Abstract
A previously established multiplex PCR that identifies to the species level Acinetobacter baumannii and Acinetobacter genomic species 13TU (GS13TU) was expanded to include Acinetobacter calcoaceticus and Acinetobacter genomic species 3.
Identification of Acinetobacter isolates to the species level is often difficult, especially in routine diagnostic laboratories (1). The clinically relevant species Acinetobacter baumannii and Acinetobacter genomic species 3 and 13TU (GS3 and GS13TU) are often grouped together alongside the environmental A. calcoaceticus species as the Acinetobacter calcoaceticus-A. baumannii complex (Acb complex) because they are genetically closely related and phenotypically very difficult to differentiate from each other (7). However, there are considerable epidemiological and clinically relevant differences among these species. A. calcoaceticus is an environmental organism that, to our knowledge, has never been involved in serious human disease, and therefore it should not be misidentified as A. baumannii. The natural habitats of A. baumannii and GS13TU are unknown, as are the differences in their epidemic behaviors, resistance mechanisms, and pathogenicities. GS3 can be found regularly on human skin, as well as in aquatic environments (5). GS3 has also been implicated in nosocomial infections, but its tendency for epidemic spread and resistance development is far less pronounced than that of A. baumannii (5, 9). For epidemiological and clinical purposes, it is therefore highly desirable to differentiate among these species correctly.
Manual and semiautomated commercial identification systems, e.g., API 20NE, Vitek 2, Phoenix, and MicroScan WalkAway, do not differentiate among these species, resulting in misidentification of up to 25% of Acinetobacter isolates belonging to the Acb complex as A. baumannii (14). Additionally, there is no recognized biochemical method to distinguish reliably between A. calcoaceticus and GS3 (3, 7). Molecular methods for species identification, such as DNA-DNA hybridization and amplified rRNA gene restriction analysis (ARDRA), are labor-intensive, difficult to interpret, and rarely used routinely (2, 13), while tRNA spacer fingerprinting does not differentiate between A. calcoaceticus and GS3 and between A. baumannii and GS13TU (6). More recently, sequencing of the rpoB gene, its flanking spacer regions, and of the 16S-23S rRNA gene spacer region has been proposed for identification of Acinetobacter isolates to the species level (4, 10), but it is unlikely that these sequencing techniques will be used routinely, except in a few specialized reference laboratories. Multiplex PCR based on species-specific gyrB primers is a simple, specific, and rapid method to reliably identify A. baumannii and GS13TU (8). This study expanded the gyrB multiplex to enable the identification of A. calcoaceticus and GS3.
A total of 146 clinical, type, and reference strains, including 23 A. calcoaceticus strains, 36 GS3 strains, 21 A. baumannii strains, and 29 GS13TU strains obtained from our own clinical culture collection (12, 14), were used in this study. Clinical isolates of the species A. beijerinckii (1), A. bereziniae (1), A. guillouiae (2), A. haemolyticus (4), A. johnsonii (1), A. junii (3), A. lwoffii (2), A. radioresistens (2), A. schindleri (1), A. ursingii (3), A. venetianus (1), and the unnamed genomic species 6 (1), 14 (4), and 15TU (1) were also included (11). All clinical strains had been previously identified to the species level by using ARDRA as well as the simplified phenotypic identification scheme for Acinetobacter species devised by Bouvet and Grimont (3, 13). In addition, the following type strains were used: A. baumannii ATCC 19606T, A. bereziniae ATCC 17924T, A. calcoaceticus ATCC 23055T, A. johnsonii ATCC 17909T, A. junii ATCC 17908T, A. lwoffii ATCC 13509T, A. radioresistens SEIP 12.81, GS3 ATCC 19004T, GS6 ATCC 17979T, and GS9 ATCC 9957T.
Template DNA for PCR was isolated by using the DNeasy kit (Qiagen, Hilden, Germany) or a 1-μl loopful of a colony from an agar plate was suspended in 100 μl PCR-grade water, boiled for 10 min, snap-cooled, and briefly centrifuged. The gyrB gene was amplified and sequenced using primer pair D14/gyrB-2 from nonduplicate A. calcoaceticus (n = 21) and GS3 (n = 24) isolates (Table 1). gyrB sequences were aligned and compared to those previously obtained for A. baumannii and GS13TU (8). Twenty-two primers were identified and evaluated for species specificity using the established multiplex PCR parameters (8). One primer pair each for A. calcoaceticus (D14/D19) and GS3 (D16/D8) was chosen and tested against the test organisms as a multiplex PCR using Taq PCR master mix (Qiagen) with a final concentration of 0.2 μM for each primer (Table 1). Using these four primers, A. calcoaceticus amplified a single 428-bp amplicon and GS3 amplified a single 194-bp amplicon (Fig. 1 and 2a). No PCR products were amplified using these primers in all other species tested.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′) | Species | Reference |
|---|---|---|---|
| gyrB-2 | CTTACGACGCGTCATTTCAC | A. calcoaceticus and GS3 reverse sequencing primer | This study |
| D14 | GACAACAGTTATAAGGTTTCAGGTG | A. calcoaceticus | This study |
| D19 | CCGCTATCTGTATCCGCAGTA | A. calcoaceticus | This study |
| D16 | GATAACAGCTATAAAGTTTCAGGTGGT | GS3 | This study |
| D8 | CAAAAACGTACAGTTGTACCACTGC | GS3 | This study |
| Sp2F | GTTCCTGATCCGAAATTCTCG | A. baumannii | 8 |
| Sp4F | CACGCCGTAAGAGTGCATTA | A. baumannii and GS13TU | 8 |
| Sp4R | AACGGAGCTTGTCAGGGTTA | A. baumannii and GS13TU | 8 |
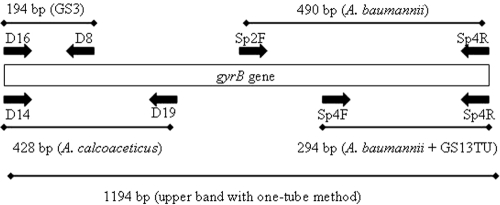
FIG. 1.
Schematic diagram showing the annealing positions, species specificities, and expected sizes of PCR products by using primer pairs shown in Table 1.
FIG. 2.
(a) Example of an agarose gel showing Acinetobacter isolates for which the species were determined by multiplex PCR using gyrB-directed primers D14, D19, D16, and D8. Lanes: 1, 100-bp marker; 2 and 3, A. calcoaceticus; 4 and 5, GS3; 6, A. baumannii; 7, GS13TU; 8, pooled DNA from A. lwoffii, A. junii, A. haemolyticus, and A. johnsonii. (b) Example of an agarose gel showing Acinetobacter isolates for which the species were determined by PCR using gyrB-directed multiplex primers (D14, D19, D16, D8, Sp2F, Sp4F, and Sp4R). Lanes: 1, 100-bp marker; 2, A. calcoaceticus; 3, A. baumannii; 4, GS3; 5, GS13TU; 6, pooled DNA from A. lwoffii, A. junii, A. haemolyticus, and A. johnsonii. In this gel, GS3 and GS13TU amplify an additional 1,194-bp product.
Primers D14, D19, D16, and D8 were also tested as a multiplex in the presence of established primers Sp2F, Sp4F, and Sp4R at a final concentration of 0.2 μM for each primer (Table 1 and Fig. 2b). With this multiplex, an additional 1,194-bp PCR product was amplified with some, but not all, GS13TU, GS3, and A. calcoaceticus isolates (Fig. 1 and Fig. 2b, lanes 4 and 5). The 1,194-bp amplicon is amplified only when the seven primers are used as a multiplex. There is no 1,194-bp amplicon with the D14, D19, D16, and D8 multiplex. The other Acinetobacter species tested (A. beijerinckii, A. bereziniae, A. guillouiae, A. haemolyticus, A. johnsonii, A. junii, A. lwoffii, A. radioresistens, A. schindleri, A. ursingii, A. venetianus, and Acinetobacter genomic species 6, 9, 14, and 15TU) did not amplify a PCR product.
The new gyrB multiplex PCR can be used as a stand-alone multiplex PCR to identify A. calcoaceticus and GS3. Alternatively, it can be performed in the presence of the previously established primers in a single PCR tube to both identify and differentiate the 4 species of the Acb complex. The method is robust, cheaper than sequencing, and reproducible, it can yield a result in <2.5 h, and it enables the reliable identification of the clinically most relevant Acinetobacter species. Its simplicity means that it can be employed readily in most laboratories, where it should contribute to a better understanding of the epidemiology and clinical significance of the most important Acinetobacter species.
Acknowledgments
The contribution of P.G.H. and H.S. was supported by a grant from Bundesministerium für Bildung und Forschung (BMBF), Germany, Klinische Forschergruppe Infektiologie (grant number 01KI0771).
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Bernards, A. T., J. van der Toorn, C. P. A. van Boven, and L. Dijkshoorn. 1996. Evaluation of the ability of a commercial system to identify Acinetobacter genomic species. Eur. J. Clin. Microbiol. Infect. Dis. 15:303-308. [DOI] [PubMed] [Google Scholar]
- 2.Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp-nov, Acinetobacter haemolyticus sp-nov, Acinetobacter johnsonii sp-nov, and Acinetobacter junii sp-nov and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240. [Google Scholar]
- 3.Bouvet, P. J. M., and P. A. D. Grimont. 1987. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur/Microbiol. 138:569-578. [DOI] [PubMed] [Google Scholar]
- 4.Chang, H. C., Y. F. Wei, L. Dijkshoorn, M. Vaneechoutte, C. T. Tang, and T. C. Chang. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 6.Ehrenstein, B., A. T. Bernards, L. Dijkshoorn, P. Gerner Smidt, K. J. Towner, P. J. M. Bouvet, F. D. Daschner, and H. Grundmann. 1996. Acinetobacter species identification by using tRNA spacer fingerprinting. J. Clin. Microbiol. 34:2414-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerner-Smidt, P., I. Tjernberg, and J. Ursing. 1991. Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 29:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins, P. G., H. Wisplinghoff, O. Krut, and H. Seifert. 2007. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin. Microbiol. Infect. 13:1199-1201. [DOI] [PubMed] [Google Scholar]
- 9.Horrevorts, A., K. Bergman, L. Kollee, I. Breuker, I. Tjernberg, and L. Dijkshoorn. 1995. Clinical and epidemiologic investigations of Acinetobacter genomospecies-3 in a neonatal intensive-care unit. J. Clin. Microbiol. 33:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.La Scola, B., V. A. K. B. Gundi, A. Khamis, and D. Raoult. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seifert, H., L. Dijkshoorn, P. Gerner Smidt, N. Pelzer, I. Tjernberg, and M. Vaneechoutte. 1997. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J. Clin. Microbiol. 35:2819-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seifert, H., and P. Gerner Smidt. 1995. Comparison of ribotyping and pulsed-field gel-electrophoresis for molecular typing of Acinetobacter isolates. J. Clin. Microbiol. 33:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaneechoutte, M., L. Dijkshoorn, I. Tjernberg, A. Elaichouni, P. Devos, G. Claeys, and G. Verschraegen. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wisplinghoff, H., M. B. Edmond, M. A. Pfaller, R. N. Jones, R. P. Wenzel, and H. Seifert. 2000. Nosocomial bloodstream infections caused by Acinetobacter species in United States hospitals: clinical features, molecular epidemiology, and antimicrobial susceptibility. Clin. Infect. Dis. 31:690-697. [DOI] [PubMed] [Google Scholar]