Abstract
We report on a case of bacteremia caused by a previously unknown urease-negative Helicobacter strain, IMMIB HP-28/08, isolated from blood cultures of a 28-year-old man with X-linked agammaglobulinemia. The identification of the isolate was based on 16S rRNA gene sequencing. In the phylogenetic tree, the isolate fell into a cluster which included Helicobacter canadensis, Helicobacter equorum, and Helicobacter pullorum. This is the first report of bacteremia caused by this fastidious organism. Further investigations are necessary to determine the potential role of this species as a pathogen of bloodstream infections.
CASE REPORT
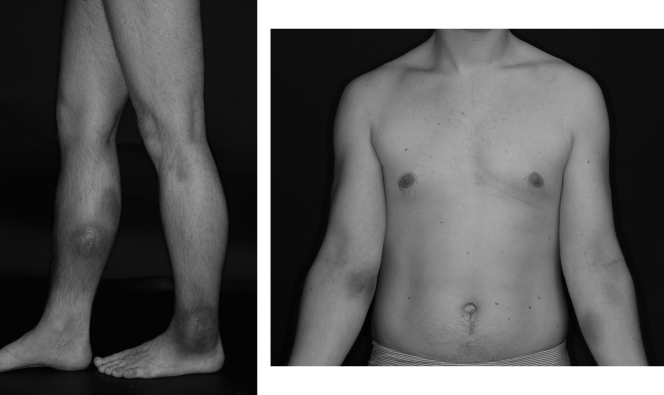
The patient was a 28-year-old man with X-linked agammaglobulinemia, which was diagnosed at age 8. He was started on monthly intravenous (i.v.) immunoglobulin therapy and remained relatively free of symptoms until 2004, when he noticed a demarcated, hyperpigmented, painful macule on the extensor surface of his right lower leg. He appeared healthy and received several empirical antibiotic therapies on suspicion of bacterial infection. He was admitted to our hospital in May 2006 with several hyperpigmented macules on the flexor surfaces of both forearms, his left ankle, and the extensor surface of his right lower leg and systemic symptoms of fatigue, night sweats, and pain in his right lower leg (Fig. 1). Laboratory analysis showed leucocytosis with a count of 23,700 leukocytes/liter with 89% neutrophils in the differential blood count, and the C-reactive protein (CRP) (115 mg/liter) and erythrocyte sedimentation rate (44 mm/h) were elevated. Several blood cultures and microbiological, virological, and autoimmune serological parameters remained negative. Next, the patient was treated empirically with multiple courses of antibiotics, including ampicillin-sulbactam, ciprofloxacin, clarithromycin, ceftazidime, and clindamycin. However, antibiotics were associated with only a brief improvement of symptoms. Skin biopsy gave the suspicion of fibrosing lymphocytic panniculitis, and thus, the patient was put on immunosuppressive therapy with tacrolimus in December 2006. Four weeks after starting immunosuppressive therapy, the patient had severe pains in his lower right leg and became septic. Blood cultures taken at this time signaled bacterial growth after 3 days of incubation. Gram staining revealed the presence of Gram-negative thin rods with a fusiform appearance. Later, this was identified by means of the 16S rRNA gene sequence analysis as a Helicobacter canadensis-like organism.
FIG. 1.
Views of upper and lower limbs showing hyperpigmented macules.
Subsequently, the patient was treated with intravenous amoxicillin and gentamicin. Laboratory results (CRP, erythrocyte sedimentation rate, leukocytes, and differential blood count) normalized rapidly; however, the hyperpigmented macula cleared only slowly. It took several courses of intravenous therapy, with amoxicillin and gentamicin switched to imipenem and fosfomycin due to ototoxicity associated with gentamicin, to prevent a recurrence of symptoms. After a total of 40 weeks of i.v. therapy, the patient was stable on an oral maintenance therapy with rifampin and doxycycline; the hyperpigmented maculae on his forearms and left ankle have resolved, and the hyperpigmentation on his right lower leg is slowly fading.
Microbiology.
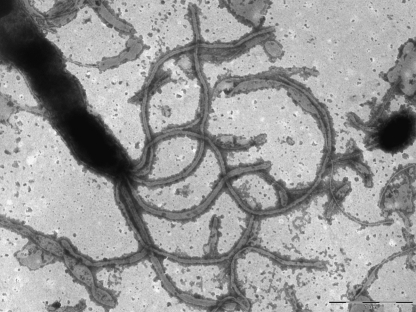
Blood cultures were incubated in an automated Bactec 9240 system (Becton Dickinson). One set of blood cultures consisted of two Bactec Plus aerobic/F bottles and one Bactec Plus anaerobic/F Bottle. The aerobic blood cultures (3 bottles out of 6) became positive after 3 days of incubation. Samples from three bottles were subcultured onto Columbia agar with 5% sheep blood (BD), Columbia agar with 5% human blood, Mueller-Hinton (MH) agar with 5% sheep blood, and chocolate agar. The plates were incubated under microaerophilic conditions at 37°C. After 6 days of incubation, moist, glassy, swarming colonies were observed on the agar plates. Microscopic examination of a Gram-stained preparation revealed the presence of Gram-negative, long, thin, straight or slightly curved to spiral-shaped rods. For examination of the ultrastructure of the cells, the isolate was grown on Columbia blood agar for 7 days, harvested from the plates, gently resuspended in Tryptone soya (TS) broth, and examined by electron microscopy using a Philips CM10 electron microscope at 80 kW. Negative staining of cells was performed with 1.0% phosphotungestic acid. Electron microscopy revealed curved- to spiral rod-shaped cells with 5 to 7 monopolar flagella (Fig. 2). Biochemical tests were performed using standard methods (2, 4). Significant morphological and biochemical reactions are shown in Table 1, along with those of the phylogenetically most-related Helicobacter species. The isolate was oxidase and catalase positive and reduced nitrate to nitrite but was negative for urease, hydrolysis of indoxyl acetate, and hippurate. The isolate was resistant to cephalothin (30 mg) and to nalidixic acid (30 mg). The isolate grew at 37°C but not at 30 or 42°C.
FIG. 2.
Transmission electron micrograph of a 7-day-old culture of strain IMMIB HP-28/08T on Columbia blood agar at 37°C. Note that the spirally curved cell has 5 monopolar flagella.
TABLE 1.
Biochemical and morphological characteristics of isolate IMMIB HP-28/08 in comparison with its phylogenetically closely related Helicobacter species
| Organism | Biochemical test result |
Distribution (no.) of flagella | Growth at 42°C | Susceptibility to: |
||||
|---|---|---|---|---|---|---|---|---|
| Catalase | Nitrate reduction | Urease | Hydrolysis of indoxyl acetate | Nalidixic acid | Cephalothin | |||
| IMMIB HP-28/08 | + | + | − | − | Monopolar (5-7) | − | Resistant | Resistant |
| H. canadensis | + | + | − | + | Bipolar (1-2) | + | Resistant | Resistant |
| H. equorum | + | + | − | − | Monopolar (1) | − | Resistant | Resistant |
| H. pullorum | + | + | − | − | Monopolar (1) | + | Resistant | Susceptible |
Identification of the organism was achieved by 16S rRNA sequence analysis. Genomic DNA extractions and PCR-mediated amplification of the 16S rRNA gene were performed on blood culture fluid (ca. 6 ml) from the positive-signaling Bactec 9240 blood culture and on pure culture of the isolate using established procedures (14). The purified PCR products were sequenced using a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems) as described by the manufacturer. The closest known relatives of the sequences were determined by performing database searches using the BLASTN 2.2.2 program. Phylogenetic analyses were performed using the ARB database (11). A phylogenetic tree was generated by the neighbor-joining method (15). Topologies of the neighbor-joining tree were evaluated using bootstrap analyses (3) based on 1,000 resamplings. It can be seen in Fig. 3 that the sequence of isolate IMMIB HP-28/08 was recovered within the Helicobacter 16S rRNA gene clade and clustered together with Helicobacter canadensis, Helicobacter equorum, and Helicobacter pullorum. However, the clustering of isolate IMMIB HP-28/08 with the aforementioned species is not statistically significant. The 16S rRNA gene sequence (1,458 nt) showed highest sequence similarities with H. pullorum (97.6%), H. canadensis (97.5%), and H. equorum (97.5%). According to the guidelines proposed by Stackebrandt and Goebel (18) and Stackbrandt and Ebers (17), a level of 97% or even a range above 98.7 to 99% sequence similarity in the 16S rRNA genes of two organisms would indicate they belong to separate species. Therefore, we believe the evidence favors the classification of our isolate as a novel species of Helicobacter.
FIG. 3.
Neighbor-joining tree showing the position of strain IMMIB HP-28/8T within the radiation of species of the genus Helicobacter. The numbers at the nodes indicate the level of bootstrap support (%) based on neighbor-joining analyses of 1,000 resampled data sets; solid circles indicate that the corresponding nodes (groupings) were also recovered in maximum-likelihood and maximum-parsimony trees. The scale bar indicates 5.0% sequence divergence.
Antimicrobial drug susceptibility patterns and MIC determination were performed using the Etest strips (AB Biodisk). Mueller-Hinton agar supplemented with 5% sheep blood was inoculated with the isolate, and Etest strips containing ampicillin (256 μg ml−1), azithromycin (256 μg ml−1), clarithromycin (256 μg ml−1), ciprofloxacin (32 μg ml−1), doxycycline (256 μg ml−1), levofloxacin (32 μg ml−1), metronidazole (256 μg ml−1), and rifampin (32 μg ml−1) were placed on the plate. After 7 days of incubation at 37°C in a microaerophilic atmosphere, MIC results showed that the isolate was susceptible to doxycycline (0.094 μg ml−1), metronidazole (6 μg ml−1), and rifampin (0.047 μg ml−1). The isolate was resistant to ampicillin (>256 μg ml−1), azithromycin >256 μg ml−1), clarithromycin (>256 μg ml−1), ciprofloxacin (>32 μg ml−1), and levofloxacin (>32 μg ml−1).
Discussion.
Helicobacter comprises a diverse group of potentially emerging pathogens in several hosts, ranging from humans and nonhuman primates to pets, household animals, and a variety of wildlife animals (23). Currently, the genus includes at least 31 formally described species. Although the majority of these species have been associated with infection of the gastrointestinal tract, several species, e.g., H. bilis, H. canis, H. cinaedi, H. canadensis, H. fennelliae, and H. pullorum, have been found to cause bacteremia and systemic diseases (5, 8, 9, 12, 13, 19). The course of our patient's illness was similar to those of other X-linked agammaglobulinemia patients (1, 6, 7, 10, 12, 16, 22). Skin lesions raised the suspicion of bacterial infection; however, the etiological agent initially remained unidentified since routine cultures were negative, leading to empirical antibiotic treatment. In most cases of bacteremia due to Helicobacter, a close association with a household pet was reported. In our case, the patient had contact with live poultry and snakes, which were fed with rodents and coney. Therefore, we hypothesize that bacteremia of our patient was related to pet exposure.
Recognition of bacteremia due to Helicobacter species is not always straightforward since organisms in this group are fastidious and may fail to grow. However, the use of enriched blood culture medium, such as Bactec Peds Plus (Becton Dickinson), provided better growth. Moreover, they are biochemically inert, and their identification by traditional biochemical methods is limited due to the paucity of useful tests. In our case, although isolate IMMIB HP-28/08 exhibited a biochemical profile similar to that of H. canadensis, H. equorum, and H. pullorum with respect to positive catalase, nitrate reduction, and negative urease (Table 1), it was readily distinguished from all currently described Helicobacter species. In particular, isolate IMMIB HP-28/08 was resistant to cephalothin, lacked growth at 42°C, and possessed distinctive morphological features, including five or more flagella on one end of the organism (monopolar).
Molecular methods have been proven to be useful in the identification of fastidious bacteria with special growth requirements. The use of 16S rRNA gene sequencing provides an alternative method for phenotypic characterization and allows the identification of novel pathogens. In our case, direct sequencing of the DNA extract from the blood culture fluid and from pure culture pointed to an H. canadensis-like organism as the etiological agent. Comparative analysis of the 16S rRNA gene sequences revealed a range of 2.4 to 2.5% sequence divergence between our isolate, IMMIB HP-28/08, and H. pullorum, H. canadensis, and H. equorum. These values are remarkably close to the 3% cutoff divergence recommended as a conservative criterion for demarcating species (18). Therefore, isolate IMMIB HP-28/08 may be considered a novel Helicobacter species. However, DNA-DNA hybridizations between isolate IMMIB HP-28/08 and H. canadensis, H. equorum, and H. pullorum are necessary to confirm that isolate IMMIB HP-28/08 represents a single genomic species as defined in taxonomic practice (21). Moreover, according to current taxonomical rules (rule 27.2 and rule 30 of the International Committee on Systematics of Prokaryotes), in order to validly publish a new species of bacteria, it is necessary to deposit the strain in at least two different culture collections in two different countries (20). Unfortunately, the organism was lost during subculture, and attempts to revive the organism by the DSM and CCUG culture collections were unsuccessful.
This is the first report of septicemia caused by a previously undescribed species of Helicobacter recovered from the blood of an X-linked agammaglobulinemia patient with painful, hyperpigmented macules and fever. In the present case, it is conceivable that the patient acquired the infection from contact with poultry and/or a household pet. This case highlights the importance of 16S rRNA sequencing as a useful tool in the rapid diagnosis of infection with fastidious organisms which are difficult to culture under routine laboratory conditions. Our present findings should stimulate further investigations to isolate and elucidate the importance of the new Helicobacter species in bloodstream infections.
Nucleotide sequence accession number.
The 16S rRNA gene sequence from isolate IMMIB HP-28/08 was submitted to GenBank and assigned the accession number AM992061.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Cuccherini, B., K. Chua, V. Gill, S. Weir, B. Wray, D. Stewart, D. Nelson, I. Fuss, and W. Strober. 2000. Bacteremia and skin/bone infections in two patients with X-linked agammaglobulinemia caused by an unusual organism related to Flexispira/Helicobacter species. Clin. Immunol. 97:121-129. [DOI] [PubMed] [Google Scholar]
- 2.Dewhirst, F. E., J. G. Fox, and S. L. On. 2000. Recommended minimal standards for describing new species of the genus Helicobacter. Int. J. Syst. Evol. Microbiol. 50:2231-2237. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 4.Fox, J. G., and F. Megraud. 2007. Helicobacter, p. 947-962. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, M. A. Pfaller, and F. Megraud (ed.), Manual of clinical microbiology, 9th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 5.Fox, J. G., C. C. Chien, F. E. Dewhirst, B. J. Paster, Z. Shen, P. L. Melito, D. L. Woodward, and F. G. Rodgers. 2000. Helicobacter canadensis sp. nov. isolated from humans with diarrhea as an example of an emerging pathogen. J. Clin. Microbiol. 38:2546-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrard, J., D. Alfredson, and I. Smith. 2001. Recurrent bacteremia and multifocal lower limb celluitis due to Helicobacter-like organisms in a patient with X-linked hypogamma-globulinemia. Clin. Infect. Dis. 33:e116-e118. [DOI] [PubMed] [Google Scholar]
- 7.Han, S. R., C. Schindel, R. Genitsariotis, E. Marker-Hermann, S. Bhakdi, and M. J. Maeurer. 2000. Identification of a unique species by 16S rRNA gene analysis in an abdominal abscess from a patient with X-linked hypogammaglobulinemia. J. Clin. Microbiol. 38:2740-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holst, H., K. Andresen, J. Blom, N. Højlyng, M. Kemp, K. A. Krogfelt, and J. J. Christensen. 2008. A case of Helicobacter cinaedi bacteraemia in a previously healthy person with cellulitis. Open Microbiol. J. 2:29-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsueh, P. R., L. J. Teng, C. C. Hung, Y. C. Chen, P. C. Yang, S. W. Ho, and K. T. Luh. 1999. Septic shock due to Helicobacter fennelliae in a non-human immunodeficiency virus-infected heterosexual patient. J. Clin. Microbiol. 37:2084-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiehlbauch, J. A., R. V. Tauxe, C. N. Baker, and I. K. Wachsmuth. 1994. Helicobacter cinaedi-associated bacteremia and cellulitis in immunocompromised patients. Ann. Intern. Med. 121:90-93. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray, P. R., A. Jain, G. Uzel, R. Ranken, C. Ivy, L. B. Blyn, D. J. Ecker, R. Sampath, C. C. Lee, and M. L. Turner. 2010. Pyoderma gangrenosum-like ulcer in a patient with X-linked agammaglobulinemia: identification of Helicobacter bilis by mass spectrometry analysis. Arch. Dermatol. 146:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prag, J., J. Blom, and K. A. Krogfelt. 2007. Helicobacter canis bacteraemia in a 7-month-old child. FEMS Immunol. Med. Microbiol. 50:264-267. [DOI] [PubMed] [Google Scholar]
- 14.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsiaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1096. [DOI] [PubMed] [Google Scholar]
- 15.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 16.Simons, E., L. A. Spacek, H. M. Lederman, and J. A. Winkelstein. 2004. Helicobacter cinaedi bacteremia presenting as macules in an afebrile patient with X-linked agammaglobulinemia. Infection 32:367-368. [DOI] [PubMed] [Google Scholar]
- 17.Stackebrandt, E., and J. Ebers. 2005. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152-155. [Google Scholar]
- 18.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 19.Tee, W., J. Montgomery, and M. Dyall-Smith. 2001. Bacteremia caused by a Helicobacter pullorum-like organism. Clin. Infect. Dis. 33:1789-1791. [DOI] [PubMed] [Google Scholar]
- 20.Tindall, B. J., P. Kämpfer, J. P. Euzéby, and A. Oren. 2006. Valid publication of names of prokaryotes according to the rules of nomenclature: past history and current practice. Int. J. Syst. Evol. Microbiol. 56:2715-2720. [DOI] [PubMed] [Google Scholar]
- 21.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, L. H. Krichevsky, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 22.Weir, S., B. Cuccherini, A. M. Whitney, M. L. Ray, J. P. MacGregor, A. Steigerwalt, M. I. Daneshvar, R. Weyant, B. Wray, J. Steele, W. Strober, and V. J. Gill. 1999. Recurrent bacteremia caused by a “Flexispira”-like organism in a patient with X-linked (Bruton's) agammaglobulinemia. J. Clin. Microbiol. 37:2439-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whary, M. T., and J. G. Fox. 2004. Natural and experimental Helicobacter infections. Comp. Med. 54:128-158. [PubMed] [Google Scholar]