Abstract
We used a whole-genome scanning technique to identify the NADH dehydrogenase gamma subunit (nuoG) primer set that is sensitive and specific enough to detect a diverse number of Bartonella species in a wide range of environmental samples yet maintains minimal cross-reactivity to mammalian host and arthropod vector organisms.
Bacteria in the genus Bartonella are found in a wide variety of mammalian hosts (2, 8, 18, 20, 21) and are thought to be transmitted by arthropod vectors, including fleas, ticks, and possibly mites (4, 7, 9, 14, 16); humans serve as accidental hosts. Of the at least 20 named Bartonella species, 10 have been shown to cause disease in humans, including Carrion's disease (13), cat scratch disease (7, 14, 25), endocarditis (6, 11), and recently a febrile illness in humans from Thailand (caused by Bartonella tamiae) (17). Because of their wide distribution and potential for frequent contact with humans, many Bartonella species are considered potential emerging pathogens (1, 26, 28).
Bartonella identification requires the ability to detect bacteria in both mammalian hosts and arthropod vectors. Although bacterial culture is considered ideal, the difficulty and time involved make it impractical for large-scale use. Additionally, nucleic acid-based detection techniques may be hindered by inhibitors in environmental and clinical samples, low sensitivity, and the absence of genus-specific primers (10, 27).
To address these issues, we used whole-genome scanning based on the complete genomes of Bartonella bacilliformis, B. henselae, and B. quintana to identify host- and vector-blind primer sets for real-time PCR detection of Bartonella in various field-collected samples. We identified a primer set based on the NADH dehydrogenase gamma subunit (nuoG) that is specific for Bartonella species and sensitive enough to detect Bartonella in both mammalian hosts and arthropod vectors.
Identification of host-blind primer sets.
A whole-genome scan was performed on complete genomic sequences from B. henselae and B. quintana and shotgun sequences from B. bacilliformis available in GenBank. Each subsequence of 16, 17, 18, and 19 nucleotides present in published Bartonella genomes was compared with subsequences from other genomes present in GenBank, including genomes for bacteria that could infect human blood and tissues and potential mammalian hosts and arthropod vectors for bartonellae. The number of base changes necessary to convert each Bartonella subsequence to the closest subsequence in the background collection was calculated to identify potential primers with a reduced probability of hybridizing to and amplifying nontarget DNA.
In total, one ultraspecific, host-blind primer pair (the nuoG primer pair) was identified that met the following conditions: the pair (i) maintained at least a 2-base specificity among the complete GenBank sequence database, (ii) amplified fragments of identical sizes in the B. henselae and B. quintana genomes, (iii) had predicted amplicon sizes of less than 400 bp, and (iv) had primer melting temperatures (Tms) within 2°C. Although they did not conform to all of these conditions, the ftsZ and gltA primer sets were included in further comparisons due to the large amount of sequence data available for these genes.
Primer pairs were tested in reaction with three Bartonella species (B. henselae, B. quintana, and B. bacilliformis) and then with the use of ∼30-fold excess competitor DNA from J774 (murine) and THP1 (human) tissue culture cells over the template DNA from B. henselae. Interestingly, despite their common use, the gltA primer set demonstrated high cross-reactivity both to potential Bartonella hosts (Rattus spp., Mus spp., and Homo sapiens) and to bacterial species, such as Ehrlichia spp., that could inhabit similar ecological niches (Table 1).
TABLE 1.
Details of primers used in this studya
| Gene and orientation | Nucleotide sequence | Primer Tm | Amplicon size (bp) | Expected amplification result | Other species carrying the gene |
|---|---|---|---|---|---|
| gltA | B. henselae, B. quintana | Legionella pneumophila, Erlichia spp., | |||
| F | GGGGACCAGCTCATGGTGG | 57.58 | 340 | Alkalilimnicola ehrlichei, Mus | |
| R | AATGCAAAAAGAACAGTAAACA | 56.68 | musculus, Rattus norvegicus, Homo sapiens | ||
| nuoG | B. henselae, B. quintana, | None | |||
| F | GGCGTGATTGTTCTCGTTA | 55.56 | 346 | B. bacilliformis | |
| R | CACGACCACGGCTATCAAT | 56.68 | |||
| ftsZ | B. henselae, B. quintana | None | |||
| F | CGCATAGAAGTATCATCCA | 50.72 | 753 | ||
| R | ACGATTAATCTGCATCGGC | 53.99 |
Expected amplification results and occurrences in other genomes were determined by whole-genome scanning versus B. henselae, B. bacilliformis, and B. quintana. F, forward; R, reverse.
Amplification performance of the nuoG, gltA, and ftsZ primer sets against reference Bartonella DNA and environmental samples.
The nuoG, gltA, and ftsZ primer sets were used to amplify reference DNAs from 11 Bartonella species, chosen for their distant phylogenetic relationships, under conditions optimized for each primer set. The amplification results differed considerably between primer sets and species of Bartonella being amplified (Table 2) and are as follows: the nuoG primer set performed best (amplifying first, with the lowest threshold cycle [CT] value) on 3 of the 11 tested species, the gltA set performed best on 7 of the 11 species, and the ftsZ set performed best on 1 of the 11 species. Although the gltA primer set performed best for the highest number of reference species, only the nuoG set successfully amplified all 11 species.
TABLE 2.
CT values for the three primer sets resulting from amplification of 11 reference DNAs derived from culture samplesa
| Species |
CT |
||
|---|---|---|---|
| nuoG1 primer set | gltA primer set | ftsZ72 primer set | |
| Cotton rat sp. A1 | 26.1 | 28.6 | 23.8 |
| Cotton rat sp. C1 | 17.1 | 18.6 | 34.5 |
| Bartonella grahamii | 21.2 | 18.3 | 21.8 |
| Bartonella phoceensis | 25.5 | 28.2 | 29.8 |
| B. rattimassiliensis | 21.9 | 21.0 | 22.0 |
| B. tamiae | 37.7 | 29.9 | NA |
| B. tribocorum | 28.3 | 11.8 | 19.8 |
| B. vinsonii subsp. arupensis | 17.9 | 15.3 | 17.7 |
| B. vinsonii subsp. berkhoffii | 26.7 | 14.9 | 18.9 |
| B. vinsonii subsp. vinsonii | 22.4 | 22.0 | 21.9 |
| Bartonella washoensis | 16.9 | NA | 19.5 |
Values for those cases with more than 3.3 cycles (1-log starting quantity) are in bold. CT values higher than 35 are considered not valid (NA), due to the potential influence of primer dimers.
The primer sets were next tested against a panel of DNA from field-collected samples (purified from liver samples from Nepalese rats and ticks from Colombia) to determine their efficacy in detecting Bartonella DNA in field-collected hosts and vectors. Consistent with the predicted specificities from the whole-genome scans, the nuoG primer set demonstrated significantly higher sensitivity and specificity for Bartonella than the other primer sets by consistently yielding more sequence-confirmed PCR-positive results (Table 3). For the 61 total ticks sampled, the nuoG primer set yielded 7 Bartonella-positive samples, compared to 1 and 0 for the ftsZ and gltA sets, respectively. Of 24 total rodent liver samples tested, 18 were found to be Bartonella positive by the nuoG primer set, compared to 10 and 2 for the ftsZ and gltA sets, respectively.
TABLE 3.
Bartonella-positive samples, as verified by sequencing, based on primer set, for field-collected samples
| Primer set | No. of Colombian ticks (%) (n = 61) | No. of Nepal rodent livers (%) (n = 24) |
|---|---|---|
| gltA set | 0 (0) | 2 (9) |
| ftsZ72 set | 1 (2) | 10 (42) |
| nuoG set | 7 (11) | 18 (75) |
Phylogenetic analysis.
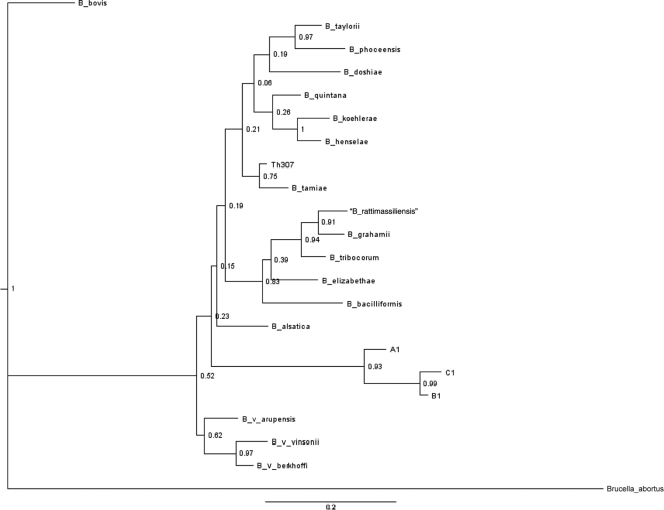
Analysis of a nuoG-derived phylogeny showed strong statistical support for the following clades: B. henselae and Bartonella koehlerae; the species found in Rattus and related hosts, including Bartonella elizabethae, B. rattimassiliensis, and B. tribocorum; 3 Bartonella vinsonii subspecies (Bartonella vinsonii subsp. arupensis, B. vinsonii subsp. vinsonii, and B. vinsonii subsp. berkhoffii); and two strains of B. tamiae, described to occur in febrile Thai patients (type strain Th239 and strain Th307) (Fig. 1). All of these species groups share high genetic similarity within their respective clades, suggesting that the nuoG primer set provides better phylogenetic estimation with closely related species. Bartonella bovis was placed extremely distant to the other Bartonella species, with strong statistical support; conversely, B. bacilliformis was placed more centrally within the phylogeny than is seen with other genes, though this placement did not have strong statistical support. These placements, which are different from those generated with multiple concatenated Bartonella sequences (Fig. 1) (17), are likely due to the genetic rearrangements and horizontal gene transfer events that commonly occur in Bartonella (3, 12, 19, 23). Because of this, care should be taken when interpreting phylogenies based solely on nuoG sequences. A much more reliable approach is to include the nuoG sequence as one of many concatenated sequences to be used for phylogenetic analysis.
FIG. 1.
Bayesian analysis of the Bartonella phylogeny based on the nuoG gene sequence. The sequences were obtained either from direct sequencing of PCR products or from GenBank. The listed node values represent posterior probabilities; Brucella was used as the outgroup.
In summary, whole-genome scanning has allowed us to identify nuoG as a sensitive and specific target gene for use in detection of Bartonella species from various clinical and environmental specimens. nuoG's superior performance in identifying Bartonella species in field-collected samples makes it an ideal candidate for complementing the use of gltA and ftsZ on culture samples.
Footnotes
Published ahead of print on 6 October 2010.
REFERENCES
- 1.Badiaga, S., D. Raoult, and P. Brouqui. 2008. Preventing and controlling emerging and reemerging transmissible diseases in the homeless. Emerg. Infect. Dis. 14:1353-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, Y., M. Y. Kosoy, J. F. Cully, T. Bala, C. Ray, and S. K. Collinge. 2007. Acquisition of nonspecific Bartonella strains by the northern grasshopper mouse (Onychomys leucogaster). FEMS Microbiol. Ecol. 61:438-448. [DOI] [PubMed] [Google Scholar]
- 3.Berglund, E. C., A. C. Frank, A. Calteau, O. Vinnere Pettersson, F. Granberg, A. S. Eriksson, K. Naslund, M. Holmberg, H. Lindroos, and S. G. Andersson. 2009. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 5:e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billeter, S. A., P. P. Diniz, J. M. Battisti, U. G. Munderloh, E. B. Breitschwerdt, and M. G. Levy. 2009. Infection and replication of Bartonella species within a tick cell line. Exp. Appl. Acarol. 49:193-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Breitschwerdt, E. B., D. L. Kordick, D. E. Malarkey, B. Keene, T. L. Hadfield, and K. Wilson. 1995. Endocarditis in a dog due to infection with a novel Bartonella subspecies. J. Clin. Microbiol. 33:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomel, B. B., R. W. Kasten, K. Floyd-Hawkins, B. Chi, K. Yamamoto, J. Roberts-Wilson, A. N. Gurfield, R. C. Abbott, N. C. Pedersen, and J. E. Koehler. 1996. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34:1952-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomel, B. B., R. W. Kasten, J. B. Henn, and S. Molia. 2006. Bartonella infection in domestic cats and wild felids. Ann. N. Y. Acad. Sci. 1078:410-415. [DOI] [PubMed] [Google Scholar]
- 9.Cotte, V., S. Bonnet, D. Le Rhun, E. Le Naour, A. Chauvin, H. J. Boulouis, B. Lecuelle, T. Lilin, and M. Vayssier-Taussat. 2008. Transmission of Bartonella henselae by Ixodes ricinus. Emerg. Infect. Dis. 14:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doring, G., K. Unertl, and A. Heininger. 2008. Validation criteria for nucleic acid amplification techniques for bacterial infections. Clin. Chem. Lab. Med. 46:909-918. [DOI] [PubMed] [Google Scholar]
- 11.Drancourt, M., J. L. Mainardi, P. Brouqui, F. Vandenesch, A. Carta, F. Lehnert, J. Etienne, F. Goldstein, J. Acar, and D. Raoult. 1995. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N. Engl. J. Med. 332:419-423. [DOI] [PubMed] [Google Scholar]
- 12.Frank, A. C., C. M. Alsmark, M. Thollesson, and S. G. Andersson. 2005. Functional divergence and horizontal transfer of type IV secretion systems. Mol. Biol. Evol. 22:1325-1336. [DOI] [PubMed] [Google Scholar]
- 13.Herrer, A., and H. A. Christensen. 1975. Implication of Phlebotomus sand flies as vectors of bartonellosis and leishmaniasis as early as 1764. Science 190:154-155. [DOI] [PubMed] [Google Scholar]
- 14.Higgins, J. A., S. Radulovic, D. C. Jaworski, and A. F. Azad. 1996. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera:Pulicidae). J. Med. Entomol. 33:490-495. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Kim, C. M., J. Y. Kim, Y. H. Yi, M. J. Lee, M. R. Cho, D. H. Shah, T. A. Klein, H. C. Kim, J. W. Song, S. T. Chong, M. L. O'Guinn, J. S. Lee, I. Y. Lee, J. H. Park, and J. S. Chae. 2005. Detection of Bartonella species from ticks, mites and small mammals in Korea. J. Vet. Sci. 6:327-334. [PubMed] [Google Scholar]
- 17.Kosoy, M., C. Morway, K. W. Sheff, Y. Bai, J. Colborn, L. Chalcraft, S. F. Dowell, L. F. Peruski, S. A. Maloney, H. Baggett, S. Sutthirattana, A. Sidhirat, S. Maruyama, H. Kabeya, B. B. Chomel, R. Kasten, V. Popov, J. Robinson, A. Kruglov, and L. R. Petersen. 2008. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J. Clin. Microbiol. 46:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. E. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 19.Lindroos, H., O. Vinnere, A. Mira, D. Repsilber, K. Naslund, and S. G. Andersson. 2006. Genome rearrangements, deletions, and amplifications in the natural population of Bartonella henselae. J. Bacteriol. 188:7426-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maggi, R. G., B. Chomel, B. C. Hegarty, J. Henn, and E. B. Breitschwerdt. 2006. A Bartonella vinsonii berkhoffii typing scheme based upon 16S-23S ITS and Pap31 sequences from dog, coyote, gray fox, and human isolates. Mol. Cell. Probes 20:128-134. [DOI] [PubMed] [Google Scholar]
- 21.Maillard, R., M. Vayssier-Taussat, C. Bouillin, C. Gandoin, L. Halos, B. Chomel, Y. Piemont, and H. J. Boulouis. 2004. Identification of Bartonella strains isolated from wild and domestic ruminants by a single-step PCR analysis of the 16S-23S intergenic spacer region. Vet. Microbiol. 98:63-69. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Philippe, H., and C. J. Douady. 2003. Horizontal gene transfer and phylogenetics. Curr. Opin. Microbiol. 6:498-505. [DOI] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Thonnard, J., F. M. Carreer, and M. Delmee. 1994. Rochalimaea henselae, Afipia felis and cat-scratch disease. Acta Clin. Belg. 49:158-167. (In French.) [DOI] [PubMed] [Google Scholar]
- 26.Vorou, R. M., V. G. Papavassiliou, and S. Tsiodras. 2007. Emerging zoonoses and vector-borne infections affecting humans in Europe. Epidemiol. Infect. 135:1231-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiley, D. M., S. B. Lambert, S. Bialasiewicz, N. Goire, M. D. Nissen, and T. P. Sloots. 2008. False-negative results in nucleic acid amplification tests-do we need to routinely use two genetic targets in all assays to overcome problems caused by sequence variation? Crit. Rev. Microbiol. 34:71-76. [DOI] [PubMed] [Google Scholar]
- 28.Wormser, G. P. 2007. Discovery of new infectious diseases—Bartonella species. N. Engl. J. Med. 356:2346-2347. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.