Abstract
We analyzed the nuclear small-subunit rRNA genes of Acanthamoeba isolates from freshwater sources (n = 16) and from patients (n = 6) in Bangkok and surrounding areas. The T10 genotype from a keratitis patient and a novel T17 genotype from water samples were diagnosed for the first time in this study.
Acanthamoebae are free-living protists capable of causing fatal granulomatous amoebic encephalitis (GAE) and amoebic keratitis (AK). Acanthamoeba has worldwide distribution and inhabits a wide range of environments (4, 11). Although Acanthamoeba species can be classified into three morphological groups based on cyst dimension and structure (12), sequence analysis of the nuclear small-subunit rRNA gene (Rns) has placed Acanthamoeba isolates into 16 genotypes (T1 to T16) (2, 3). Of these, genotype T4 predominates among both clinical specimens and environmental sources (2). Acanthamoeba genotypes T2 to T4 and T11 have been found to cause keratitis, while genotypes T1, T4, T10, and T12 are responsible for GAE (2). Although both GAE and AK occurred in Thai patients and the organisms are widely distributed in water sources, little is known about genotypic distribution of acanthamoebae in this country (7, 10, 13).
We conducted a survey of Acanthamoeba in 23 freshwater ponds and 5 irrigation canals in Bangkok and surrounding areas (13°39′39"N to 14°3′54"N, 100°26′9"E to 100°47′3"E) from February to May 2009. These water sources were near residential areas or in public places. Water samples were taken ∼20 to 50 cm from the shore and ∼10 to 20 cm in depth. Samples were collected at least twice for each place at 1- or 2-month intervals. Before cultivation, 200 ml of sample was centrifuged at 750 × g for 10 min. After the supernatant was discarded, the pellet was resuspended in 1 ml of sterile water and applied onto 1.5% nonnutrient agar overlaid with 2 ml of heat-inactivated (60°C, 30 min) Escherichia coli. The culture was incubated at room temperature (25°C to 30°C), 37°C, and 40°C and examined every 24 h for 10 days (Table 1). Acanthamoebae were successfully isolated from 7 places (25%). Of these, acanthamoebae were detected on ≥2 occasions at four places, yielding 16 isolates out of 84 samples collected (Table 1). Assessment of cyst structure revealed that 13 isolates belonged to Pussard and Pons' group II while the remaining 3 isolates were group I.
TABLE 1.
Morphological group, number of gene clones and genotypes of Acanthamoeba from freshwater sources and clinical isolates
| Isolate | Source/condition of the sample during collection | Groupa | No. of haplotypesb |
Genotype | Growth at °Cc |
|||
|---|---|---|---|---|---|---|---|---|
| Rns | DF3 | 25-30 | 37 | 40 | ||||
| AE1 | Lampangpui pond, Bangkok/29.5°C, pH 8.0 | I | 3 | 2 | T17 | + | − | − |
| AE2 | Lampangpui pond, Bangkok/28.5°C, pH 7.4 | I | 3 | 2 | T9 | + | − | − |
| AE3 | Freshwater pond, Kasetsart University, Bangkok/31.5°C, pH 7.9 | II | 1 | 1 | T4 | + | + | + |
| AE4 | Freshwater pond, Kasetsart University, Bangkok/30°C, pH 7.3 | II | 5 | 3 | T4 | + | + | + |
| AE5 | Muangthong Thani lake, Nonthaburi/34°C, pH 9.4 | II | 2 | 1 | T4 | + | + | + |
| AE6 | Rangsit distribution canal, Pathumthani/24°C, pH 8.0 | II | 5 | 2 | T4 | + | + | + |
| AE7 | Freshwater pond at National Stadium, Bangkok/32.5°C, pH 8.0 | II | 2 | 1 | T4 | + | + | + |
| AE8 | Freshwater pond at National Stadium, Bangkok/34.5°C, pH 8.2 | II | 3 | 1 | T4 | + | + | + |
| AE9 | Freshwater pond at National Stadium, Bangkok/32°C, pH 8.0 | I | 2 | 1 | T17 | + | − | − |
| AE10 | Freshwater pond at National Stadium, Bangkok/28.5°C, pH 7.7 | II | 3 | 1 | T4 | + | + | + |
| AE11 | Sammakorn pond, Bangkok/32°C, pH 8.2 | II | 1 | 1 | T4 | + | + | + |
| AE12 | Sammakorn pond, Bangkok/30°C, pH 8.0 | II | 4 | 1 | T4 | + | + | + |
| AE13 | Sammakorn pond, Bangkok/28°C, pH 8.5 | II | 1 | 1 | T3 | + | − | − |
| AE14 | Lampangpui pond, Bangkok/28.5°C, pH 8.3 | II | 5 | 4 | T4 | + | + | + |
| AE15 | Lampangpui pond, Bangkok/30°C, pH 7.8 | II | 1 | 1 | T4 | + | + | + |
| AE16 | Perfect Place pond, Bangkok/29°C, pH 8.3 | II | 1 | 1 | T4 | + | + | + |
| APC1′ | Corneal scrape, 16-year-old female/keratitis, contact lens user | II | 1 | 1 | T4 | + | + | + |
| APC2 | Corneal scrape, 44-yr-old male/keratitis, glaucoma | III | 1 | 1 | T10 | + | + | + |
| APC3 | Corneal scrape, 31-yr-old female/keratitis, contact lens user | II | 4 | 2 | T4 | + | + | + |
| APC4 | Corneal scrape, 42-yr-old female/keratitis, exposed to fish farm water | II | 3 | 1 | T4 | + | + | + |
| APC5 | Corneal scrape, 21-yr-old female/keratitis, contact lens user | II | 1 | 1 | T4 | + | + | + |
| APB1 | Cerebrospinal fluid, 31-yr-old female/meningoencephalitis | II | 1 | 1 | T4 | + | + | + |
Pussard and Pons' morphological group (12).
Number of haplotypes based on near-complete Rns sequences (Rns) and diagnostic fragment 3 (DF3).
Growth determined within 10 days of incubation. +, growth; −, absence of growth.
From January 2008 to December 2009, single corneal scrapes from 19 keratitis patients grew positive cultures for Acanthamoeba in five patients (five isolates). A single cerebrospinal fluid sample from a meningoencephalitis patient submitted for Acanthamoeba culture gave a positive result (isolate APB1) (Table 1). All but one clinical isolate had cyst structure consistent with group II, whereas isolate APC2 from a keratitis patient belonged to group III (Table 1).
The Rns gene of each isolate was amplified by PCR encompassing >2 kb using primers ACAN18SF0 (5′-TCCTGCCAGTAGTCATATGC-3′) and ACAN18SR0 (5′-CTTCTCCTTCCTCTAAATGGT-3′). An aliquot of purified PCR product was subcloned into pGEM-T-Easy vector (Promega, Madison, WI). Sequences were determined by both direct sequencing of purified PCR products and ≥10 subclones for each isolate. Results revealed that 9 isolates contained single haplotypes while 13 isolates contained 2 to 5 haplotypes. In total, 53 distinct haplotypes were obtained, with sequence lengths varying from 2,156 to 2,581 bp. Meanwhile, Acanthamoeba has a polyploid genome (25n) and each genome contains 24 copies of Rns; therefore, each cell possesses ∼600 gene copies (18). The presence of mixed Rns sequences in most isolates could stem from heterogenous copies of Rns within each cell, as previously noted by Stothard and colleagues, who observed multiple Rns haplotypes within cloned Acanthamoeba samples (15). Alternatively, clonal mixtures of acanthamoebae carrying different Rns haplotypes could frequently occur among isolates. Despite the complexity of sequence variation in Rns, current classification based on this locus remains useful for genotypic determinations of Acanthamoeba unless more appropriate markers are identified. Meanwhile, a 113-bp polymorphic region of the Rns gene designated ASA.S1 or diagnostic fragment 3 (DF3) has been deployed for tracking Acanthamoeba strains from clinical and water samples (1, 14). However, acanthamoebae that shared identical DF3 sequences may not be identical strains because sequence variation may occur in other regions of Rns. It is noteworthy that haplotype assignment based on the DF3 sequences per se of 13 isolates carrying multiple Rns sequences in this study failed to detect mixed haplotypes in 7 isolates and underestimated the number of mixed haplotypes in 6 isolates. Hence, for a better understanding of genotype-related differences in pathogenicity of acanthamoebae and a more precise tracking of Acanthamoeba strains from various sources, haplotype assignment based on the complete or near-complete Rns sequences could be more accurate.
Phylogenetic reconstruction of all isolates and known genotypes inferred from the near-complete Rns sequences using neighbor-joining and maximum likelihood methods yielded similar tree topologies (6, 16). Of 13 environmental isolates in group II, 12 isolates had genotype T4 comprising 33 haplotypes and 1 isolate (AE13) belonged to genotype T3 (Table 1). One of the three environmental isolates belonging to group I had genotype T9 (AE2), while two isolates (AE1 carrying three haplotypes and AE9 carrying two haplotypes) could not be assigned to any known genotypes. The Rns sequences of isolates AE1 and AE9 were phylogenetically distinct from those of genotypes T7 and T8 with 70% bootstrap support, although they clustered in the same monophyletic group as genotypes T7/T8/T9 (Fig. 1). All clones of isolates AE1 and AE9 shared >97.8% sequence similarity, indicating the same genotype (data not shown). Importantly, sequence comparison between these gene clones and genotypes T1 to T16 revealed a range of sequence similarity from 57.8% (between isolate AE1 and genotype T5) to 88.4% (between isolate AE1 and genotype T8). An arbitrary figure of ≥5% sequence dissimilarity in Rns has been used to define distinct genotypes of Acanthamoeba (5, 15). Hence, both isolates AE1 and AE9 represent a novel “T17” genotype. Our phylogenetic inference revealed that all the members of nonpathogenic morphological group I consisting of genotypes T7, T8, and T9 formed a distinct monophyletic clade. On the other hand, genotypes T1 to T6 and T10 to T16 belonging to morphological groups II and III comprising both pathogenic and nonpathogenic genotypes were paraphyletic. Therefore, phylogenetic inference based on the near-complete Rns sequences seems to relate to morphological groups and pathogenic potential of Acanthamoeba (Fig. 1).
FIG. 1.
The Rns genealogy of Acanthamoeba isolates from Thailand inferred by the neighbor-joining method using the maximum composite likelihood model. Bootstrap values ≥70% from 1,000 pseudoreplicates are shown next to the branches. The GenBank accession numbers of known T1 to T16 genotypes are in parentheses. Filled circles and filled diamonds represent clinical isolates and genotype “T17,” respectively.
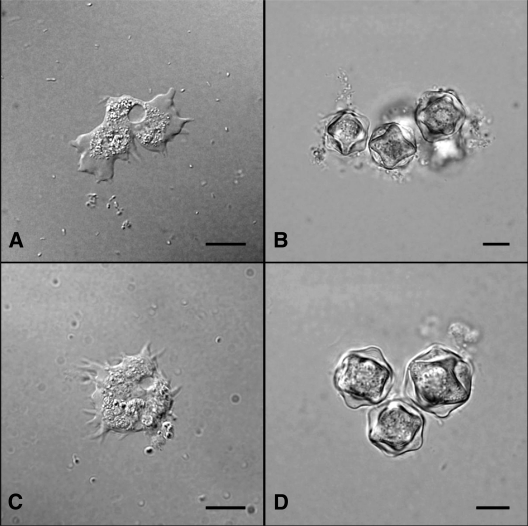
The average large and small diameters with their standard deviations of genotype “T17” cysts measured 19.4 ± 2.9 μm and 17.9 ± 2.1 μm, respectively (n = 100 cysts). A wide separation of endocysts and ectocysts with 3 to 6 arms were observed (Fig. 2). Although we have not elucidated the pathogenic potential of genotype “T17,” our initial study has shown that both isolates were not thermotolerant because they could not grow at 40°C (Table 1), a temperature that is shared among pathogenic genotypes (8, 9, 17). Furthermore, phylogenetic analysis has placed the “T17” genotype within the same monophyletic group as the nonpathogenic genotypes T7/T8/T9. Taken together, isolates of genotype “T17” could be nonpathogenic. Nevertheless, pathogenicity of Acanthamoeba depends on a multifactoral process; therefore, further studies are required (8).
FIG. 2.
Photomicrograph of Acanthamoeba genotype “T17” depicting trophozoites of isolates ACE1 (A) and ACE9 (C) and their respective cysts (B and D). Bars, 10 μm.
For clinical samples, all isolates belonging to group II exhibited genotype T4 sequences. Interestingly, isolate APC2, from a 44-year-old Thai patient who suffered from chronic keratitis and glaucoma, had genotype T10 (Table 1). The cyst structure of this isolate also displayed group III characteristics like those reported for genotype T10 (2). Importantly, genotype T10 has been reportedly isolated from patients with GAE but none with AK (2). Despite the fact that genotype T4 contains both pathogenic and nonpathogenic strains, the predominant occurrence of the T4 genotypes among water sources around human communities and their abilities to grow well at 37°C, a characteristic of pathogenic phenotypes, could pose a risk to human infections in Thailand, where the majority of clinical isolates belong to this genotype. Although not all T4 isolates in this study shared identical Rns sequences, environmental isolates AE3, AE10, AE12, and AE15 seem to be closely related to clinical isolates APC1, APC4, APC5, and APB1, respectively, suggesting that they could potentially be pathogenic ones (Fig. 1).
In conclusion, molecular genotype analysis of Acanthamoeba in this study has led to the identification of genotype T10 as a causative genotype of AK, thereby extending the pathological range of this genotype in human infections. Furthermore, isolation of samples carrying a novel “T17” genotype from a limited number of samples analyzed suggests that this genotype may not be uncommon in nature. It is likely that more novel genotypes of Acanthamoeba await discovery.
Nucleotide sequence accession numbers.
Nucleotide sequence data from this study are available in the GenBank database under accession numbers GU808277 to GU808329.
Acknowledgments
We are grateful to Takuro Endo for advice on the cultivation method and to Pannadhat Areekul, Rattiporn Kosuwin, and Pattakorn Buppan for assistance with the sample collection.
This study was supported by a Graduate School Thesis Grant (academic year 2008) to W.N. and the Molecular Biology of Malaria and Opportunistic Parasites Research Unit, Chulalongkorn University.
Footnotes
Published ahead of print on 13 October 2010.
REFERENCES
- 1.Booton, G. C., D. J. Kelly, Y. W. Chu, D. V. Seal, E. Houang, D. S. C. Lam, T. J. Byers, and P. A. Fuerst. 2002. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lens, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J. Clin. Microbiol. 40:1621-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booton, G. C., G. S. Visvesvara, T. J. Byers, D. J. Kelly, and P. A. Fuerst. 2005. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J. Clin. Microbiol. 43:1689-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corsaro, D., and D. Venditti. 2010. Phylogenetic evidence for a new genotype of Acanthamoeba (Amoebozoa, Acanthamoebida). Parasitol. Res. 107:233-238. [DOI] [PubMed] [Google Scholar]
- 4.De Jonckheere, J. F. 1991. Ecology of Acanthamoeba. Rev. Infect. Dis. 13(Suppl. 5):S385-S387. [DOI] [PubMed] [Google Scholar]
- 5.Gast, R. J., D. R. Ledee, P. A. Fuerst, and T. J. Byers. 1996. Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J. Eukaryot. Microbiol. 43:498-504. [DOI] [PubMed] [Google Scholar]
- 6.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. System Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 7.Jongwutiwes, S., L. Pariyakanok, M. Charoenkorn, K. Yagita, and T. Endo. 2000. Heterogeneity in cyst morphology within isolates of Acanthamoeba from keratitis patients in Thailand. Trop. Med. Int. Health 5:335-340. [DOI] [PubMed] [Google Scholar]
- 8.Khan, N. A. 2003. Pathogenesis of Acanthamoeba infections. Microb. Pathog. 34:277-285. [DOI] [PubMed] [Google Scholar]
- 9.Khan, N. A. 2006. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol. Rev. 30:564-595. [DOI] [PubMed] [Google Scholar]
- 10.Lekkla, A., C. Sutthikornchai, S. Borvornkitti, and Y. Sukthana. 2005. Free-living ameba contamination in natural hot springs in Thailand. Southeast Asian J. Trop. Med. Public Health 36(Suppl. 4):5-9. [PubMed] [Google Scholar]
- 11.Marciano-Cabral, F., and G. Cabral. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16:273-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pussard, M., and R. Pons. 1977. Morphologie de la paroi kystique et taxonomie du genre Acanthamoeba (Protozoa: Amoeba). Protistologica 8:557-598. [Google Scholar]
- 13.Sangruchi, T., A. J. Martinez, and G. S. Visvesvara. 1994. Spontaneous granulomatous amebic encephalitis: report of four cases from Thailand. Southeast Asian J. Trop. Med. Public. Health 25:309-313. [PubMed] [Google Scholar]
- 14.Schroeder, J. M., G. C. Booton, J. Hay, I. A. Niszl, D. V. Seal, M. B. Markus, P. A. Fuerst, and T. J. Byers. 2001. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 39:1903-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stothard, D. R., J. M. Schroeder-Diedrich, M. H. Awwad, R. J. Gast, D. R. Ledee, S. Rodriguez-Zaragoza, C. L. Dean, P. A. Fuerst, and T. J. Byers. 1998. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J. Eukaryot. Microbiol. 45:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 17.Visvesvara, G. S., H. Moura, and F. L. Schuster. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 50:1-26. [DOI] [PubMed] [Google Scholar]
- 18.Yang, Q., M. G. Zwick, and M. R. Paule. 1994. Sequence organization of the Acanthamoeba rRNA intergenic spacer: identification of transcriptional enhancers. Nucleic Acids Res. 22:4798-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]