Abstract
Eighty-two percent of 320 clinical methicillin-resistant Staphylococcus aureus (MRSA) isolates from various infection sites collected throughout South Africa were separated into five major globally prevalent clusters by SmaI pulsed-field gel electrophoresis, spa, and SCCmec typing. Only one Panton-Valentine leukocidin-positive isolate was detected. This is the first detailed MRSA epidemiology study for the whole country.
Methicillin-resistant Staphylococcus aureus (MRSA) is a serious public health concern and an economic burden to national health care systems (13, 15). A recent survey of S. aureus bacteremia at two academic hospitals in Gauteng Province, South Africa, reported a MRSA prevalence of 23% (11). A similar prevalence, 27%, was reported in the KwaZulu-Natal (KZN) Province (17). The latter study also describes MRSA strain types in KZN (16), but no information is available on the clonal types circulating in the other eight provinces of South Africa. We describe for the first time the population structure of MRSA in South Africa using pulsed-field gel electrophoresis (PFGE) (8), spa typing (3), SCCmec typing (4, 10), multilocus sequence typing (MLST) (2), and detection of Panton-Valentine leukocidin (PVL) by PCR (5).
The 320 MRSA isolates cultured were from 187 males (median age, 42 years; range, newborn to 91 years), 119 females (median age, 35 years; range, newborn to 87 years), and 14 persons of unknown gender. Strains were isolated from the following clinical infections or sources: bacteremia (n = 123), skin and soft tissue infections (n = 144), cerebrospinal fluid (n = 3), urine (n = 4), catheter tip (n = 3), drainage fluid (e.g., tracheal aspirate; n = 17), and unknown sources (n = 26). Isolates were collected between August 2005 and November 2006 at 15 state and eight private diagnostic microbiology laboratories in the nine provinces of South Africa. Only the first isolate from a patient was included in this study. Ethical clearance was obtained from the University of the Witwatersrand Human Research Ethics Committee.
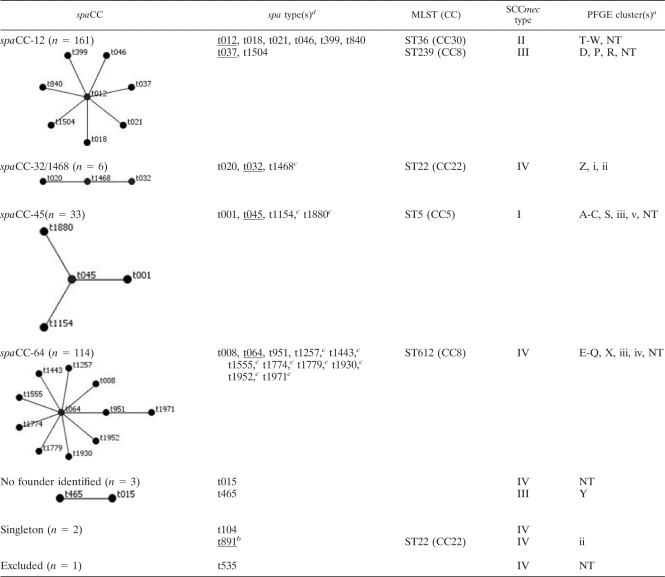
SmaI PFGE was used to investigate genetic diversity among the 310 typeable isolates. S. aureus NCTC 8325, BK2464, ANS46, MW2, E2125, and HPV107 were included for normalization between gels. Fragments ranging from 50 to 1,000 kb were included in the analysis. PFGE cluster analysis (unweighted-pair group method using average linkages based on the Dice similarity coefficient with optimization and position tolerance set at 0.5% and 1.0%, respectively) was done using GelCompar II (Applied Maths, Kortrijk, Belgium) and resulted in 31 PFGE types displaying less than 70% similarity. Eighty-two percent of the typeable isolates clustered into five major PFGE types, A (n = 21), D (n = 68), F (n = 20), K (n = 67), and T (n = 77). spa typing of all of the MRSA isolates revealed 31 different spa types, 11 of which were novel. The five most common spa types were t012 (n = 68), t037 (n = 77), t045 (n = 25), t064 (n = 68), and t1257 (n = 31), which made up 84% of the isolates. The spa types were clustered into five spa clonal complexes (spaCC) (Table 1) using the Based Upon Repeat Pattern (BURP) algorithm at a cost setting of ≤5 (14) and excluding spa types with four or fewer repeats. spaCC12 and -64 were the two largest CCs. SCCmec type IV was the most common cassette type (n = 124, 38%), followed by type II (n = 83), type III (n = 80), and type I (n = 33). SCCmec types clustered in congruence with PFGE and spa types (Table 1).
TABLE 1.
Genotypes of the 320 MRSA isolates used in this study
PFGE clustering based on a 70% cutoff value.
PVL positive.
Novel spa type.
Underlined spa types were subjected to MLST.
Based on PFGE, spaCC, and SCCmec types, 246/310 isolates (77%) could be grouped into five major clonal types. Two isolates belonging to each of the five clonal types were further subjected to MLST (Table 1). Five major MRSA clonal lineages were identified: PFGE type A-spa type t045-SCCmecI-ST5 (CC5) (n = 21), PFGE type D-spa type t037-SCCmecIII-ST239 (CC8) (n = 67), PFGE type T-spa type t012-SCCmecII-ST36 (CC30) (n = 76), and PFGE types F (n = 20) and K (n = 61), both having spa type t064-SCCmecIV and belonging to ST612, which is a double-locus variant of ST8 (CC8). Only one PVL-positive MRSA was detected, which belonged to spa type t891, ST22 (CC22), carrying SCCmec type IV.
This is the first report on MRSA clonal types circulating in the nine provinces of South Africa. The lineages identified in the present study are major clones that are disseminated worldwide (1). There were two major clones associated with spaCC64-SCCmecIV-CC8: HA-MRSA USA500 (UK-EMRSA-2/-6) and CA-MRSA USA 300 (1). The arginine catabolic mobile element associated with USA300 was not detected by PCR, suggesting that our isolates are likely to be USA500. It was noted that spa types t037 and t012 grouped together in spaCC12 by BURP analysis but clustered independently of each other based on PFGE, SCCmec, and MLST. The presence of these two distinct MRSA clones in spaCC12 can be explained by the recombination of a 557-kb fragment which includes spa from ST30 (CC30) into CC8, resulting in the evolution of ST239 (CC8) (12).
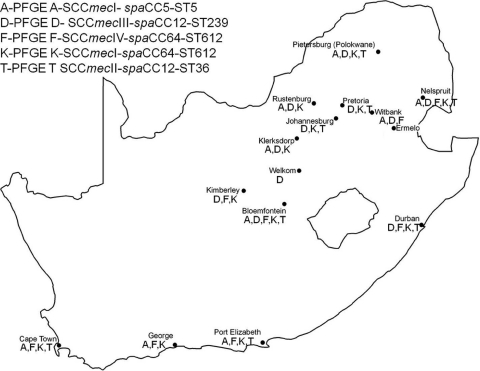
The five major MRSA clones identified in this study were widespread in South Africa (Fig. 1). However, there appeared to be some geographic associations with specific clones. Type K-spaCC64-SCCmecIV-ST612 was the most widespread clone. Type D-spaCC12-SCCmecIII-ST239 was not detected in the Eastern and Western Cape Provinces. Similarly, type F-spaCC64-SCCmecIV-ST612 was not detected in the Limpopo, Gauteng, and North West Provinces. Only two recent studies provide some insight into the clonal types circulating in KZN (16) and Pretoria, a major city in Gauteng Province (6). In Durban, KZN, we identified four clones. One of the three clonal types identified by Shittu et al. (16), namely, t037-ST239-SCCmecIII, was also identified in the present study. In contrast, all our t045-ST5 isolates carried SCCmec type I cassettes, as opposed to the type III cassettes identified by Shittu et al. This suggests two independent SCCmec acquisitions by this clonal lineage within a defined region. Nübel et al. (9), using single-nucleotide polymorphism analysis, demonstrated that within the MRSA ST5 lineage, there have been multiple independent introductions of SCCmec and that the lineage has been associated with at least six SCCmec types, including types I and III. The t064 clone identified in the present study belonged to a different ST (ST612) but is a single-locus variant of ST1338 and a double-locus variant of ST1173, which were identified by Shittu et al. (16). Makgotlho et al. (6) published molecular typing data for MRSA isolates from a hospital in Pretoria, but no spa or MLST data were provided; therefore, it is difficult to compare our data. However, they found that SCCmec type II was the predominant cassette type (64%), followed by type III (14%) and type IV (4%). We found three of the five major clones identified this study in Pretoria, which is in agreement with the SCCmec results of Makgotlho et al. (6).
FIG. 1.
Locations of participating health care laboratories and distribution of the five main MRSA clonal types identified in the present study. (The map of South Africa was produced using ArcGIS software [ESRI, Redlands, CA].)
Marais et al. (7) previously published antimicrobial susceptibility data on 247 of the 320 MRSA isolates in this study. The results showed that all were sensitive to vancomycin, teicoplanin, linezolid, quinupristin-dalfopristin, and fusidic acid, whereas 80% were multidrug resistant (resistant to β-lactams and three or more different antimicrobial classes). Ninety-three of the 247 isolates tested by Marais et al. (7) were from blood cultures, and 74 (80%) of them were multidrug resistant. The most common antimicrobial resistance pattern observed was resistance to β-lactams, macrolides, lincosamides, and fluoroquinolones (n = 45, 18%) and was mainly associated with spa type t012-SCCmecII-ST36. This antimicrobial resistance pattern is similar to that observed among South African MRSA isolates from 1996 (18). These four antimicrobial classes are commonly used for the treatment of staphylococcal infections.
The occurrence of indistinguishable and closely related MRSA strains in different hospitals and across different provinces strongly indicates interhospital spread and further demonstrates the clonal nature of MRSA. Shittu et al. (16) found similar results when comparing MRSA isolates from 14 hospitals within the KZN Province. This is the first comprehensive molecular characterization study performed with South African MRSA isolates and demonstrates the presence of a variety of hospital-acquired MRSA clones. These data can serve as a baseline for future MRSA surveillance to study the evolution of MRSA clonal types.
Acknowledgments
The members of the South African MRSA Surveillance Group are W. F. Oosthuysen, E. Marais, A. Dusé (University of the Witwatersrand, Johannesburg), A. Brink (Ampath, Johannesburg), G. Coetzee, J. Mogale, S. Mahlati (National Health Laboratory Service [NHLS]), L. Badenhorts, K. Fick (Ampath Bloemfontein), E. Botha (NHLS, Welkom Laboratory), C. Jordaan, M. N. Janse van Rensburg (NHLS Universitas Laboratory), M. Botha, L. Ho (Ampath, Johannesburg), F. Botha, D. Hari-Makkan (Ampath Pretoria), O. Perovic, D. Antelme (NHLS Charlotte Maxeke Hospital, K. Lindeque (NHLS Tshwane Academic Complex, Pretoria), R. van der Linde, P. Nickleson (Ampath Nelspruit), G. Hoyland (NHLS Nelspruit Laboratory), J. Kruger (NHLS Ermelo Laboratory), A. Pumeza, C. Hammans (NHLS Witbank Laboratory), D. Bothes, S. de Wit (Ampath Limpopo), Z. Kola (NHLS Polokwane Laboratory), J. Weenink (NHLS Kimberley Laboratory), M. Enslin, H. Kuhn (Lancet Johannesburg for North West Province), D. Cilliers (NHLS Rustenburg Laboratory), E. du Plessis, I. Khantsi (NHLS Potchefstroom Laboratory), W. Swart, C. Kuhn (Pathcare Laboratory Port Elizabeth), V. Pearce, O. Bosch (NHLS Port Elizabeth Laboratory), M. Senekal, S. Lalloo (Pathcare Laboratory Cape Town), S. Oliver, Iva Shankland (NHLS Groote Schuur Hospital), B. Jarvis (NHLS George Laboratory), N. Miller, J. Bredenkamp (Ampath Durban), N. Mkhize, N. Khedzie (KwaZulu-Natal Health), T. Thembe, and J. Mackenza (R. K. Khan Hospital, KwaZulu-Natal).
We thank H. de Lencastre for control strains HPV107, BK2464, ANS46, and MW2.
This study was funded by the South African Medical Research Council, the National Health Laboratory Research Trust, and the University of the Witwatersrand Health Faculty Research Committee.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Deurenberg, R. H., and E. E. Stobberingh. 2009. The molecular evolution of hospital- and community-associated methicillin-resistant Staphylococcus aureus. Curr. Mol. Med. 9:100-115. [DOI] [PubMed] [Google Scholar]
- 2.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 6.Makgotlho, P. E., M. M. Kock, A. Hoosen, R. Lekalakala, S. Omar, M. Dove, and M. M. Ehlers. 2009. Molecular identification and genotyping of MRSA isolates. FEMS Immunol. Med. Microbiol. 57:104-115. [DOI] [PubMed] [Google Scholar]
- 7.Marais, E., N. Aithma, O. Perovic, W. F. Oosthuysen, E. Musenge, and A. G. Dusé. 2009. Antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates from South Africa. S. Afr. Med. J. 99:170-173. [PubMed] [Google Scholar]
- 8.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nübel, U., P. Roumagnac, M. Feldkamp, J. H. Song, K. S. Ko, Y. C. Huang, G. Coombs, M. Ip, H. Westh, R. Skov, M. J. Struelens, R. V. Goering, B. Strommenger, A. Weller, W. Witte, and M. Achtman. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 105:14130-14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perovic, O., H. Koornhof, V. Black, I. Moodley, A. Duse, and J. Galpin. 2006. Staphylococcus aureus bacteraemia at two academic hospitals in Johannesburg. S. Afr. Med. J. 96:714-717. [PubMed] [Google Scholar]
- 12.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubin, R. J., C. A. Harrington, A. Poon, K. Dietrich, J. A. Greene, and A. Moiduddin. 1999. The economic impact of Staphylococcus aureus infection in New York City hospitals. Emerg. Infect. Dis. 5:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sammeth, M., and J. Stoye. 2005. Alignment of tandem repeats with excision, duplication, substitution and indels (EDSI). Report 2005-05, Technische Fakultät, Universität Bielefeld, Bielefeld, Germany. (http://bieson.ub.uni-bielefeld.de/volltexte/2005/764/pdf/report.pdf.
- 15.Seybold, U., E. V. Kourbatova, J. G. Johnson, S. J. Halvosa, Y. F. Wang, M. D. King, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42:647-656. [DOI] [PubMed] [Google Scholar]
- 16.Shittu, A., U. Nübel, E. Udo, J. Lin, and S. Gaogakwe. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates from hospitals in KwaZulu-Natal Province, Republic of South Africa. J. Med. Microbiol. 58:1219-1226. [DOI] [PubMed] [Google Scholar]
- 17.Shittu, A. O., and J. Lin. 2006. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal Province, South Africa. BMC Infect. Dis. 6:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zinn, C. S., H. Westh, V. T. Rosdahl, and the Sarisa Study Group. 2004. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb. Drug Resist. 10:160-168. [DOI] [PubMed] [Google Scholar]