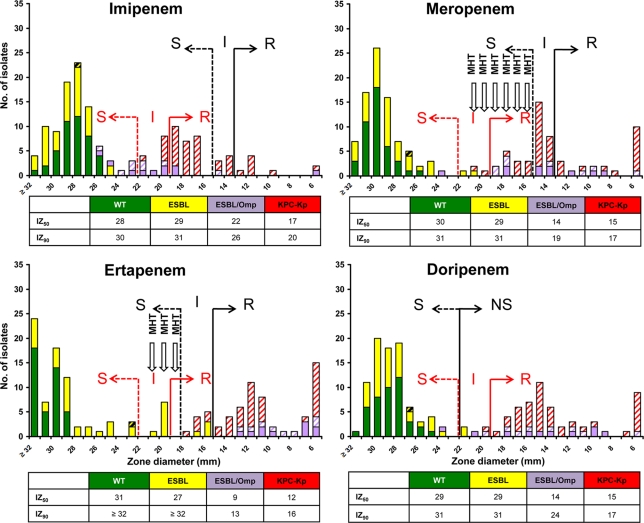
Histograms of MIC distributions of the four carbapenems by BMD and Etest and of zone diameters by DD are shown in Fig. 1, 2, and 3. These show bimodal distributions for all four carbapenems, with the more-susceptible peak including the WT and ESBL groups and the less-susceptible peak including the ESBL/Omp and KPC-Kp groups, with some overlap between the two distributions that varied by method and carbapenem.
FIG. 1.
Histograms of broth microdilution MICs (μg/ml) of the four carbapenems studied against the 145 K. pneumoniae isolates. MIC50 and MIC90 values (μg/ml) are also shown. The original (black) and updated (red) susceptibility breakpoints for imipenem, meropenem, doripenem, and ertapenem are indicated (6, 7). S, susceptible; I, intermediate; R, resistant; NS, nonsusceptible; MHT, modified Hodge test. Arrows labeled MHT indicate when the MHT should be performed. WT K. pneumoniae isolates are shown in green, ESBL-positive (ESBL) isolates are shown in yellow, ESBL-positive isolates with Omp loss (ESBL/Omp) are shown in lavender, and blaKPC-possessing (KPC-Kp) isolates are shown in red. All isolates that tested positive with the MHT are designated by diagonal hatching.
FIG. 2.
Histograms of Etest MICs (μg/ml) of the four carbapenems studied against the 145 K. pneumoniae isolates. Abbreviations and colors used are the same as those in Fig. 1.
FIG. 3.
Histograms of disk diffusion inhibitory zones (IZ) (mm) of the four carbapenems studied against the 145 K. pneumoniae isolates. IZ50 and IZ90 values (mm) are also shown. Abbreviations and colors used are the same as those in Fig. 1.
Application of previous interpretative criteria showed that all WT isolates were categorized as susceptible to IPM, MEM, ERT, and DOR, regardless of the phenotypic test used. In contrast, differences among methodologies were observed for the remaining three groups of K. pneumoniae isolates. By BMD, all ESBL-positive isolates were susceptible to IPM and MEM, whereas two strains (4.8%) were nonsusceptible to ERT and DOR (Fig. 1). Six (33.3%) ESBL/Omp isolates were nonsusceptible to IPM, and 10 (55.6%) were nonsusceptible to MEM; all were resistant to ERT and nonsusceptible to DOR. One (2.4%) KPC-Kp isolate was susceptible to IPM, and two (4.8%) were nonsusceptible to MEM, whereas all isolates were nonsusceptible to ERT and DOR. By Etest, all ESBL isolates were susceptible to IPM, MEM, and DOR, whereas one (2.4%) strain was intermediate for ERT (Fig. 2). Two (11.1%) ESBL/Omp isolates were nonsusceptible to IPM, and seven (38.9%) were nonsusceptible to MEM; all were resistant to ERT and nonsusceptible to DOR. Twenty-nine (69.0%) KPC-Kp isolates were susceptible to IPM, 26 (61.9%) were susceptible to MEM, 5 (11.9%) were susceptible to ERT, and 1 (2.4%) was susceptible to DOR. By DD, all ESBL isolates were susceptible to IPM and MEM, whereas four (9.5%) isolates were nonsusceptible to ERT and two (4.8%) were nonsusceptible to DOR (Fig. 3). Two (11.1%) ESBL/Omp isolates were nonsusceptible to IPM, 11 (61.1%) were nonsusceptible to MEM, all were nonsusceptible to ERT, and 1 (5.6%) was nonsusceptible to DOR. Twenty-seven (62.3%) KPC-Kp isolates were susceptible to IPM, nine (21.4%) were susceptible to MEM, and none were susceptible to ERT or DOR.
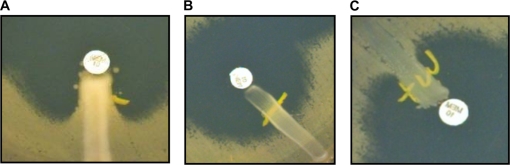
The MHT performed with IPM was positive for all KPC-Kp isolates and for 14 (9.7%) non-KPC-Kp isolates (11 ESBL/Omp strains and 3 ESBL-positive strains). The MHT with MEM was positive for all KPC-Kp isolates and for 10 (6.9%) non-KPC-Kp isolates (9 ESBL/Omp strains and 1 ESBL-positive strain). Examples of MHT results are shown in Fig. 4. Further testing to detect other carbapenemases was then performed on the 10 blaKPC-negative strains with positive MHT results for both IPM and MEM; all were negative for blaVIM, blaIPM, blaSPM, blaSME, blaOXA-48, blaGIM, blaSIM, blaIMI, blaNMC, and blaGES genes by PCR. Furthermore, none of them showed a decrease in the zone of inhibition of IPM plus crude β-lactamase extract compared to that of IPM alone.
FIG. 4.
Representative pictures of MHT performed with MEM. (A) Strongly positive MHT for a blaKPC-possessing isolate. (B) Negative MHT for a WT isolate. (C) Weakly positive MHT for an isolate producing ESBL and showing Omp loss.
Sensitivities and specificities of original and updated criteria in differentiating KPC-Kp isolates from the other three groups.
The ability of the 2009, 2010, and updated 2010 CLSI criteria to differentiate between the KPC-Kp group and the other three groups was evaluated with the 145 study isolates. For these analyses, the strains in the KPC-Kp group were classified as true positive strains, whereas strains in the other three groups were classified as true negative strains if characterized as carbapenem susceptible and as false-negative strains if not. Results of these analyses are presented in Table 4. When the original CLSI/FDA criteria were used, testing by the BMD method showed good sensitivity (100%) but limited specificity (83 to 92%), regardless of the carbapenem tested. Similar results were obtained by the Etest and DD methods. Applying the updated 2010 CLSI criteria resulted in 100% sensitivity for all four carbapenems by BMD and DD but in a lower sensitivity by Etest (84 to 90%); specificities for all three methods ranged from 84 to 90% for IPM, MEM, and DOR and from 76 to 77% for ERT. The sensitivity and specificity of MHT alone, performed with IPM and MEM, were 100% and 88 to 91%, respectively.
TABLE 4.
Statistical analysis of the different phenotypic tests used to differentiate carbapenemase-producing K. pneumoniae isolates (n = 42) from non-carbapenemase producers (n = 103)a
| Method | Interpretative criteria | Sensitivity (%)b |
Specificity (%)b |
||||||
|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ERT | DOR | IPM | MEM | ERT | DOR | ||
| BMD | Original CLSI/FDA criteriac | 100 | 100 | 100 | 100 | 92.0 | 89.6 | 83.1 | 83.7 |
| Updated 2010 CLSI criteriad | 100 | 100 | 100 | 100 | 86.6 | 86.6 | 76.3 | 85.1 | |
| Etest | Original CLSI/FDA criteriac | 100 | 100 | 100 | 97.7 | 95.4 | 92.8 | 84.4 | 85.1 |
| Updated 2010 CLSI criteriad | 80.8 | 91.3 | 97.7 | 95.5 | 88.8 | 85.8 | 77.4 | 88.8 | |
| DD | Original CLSI/FDA criteriac | NA | 100 | 100 | 100 | NA | 87.3 | 82.4 | 84.4 |
| Updated 2010 CLSI criteriad | 100 | 100 | 100 | 100 | 90.4 | 84.4 | 77.4 | 84.4 | |
| MHT alonee | NA | 100 | 100 | NA | NA | 88.0 | 91.2 | NA | NA |
BMD, broth microdilution; MHT, modified Hodge test; DD, disk diffusion; IPM, imipenem; MEM, meropenem; ERT, ertapenem; DOR, doripenem.
Sensitivity = TP/(TP + FN); specificity = TN/(TN+FP). TP, number of carbapenemase producers (n = 42); TN, number of non-carbapenemase producers (n = 103); TP, number of true positive results; TN, number of true negative results; FP, number of false-positive results; FN, number of false-negative results.
Based on original CLSI criteria for IPM, MEM, and ERT, with MHT performed with meropenem, and on FDA criteria for DOR (see Table 1).
Based on updated CLSI criteria (see Table 2).
Ability of MHT performed on all strains (n = 145), using IPM and MEM disks, to identify carbapenemase-producing isolates correctly.
Differentiation of carbapenem-susceptible from carbapenem-nonsusceptible groups.
The clinical significance of the above sensitivity and specificity analyses, which address differentiation of the KPC-Kp group from the other three groups, is confounded by the carbapenem nonsusceptibility of the ESBL/Omp group. A more clinically relevant analysis is differentiation of the two carbapenem-susceptible groups (WT and ESBL-producing strains) from the two carbapenem-nonsusceptible groups (ESBL/Omp and KPC-Kp isolates). Application of updated CLSI criteria accurately characterized all WT isolates by all three methods (Table 5). The ESBL group was also accurately characterized by all three methods with IPM, MEM, and DOR, but not with ERT, where one-half to one-third of isolates were characterized as nonsusceptible. All ESBL/Omp strains were characterized as nonsusceptible to ERT by all three methods and as nonsusceptible to DOR by BMD; up to 7 of the 18 strains in this group were susceptible with other combinations. All KPC isolates were categorized as nonsusceptible by BMD and DD. However, this was not the case by Etest, where between 1 and 10 strains were classified as susceptible, with most of the misclassified strains falling at the upper limit of the susceptible range.
TABLE 5.
Accuracy of updated 2010 susceptibility breakpoints in differentiating between carbapenem-susceptible groups (WT strains and ESBL producers) and groups with decreased carbapenem susceptibility (ESBL/Omp and KPC-Kp isolates)
| Group (n) | Method | No. of susceptible isolates/no. of resistant isolates |
|||
|---|---|---|---|---|---|
| Imipenem | Meropenem | Ertapenem | Doripenem | ||
| WT (43) | BMDa | 43/0 | 43/0 | 43/0 | 43/0 |
| Etestb | 43/0 | 43/0 | 43/0 | 43/0 | |
| DDc | 43/0 | 43/0 | 43/0 | 43/0 | |
| ESBL producers (42) | BMDa | 40/2 | 40/2 | 28/14 | 40/2 |
| Etestb | 40/2 | 40/2 | 30/12 | 42/0 | |
| DDc | 42/0 | 40/2 | 30/12 | 40/2 | |
| ESBL/Omp strains (18) | BMDa | 4/14 | 2/16 | 0/18 | 0/18 |
| Etestb | 5/13 | 1/17 | 0/18 | 5/13 | |
| DDc | 7/11 | 1/17 | 0/18 | 1/17 | |
| KPC-Kp isolates (42) | BMDa | 0/42 | 0/42 | 0/42 | 0/42 |
| Etestb | 10/32 | 4/38 | 1/41 | 2/40 | |
| DDc | 0/42 | 0/42 | 0/42 | 0/42 | |
Based on MIC breakpoints of ≤1 μg/ml (susceptible) and ≥2 μg/ml (nonsusceptible) for IPM, MEM, and DOR and ≤0.25 μg/ml (susceptible) and ≥0.5 μg/ml (nonsusceptible) for ERT.
Based on MIC breakpoints of ≤1 μg/ml (susceptible) and ≥1.5 μg/ml (nonsusceptible) for IPM, MEM, and DOR and ≤0.25 μg/ml (susceptible) and ≥0.38 μg/ml (nonsusceptible) for ERT.
Based on disk diffusion breakpoints of ≥23 mm (susceptible) and ≤22 mm (nonsusceptible) for IPM, MEM, ERT, and DOR.
DISCUSSION
Production of carbapenemases is an increasing problem among clinical isolates of Gram-negative bacilli (40). In particular, the spread of KPC-Kp isolates represents a serious threat to our therapeutic armamentarium, as these isolates are resistant to all currently available β-lactams and β-lactam-β-lactamase inhibitor combinations. Prevalence rates of KPC-Kp isolates of >30% have been recorded in some institutions in the eastern United States, in association with nosocomial outbreaks (24, 33). Detection and reporting of KPC-Kp isolates pose a significant challenge to clinical microbiologists, since these organisms may be difficult to detect using standard methods (33, 39). Endimiani et al. reported that approximately 60% of KPC-Kp isolates have IPM or MEM MICs in the susceptible range (15). This low-level expression of the resistance mechanism is the main reason that automated and nonautomated phenotypic tests have difficulty in detecting KPC-Kp isolates (1, 28). In contrast to the case for IPM and MEM, KPC-Kp strains frequently have ERT MICs in the resistant range (2, 15). Thus, the use of ERT has been suggested to screen for KPC production among Enterobacteriaceae (1, 2, 33). Nevertheless, recent papers have reported increased numbers of ERT-resistant but IPM- or MEM-susceptible K. pneumoniae isolates that are not carbapenemase producers (22, 36). These isolates are associated with loss or decreased expression of outer membrane porins combined with production of ESBLs (11, 25). These strains are responsible for the low specificity recorded for ERT and the MHT in detecting KPC-Kp isolates (4, 28).
CLSI modified the carbapenem interpretations for Enterobacteriaceae in 2009, recommending performance of the MHT, using either MEM or ERT disks, for strains with susceptibilities near susceptibility breakpoints (Table 1) (5). However, performing the MHT is time-consuming and delays the availability of results by at least 24 h. Additionally, the MHT is sometimes difficult to interpret, and false-positive results have been reported (4, 26, 34). Use of a phenotypic test using boronic acid distinguished between KPC-Kp isolates and strains not harboring KPC enzymes (10, 34, 42). Boronic acid may offer some advantage in accuracy over the MHT but offers no time advantage, as an additional day is needed before results are available. CLSI updated the carbapenem breakpoints for Enterobacteriaceae in June 2010, eliminating the need for the MHT and further lowering the ERT breakpoints (7).
In the present work, we evaluated the original and updated criteria for determining carbapenem susceptibility, testing a large collection of well-characterized K. pneumoniae isolates. Along with WT strains, KPC-Kp isolates, and ESBL producers, a fourth group of K. pneumoniae isolates was investigated. These isolates were categorized as ESBL/Omp strains on the basis of their carbapenem-resistant phenotype and molecular analysis of ompK genes.
Based on our results, standard phenotypic susceptibility testing by BMD and DD differentiated most carbapenem-susceptible from carbapenem-nonsusceptible K. pneumoniae isolates, based on the updated CLSI criteria (Fig. 1 to 3 and Table 5). However, the Etest method characterized many KPC-Kp isolates as susceptible, and breakpoints may need to be lowered for this method.
However, several issues are presented by these findings. First, if susceptibility testing is performed by currently available breakpoint MIC methods, including automated systems, such as MicroScan and Vitek methods, many KPC-Kp isolates will be misclassified as susceptible and additional testing will continue to be needed, significantly increasing the turnaround time. Second, many KPC-Kp and ESBL/Omp strains are not distinguished from each other, which may unnecessarily limit the use of carbapenems with ESBL/Omp strains with MICs within pharmacokinetic targets. Third, the MHT does not accurately distinguish between KPC-Kp and ESBL/Omp strains, as many in the latter group are MHT positive. In our study, half of the ESBL/Omp isolates were MHT positive when MEM was employed as suggested by the CLSI. These strains were confirmed as non-carbapenemase producers on the basis of molecular and disk enzymatic assay analyses. This finding has been reported previously, but a clear explanation for this phenomenon is still lacking (4, 34).
The importance of these issues depends on the clinical utility of carbapenems in treating infections due to KPC-Kp and ESBL/Omp isolates. Based upon our findings, an understanding of the enzymatic mechanism of resistance mediated by KPC, and the initial limited expression of KPC by many strains, resulting in MICs that are raised but still in the susceptible range, patients infected with KPC-producing K. pneumoniae are unlikely to respond to carbapenems, even if these antibiotics are dosed according to the level of susceptibility manifested by the pathogen and appropriate pharmacokinetic/pharmacodynamic targets are achieved (37, 43). On the other hand, infections due to isolates with Omp loss and coproduction of ESBLs might be treatable with carbapenems if MICs are in the susceptible range based on pharmacokinetic/pharmacodynamic targets (17, 18, 21, 23, 29-32, 38). Further studies are needed to define these relationships and to determine if there is a clinical need to differentiate between the KPC-Kp and ESBL/Omp groups.
Acknowledgments
This work was supported in part by the Veterans Affairs Merit Review Program (R.A.B.), the National Institutes of Health (grant RO1-AI063517 to R.A.B.), the Geriatric Research Education and Clinical Center (VISN 10 to R.A.B.), and GlaxoSmithKline (M.R.J.).
We thank David L. Paterson, Stephen G. Jenkins, and Geraldine S. Hall for the kind gift of isolates and Ortho McNeil for providing microdilution trays.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Anderson, K. F., D. R. Lonsway, J. K. Rasheed, J. Biddle, B. Jensen, L. K. McDougal, R. B. Carey, A. Thompson, S. Stocker, B. Limbago, and J. B. Patel. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 45:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bratu, S., M. Mooty, S. Nichani, D. Landman, C. Gullans, B. Pettinato, U. Karumudi, P. Tolaney, and J. Quale. 2005. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob. Agents Chemother. 49:3018-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrer, A., L. Poirel, H. Eraksoy, A. A. Cagatay, S. Badur, and P. Nordmann. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52:2950-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalhaes, C. G., R. C. Picao, A. G. Nicoletti, D. E. Xavier, and A. C. Gales. 2010. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false-positive results. J. Antimicrob. Chemother. 65:249-251. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. 2009. Performance standards for antimicrobial susceptibility testing: 19th informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.CLSI. 2010. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.CLSI. 2010. Performance standards for antimicrobial susceptibility testing; update. CLSI document M100-S20 June 2010 update. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Deshpande, L. M., R. N. Jones, T. R. Fritsche, and H. S. Sader. 2006. Occurrence and characterization of carbapenemase-producing Enterobacteriaceae: report from the SENTRY Antimicrobial Surveillance Program (2000-2004). Microb. Drug Resist. 12:223-230. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande, L. M., P. R. Rhomberg, H. S. Sader, and R. N. Jones. 2006. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 56:367-372. [DOI] [PubMed] [Google Scholar]
- 10.Doi, Y., B. A. Potoski, J. M. Adams-Haduch, H. E. Sidjabat, A. W. Pasculle, and D. L. Paterson. 2008. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type beta-lactamase by use of a boronic acid compound. J. Clin. Microbiol. 46:4083-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doumith, M., M. J. Ellington, D. M. Livermore, and N. Woodford. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659-667. [DOI] [PubMed] [Google Scholar]
- 12.Endimiani, A., L. L. Carias, A. M. Hujer, C. R. Bethel, K. M. Hujer, F. Perez, R. A. Hutton, W. R. Fox, G. S. Hall, M. R. Jacobs, D. L. Paterson, L. B. Rice, S. G. Jenkins, F. C. Tenover, and R. A. Bonomo. 2008. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob. Agents Chemother. 52:2680-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endimiani, A., Y. Choudhary, and R. A. Bonomo. 2009. In vitro activity of NXL104 in combination with beta-lactams against Klebsiella pneumoniae isolates producing KPC carbapenemases. Antimicrob. Agents Chemother. 53:3599-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endimiani, A., J. M. Depasquale, S. Forero, F. Perez, A. M. Hujer, D. Roberts-Pollack, P. D. Fiorella, N. Pickens, B. Kitchel, A. E. Casiano-Colon, F. C. Tenover, and R. A. Bonomo. 2009. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J. Antimicrob. Chemother. 64:1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endimiani, A., A. M. Hujer, F. Perez, C. R. Bethel, K. M. Hujer, J. Kroeger, M. Oethinger, D. L. Paterson, M. D. Adams, M. R. Jacobs, D. J. Diekema, G. S. Hall, S. G. Jenkins, L. B. Rice, F. C. Tenover, and R. A. Bonomo. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern U.S.A. J. Antimicrob. Chemother. 63:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endimiani, A., K. M. Hujer, A. M. Hujer, E. S. Armstrong, Y. Choudhary, J. B. Aggen, and R. A. Bonomo. 2009. ACHN-490, a neoglycoside with potent in vitro activity against multidrug-resistant Klebsiella pneumoniae isolates. Antimicrob. Agents Chemother. 53:4504-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frasca, D., S. Marchand, F. Petitpas, C. Dahyot-Fizelier, W. Couet, and O. Mimoz. 2010. Pharmacokinetics of ertapenem following intravenous and subcutaneous infusions in patients. Antimicrob. Agents Chemother. 54:924-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frei, C. R., N. P. Wiederhold, and D. S. Burgess. 2008. Antimicrobial breakpoints for gram-negative aerobic bacteria based on pharmacokinetic-pharmacodynamic models with Monte Carlo simulation. J. Antimicrob. Chemother. 61:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoban, D. J., S. K. Bouchillon, S. P. Hawser, and R. E. Badal. 2010. Trends in the frequency of multiple drug-resistant Enterobacteriaceae and their susceptibility to ertapenem, imipenem, and other antimicrobial agents: data from the Study for Monitoring Antimicrobial Resistance Trends 2002 to 2007. Diagn. Microbiol. Infect. Dis. 66:78-86. [DOI] [PubMed] [Google Scholar]
- 20.Jacoby, G. A., D. M. Mills, and N. Chow. 2004. Role of beta-lactamases and porins in resistance to ertapenem and other beta-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaruratanasirikul, S., and T. Sudsai. 2009. Comparison of the pharmacodynamics of imipenem in patients with ventilator-associated pneumonia following administration by 2 or 0.5 h infusion. J. Antimicrob. Chemother. 63:560-563. [DOI] [PubMed] [Google Scholar]
- 22.Jones, R. N., H. S. Sader, and T. R. Fritsche. 2005. Comparative activity of doripenem and three other carbapenems tested against Gram-negative bacilli with various beta-lactamase resistance mechanisms. Diagn. Microbiol. Infect. Dis. 52:71-74. [DOI] [PubMed] [Google Scholar]
- 23.Keam, S. J. 2008. Doripenem: a review of its use in the treatment of bacterial infections. Drugs 68:2021-2057. [DOI] [PubMed] [Google Scholar]
- 24.Landman, D., S. Bratu, S. Kochar, M. Panwar, M. Trehan, M. Doymaz, and J. Quale. 2007. Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. J. Antimicrob. Chemother. 60:78-82. [DOI] [PubMed] [Google Scholar]
- 25.Leavitt, A., I. Chmelnitsky, R. Colodner, I. Ofek, Y. Carmeli, and S. Navon-Venezia. 2009. Ertapenem resistance among extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae isolates. J. Clin. Microbiol. 47:969-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Yong, and J. H. Yum. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88-91. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Martinez, L. 2008. Extended-spectrum beta-lactamases and the permeability barrier. Clin. Microbiol. Infect. 14(Suppl. 1):82-89. [DOI] [PubMed] [Google Scholar]
- 28.McGettigan, S. E., K. Andreacchio, and P. H. Edelstein. 2009. Specificity of ertapenem susceptibility screening for detection of Klebsiella pneumoniae carbapenemases. J. Clin. Microbiol. 47:785-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moczygemba, L. R., C. R. Frei, and D. S. Burgess. 2004. Pharmacodynamic modeling of carbapenems and fluoroquinolones against bacteria that produce extended-spectrum beta-lactamases. Clin. Ther. 26:1800-1807. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, T., C. Shimizu, M. Kasahara, K. Okuda, C. Nakata, H. Fujimoto, H. Okura, M. Komatsu, K. Shimakawa, N. Sueyoshi, T. Ura, K. Satoh, M. Toyokawa, Y. Wada, T. Orita, T. Kofuku, K. Yamasaki, M. Sakamoto, H. Nishio, S. Kinoshita, and H. Takahashi. 2009. Monte Carlo simulation for evaluation of the efficacy of carbapenems and new quinolones against ESBL-producing Escherichia coli. J. Infect. Chemother. 15:13-17. [DOI] [PubMed] [Google Scholar]
- 31.Nicolau, D. P. 2008. Carbapenems: a potent class of antibiotics. Expert Opin. Pharmacother. 9:23-37. [DOI] [PubMed] [Google Scholar]
- 32.Nicolau, D. P. 2008. Pharmacodynamic optimization of beta-lactams in the patient care setting. Crit. Care 12(Suppl. 4):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 34.Pasteran, F., T. Mendez, L. Guerriero, M. Rapoport, and A. Corso. 2009. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J. Clin. Microbiol. 47:1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, M. D. Bonomo, L. B. Rice, and R. A. Bonomo. 2003. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson, D. L., L. B. Rice, and R. A. Bonomo. 2001. Rapid method of extraction and analysis of extended-spectrum beta-lactamases from clinical strains of Klebsiella pneumoniae. Clin. Microbiol. Infect. 7:709-711. [PubMed] [Google Scholar]
- 37.Perez, F., A. Endimiani, A. J. Ray, B. K. Decker, C. J. Wallace, K. M. Hujer, D. J. Ecker, M. D. Adams, P. Toltzis, M. J. Dul, A. Windau, S. Bajaksouzian, M. R. Jacobs, R. A. Salata, and R. A. Bonomo. 2010. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J. Antimicrob. Chemother. 65:1807-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrott, J., V. H. Mabasa, and M. H. Ensom. 2010. Comparing outcomes of meropenem administration strategies based on pharmacokinetic and pharmacodynamic principles: a qualitative systematic review. Ann. Pharmacother. 44:557-564. [DOI] [PubMed] [Google Scholar]
- 39.Quale, J. 2008. Global spread of carbapenemase-producing Klebsiella pneumoniae. Microbe 3:516-520. [Google Scholar]
- 40.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robledo, I. E., E. E. Aquino, M. I. Sante, J. L. Santana, D. M. Otero, C. F. Leon, and G. J. Vazquez. 2009. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsakris, A., I. Kristo, A. Poulou, K. Themeli-Digalaki, A. Ikonomidis, D. Petropoulou, S. Pournaras, and D. Sofianou. 2009. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J. Clin. Microbiol. 47:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weisenberg, S. A., D. J. Morgan, R. Espinal-Witter, and D. H. Larone. 2009. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn. Microbiol. Infect. Dis. 64:233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]