Abstract
Light is a ubiquitous environmental signal that many organisms sense and respond to by modulating their physiological responses accordingly. While this is an expected response among phototrophic microorganisms, the ability of chemotrophic prokaryotes to sense and react to light has become a puzzling and novel issue in bacterial physiology, particularly among bacterial pathogens. In this work, we show that the opportunistic pathogen Acinetobacter baumannii senses and responds to blue light. Motility and formation of biofilms and pellicles were observed only when bacterial cells were incubated in darkness. In contrast, the killing of Candida albicans filaments was enhanced when they were cocultured with bacteria under light. These bacterial responses depend on the expression of the A. baumannii ATCC 17978 A1S_2225 gene, which codes for an 18.6-kDa protein that contains an N-terminal blue-light-sensing-using flavin (BLUF) domain and lacks a detectable output domain(s). Spectral analyses of the purified recombinant protein showed its ability to sense light by a red shift upon illumination. Therefore, the A1S_2225 gene, which is present in several members of the Acinetobacter genus, was named blue-light-sensing A (blsA). Interestingly, temperature plays a role in the ability of A. baumannii to sense and respond to light via the BlsA photoreceptor protein.
Acinetobacter baumannii is a Gram-negative opportunistic human pathogen that has been recognized as the etiological agent of severe nosocomial infections in compromised patients and wounded military personnel (14, 32). More recently, it was identified as the sole causative agent of necrotizing fasciitis (11). This pathogen survives in hospital environments despite unfavorable conditions such as desiccation, nutrient starvation, and antimicrobial treatments (6, 52). These remarkable properties could be due to the ability of this pathogen to attach to and form biofilms on abiotic and biotic surfaces (19, 41). In spite of extensive work on antibiotic resistance and the epidemiology of infections caused by A. baumannii, little is known about the factors and environmental signals modulating the physiology of this bacterium and its capacity to cause diseases in humans.
Pathogens sense and respond to extracellular cues that play a role in host-pathogen interactions and their ability to persist in the environment. Accordingly, A. baumannii responds to iron limitation, a condition that is central to human defense against microbial infections, by sensing this condition via the Fur repressor and expressing active iron acquisition systems (12, 15, 54). This pathogen could also sense other environmental signals that modulate functions supporting its persistence in clinical settings, such as biofilm formation on abiotic surfaces (42). Among these signals is light, a ubiquitous environmental stimulus that plays an obvious role in photosynthetic eukaryotes and prokaryotes. However, it is becoming apparent that this signal also plays a role in the physiology of environmental and pathogenic chemotrophic nonphototrophic prokaryotes (30, 33, 49). This is due to the production of light-sensing photoreceptors, with those harboring a blue-light-sensing-using flavin (BLUF); light, oxygen, or voltage (LOV); or photoactive yellow protein (PYP) domain being the most prevalent, as predicted by in silico analysis of bacterial genomes (30, 49). While some of the bacterial photoreceptors are single-domain proteins, others are coupled to output domains that regulate gene expression. The structure of the latter type of photoreceptors, together with studies such as those done with the Rhodobacter sphaeroides AppA (23), Escherichia coli YcgF (45), and Bacillus subtilis YtvA (20) photoreceptors, supports the idea that these systems regulate a wide variety of cell functions, including development, stress response, and virulence in response to light (30, 33). Regarding the latter cell function, the report by Swartz et al. (40) is so far the only study that links blue-light regulation to the virulence of a human bacterial pathogen—Brucella abortus. Here, we report the observation that A. baumannii senses and responds to blue light through a temperature-dependent process that involves a gene coding for an 18.6-kDa single-BLUF-domain-containing photoreceptor protein, which regulates cell motility, biofilm formation, and killing of fungal filaments.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this work are listed in Table 1. Luria-Bertani (LB) broth and agar (37) were used to grow and maintain bacterial strains. Broth cultures were incubated either statically or with shaking at 200 rpm at 24°C or 37°C. The growth rates of the parental strain and isogenic derivatives were determined as described before (16). Growth curves were determined three times with fresh overnight inocula each time at 24°C or 37°C using LB. The Candida albicans tup1 mutant was cultured as described previously (19).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| A. baumannii | ||
| ATCC 19606T | Clinical isolate, type strain | ATCC |
| ATCC 17978 | Clinical isolate | ATCC |
| ATCC 17978.OR | blsA::aph derivative of 17978; Kmr | This work |
| ATCC 17978.ORc | 17978.OR harboring pWHBLSA; Kmr Ampr | This work |
| ATCC 17978.ORp | 17978.OR harboring pWH1266; Kmr Tetr Ampr | This work |
| AYE | Clinical isolate | ATCC |
| LUH 07672 | Clinical isolate, EU clone III | 47 |
| LUH 8809 | Clinical isolate, EU clone I | 48 |
| LUH 5875 | Clinical isolate, reference strain, EU clone III | 51 |
| LUH 13000 | Clinical isolate, EU clone II | L. Dijkshoorn |
| RUH 134 | Clinical isolate, reference strain, EU clone II | 13 |
| RUH 875 | Clinical isolate, reference strain, EU clone I | 13 |
| C. albicans tup1 | Constitutive filamentous derivative of SC5314 | 9 |
| E. coli | ||
| DH5α | Used for DNA recombinant methods | Gibco-BRL |
| Top10 | Used for DNA recombinant methods | Invitrogen |
| EC100D+ | pir+, host for pKNOCK-Amp maintenance | Epicentre |
| HB101 | Conjugation helper strain harboring pRK2073; Tpr Str | 7 |
| BL21 (DE) | Overexpression of His-tagged BlsA | Novagen |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR cloning vector; Kmr Zeor | Invitrogen |
| pGEM-T Easy | PCR cloning vector; Ampr | Promega |
| pUC4K | Source of the Kmr cassette; Kmr Ampr | Pharmacia Biotech |
| pET-TEV | pET28a (Novagen) containing TEV protease cleavage site; Kmr | 25 |
| pKNOCK-Amp | Suicide vector for allelic exchange; Ampr | 2 |
| pRK2073 | Used as helper in plasmid conjugation; Tpr | 29 |
| pWH1266 | E. coli-A. baumannii shuttle vector; Ampr Tcr | 26 |
| pBLSA | blsA cloned into pCR-Blunt II-TOPO; Kmr Zeor | This work |
| pKABLSA | pKNOCK-Amp harboring blsA; Ampr | This work |
| pKABLSA-Km | pKABLSA with the pUC4K Kmr cassette inserted into NsiI restriction site; Ampr Kmr | This work |
| pGBLSA1 | Amplicon harboring blsA promoter and coding region cloned into pGEM-T Easy; Ampr | This work |
| pWHBLSA | pWH1266 harboring wild-type copy of blsA expressed under its own promoter; Ampr | This work |
| pGBLSA2 | Amplicon harboring blsA coding region cloned into pGEM-T Easy; Ampr | This work |
| pEBLSA | pET-TEV harboring a parental copy of blsA; Kmr | This work |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; Str, streptomycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance; Zeor, zeocin resistance.
Cell motility experiments and biofilm assays.
Swimming plates containing 0.3% agarose (35) were used as a tool to detect cell motility on a semisolid surface rather than swimming motility. A. baumannii is not capable of flagellum-mediated swimming motility because members of this genus do not produce this type of cell appendage (44). Therefore, to avoid confusion, we refer to these plates here as motility plates. The plates were inoculated on the surface with bacteria lifted from overnight LB agar cultures using flat-ended sterile wooden sticks or depositing 0.003 ml of LB cultures grown to an optical density at 600 nm (OD600) of 0.3. Plates were incubated for 10 to 12 h (overnight) or 24 h at 24°C or 37°C in the dark or under light emitted by nine-LED (light-emitting diode) arrays with an intensity of 6 to 10 μmol photons/m2/s. Each array was built using three-LED module strips emitting blue, green, or red light with emission peaks centered at 462 nm, 514 nm, and 636 nm, respectively, as determined using a LI-COR LI-1800 spectroradiometer (see Fig. S1B in the supplemental material). For biofilm assays, one milliliter of fresh LB broth medium was inoculated into glass tubes (1 by 10 cm) with 0.01 ml of an overnight shaking culture grown at 37°C. The cultures were then incubated for 4 days stagnantly at 24°C either in darkness or under blue light. Biofilms that formed on the walls of the glass tubes were visualized by crystal violet staining as previously described (41). Pellicles that formed on the culture surfaces were detected by visual inspection. To estimate total biofilm formation, biofilms attached to the wall tubes plus pellicles on the liquid surfaces, the culture medium was removed gently with a Pasteur pipette without losing pellicles. The cells that remained in the tubes were resuspended in 1 ml of sterile phosphate-buffered saline (PBS) solution, sonicated for 10 s at low power with a thin probe, and then suspended vigorously for 1 min with a vortex mixer. The suspended cells were transferred to a spectrophotometer cuvette, and the cell density was determined by measurement of OD600. Full cell suspension was confirmed by staining the tubes with crystal violet after the samples were transferred to spectrophotometer cuvettes. The amount of biofilm formed by each sample was normalized to its total biomass, which was determined by measuring the OD600 as described before (41). Accordingly, the biofilm results are reported as a percentage of the total biomass of each sample. Triplicate assays were done at least three times using fresh samples each time.
Killing of C. albicans filaments.
Assays were performed as described before (19), with the modification of incubating 1-ml cocultures without shaking at 24°C from 24 h to 120 h under the dark or blue light conditions used in the cell motility and biofilm experiments. CFU counts were determined after the plates were incubated at 28°C for 48 h. These experiments were repeated at least three times using fresh samples each time.
General DNA procedures.
Genomic and plasmid DNAs were isolated as described before (41). DNA restriction and Southern blot analyses were conducted using standard protocols (37). DNA was sequenced by standard automated sequencing methods with appropriate primers. Sequences were assembled and analyzed as described before (41). The SABLE server (http://sable.cchmc.org/sable_doc.html) was used for protein structure predictions. The presence of the chromosomal region harboring blsA in A. baumannii clinical isolates was determined by PCR using as the template total DNA, which was obtained after lysing bacterial cells in 0.050 ml distilled water at 98°C for 7 min, and primers BlsA.R/1 and BlsA.F/2 (Table 2).
TABLE 2.
Primers used in this study
| Name/no. | Nucleotide sequence |
|---|---|
| BlsA.R/1 | 5′-GCAATGTCTCACAATTATGT-3′ |
| BlsA.F/2 | 5′-ATGACCATACAAACATCTAG-3′ |
| PblsA.R/3 | 5′-TGATATGGATCCTGTCGATTTCAGT-3′ |
| PblsA.F/4 | 5′-ATGTGAGGATCCAGTATTACAAATT-3′ |
| EblsA.R/5 | 5′-GGATCCCTAGAACGGGTTTAC-3′ |
| EblsA.F/6 | 5′-CATATGAACGTTCGCCTGTGT-3′ |
| BlsAR.rt/7 | 5′-TCAACGACCTCTTGTTCACC-3′ |
| BlsAF.rt/8 | 5′-GAACGTTCGCCTGTGTTATG-3′ |
| RecAF.rt | 5′-TACAGAAAGCTGGTGCATGG-3′ |
| RecAR.rt | 5′-TGCACCATTTGTGCCTGTAG-3′ |
Construction of the A. baumannii ATCC 17978.OR isogenic insertion derivative.
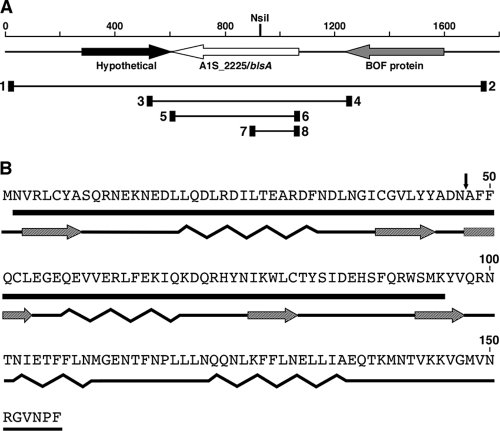
A 1,738-bp fragment containing the blsA gene and flanking sequences was PCR amplified using primers BlsA.R/1 and BlsA.F/2 (see Fig. 2 and Table 2). The amplicon was cloned with the Invitrogen Zero Blunt TOPO PCR cloning kit to generate pBLSA. This fragment was subsequently subcloned into the EcoRI sites of pKNOCK-Amp, and the resulting plasmid (pKABLSA) was used to construct pKABLSA-Km, in which the pUC4K PstI restriction fragment harboring the DNA kanamycin resistance (Kmr) cassette was inserted into a unique NsiI site located at nucleotide 145 of blsA that maps to the BLUF protein domain. E. coli EC100D+ cells harboring pKABLSA-Km, E. coli HB101 cells harboring pRK2073, and A. baumannii ATCC 17978 cells were used as donor, helper, and recipient strains, respectively, in triparental conjugations. Transconjugants were selected on Simmons citrate agar plates containing 40 μg/ml Km. Total DNA was isolated from a putative A. baumannii ATCC 17978.OR transconjugant derivative, which was resistant to Km and sensitive to 200 μg/ml ampicillin (Amp), and used to confirm the nature of the site-directed insertion mutation by PCR with primers BlsA.R/1 and BlsA.F/2 and Southern blotting hybridization of HincII- or HindIII-digested DNA. The blots were hybridized with [32P]dCTP-labeled probes (17) harboring either the pUC4K aph or the pBLSA blsA gene.
Construction of a complementation plasmid.
A 747-bp fragment harboring blsA and its predicted promoter was PCR amplified using A. baumannii ATCC 17978 total DNA and primers PblsA.R/3 and PblsA.F/4 (see Fig. 2 and Table 2), both of which were tailed with BamHI restriction sites. The amplicon was cloned into pGEM-T Easy (pGBLSA1) and then subcloned as a BamHI fragment into the cognate site of pWH1266. Proper construction of the complementing pWHBLSA plasmid was confirmed by automated DNA sequencing. Plasmid DNA was electroporated into A. baumannii ATCC 17978.OR as described before (16).
Overexpression and purification of BlsA.
The blsA coding sequence was PCR amplified from A. baumannii ATCC 17978 genomic DNA using primers EblsA.R/5 and EblsA.F/6 (see Fig. 2 and Table 2), which were tailed with NdeI and BamHI restriction sites, respectively, and cloned into pGEM-T Easy (pGBLSA2). The NdeI-BamHI restriction fragment was subcloned from pGBLSA2 into the corresponding sites of the pET-TEV expression plasmid to generate pEBLSA, a derivative coding for a His6 tag fused to the BlsA amino-terminal end. Proper construction of this derivative was confirmed by automated DNA sequencing, and the plasmid was transformed into E. coli BL21(DE3) cells, which were cultured in LB broth at 37°C until they reached an OD600 of 0.6 to 0.7. Overexpression of the His-tagged BlsA derivative was induced with 0.5 mM IPTG. After incubation for 5 h at 15°C to avoid the formation of inclusion bodies, the cells were collected by centrifugation, suspended in 20 ml of lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM β-mercaptoethanol), and disrupted at 2,000 lb/in2 using a French pressure cell press. Cell debris was removed by centrifugation at 20,000 × g for 30 min at 4°C, and the supernatant was loaded onto a nickel nitrilotriacetic acid (NiNTA)-agarose column (Qiagen). The column was washed sequentially with lysis buffer containing 20 mM and 40 mM imidazole, and the His-tagged protein was eluted with the same buffer containing 80 mM imidazole (elution buffer). Amicon Ultra-4 centrifugal filter units (Millipore) with a molecular mass cutoff of 10 kDa were used to concentrate purified BlsA and exchange the elution buffer to a buffer containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 3 mM imidazole. The His-tagged BlsA recombinant derivative was digested with His-tagged tobacco etch virus (TEV) protease (1:100 mass ratio) overnight at 4°C. After digestion, untagged BslA was separated from the protease and the His tag by a further Ni-NTA column purification step. The flowthrough fraction containing the digested protein was concentrated and used for photoactivation kinetic experiments. The purity of the overexpressed His-tagged protein and the TEV protease-digested product was confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using a 15% polyacrylamide gel (28). Protein concentration was determined by the Bradford method (8).
Spectroscopy.
UV and visible light absorption spectra were recorded using protein samples in a buffer containing 20 mM Tris-HCl (pH 7.0) and 200 mM NaCl with a Jasco V550 spectrophotometer at 20°C. Spectra were recorded before and after illumination for 10, 40, 100, or 220 s with a nine-blue-LED array at a constant light intensity of 10 μmol photons/m2/s. The kinetics of recovery to the initial darkness receptor state was determined by absorbance at 510 nm after a protein sample was illuminated for 3 min with blue light at the aforementioned intensity.
Transcriptional analysis.
Cells were scraped from motility plates after overnight incubation at 24°C or 37°C in the presence or absence of blue light, quickly resuspended in 4 ml of diethyl pyrocarbonate (DEPC)-treated deionized water and immediately mixed with 2 ml lysis buffer (0.3 M Na acetate, 30 mM EDTA, 3% SDS) in a boiling-water bath. Each motility plate incubated at 24°C under illumination was inoculated several times at different positions to scrape enough cells to obtain appropriate amounts of total RNA. Cell lysates were extracted twice at 60°C with phenol, which was equilibrated to pH 4.0 with 50 mM Na acetate, and then once with chloroform at room temperature. The RNA precipitated overnight at −20°C with ethanol was collected by centrifugation, washed with 70% ethanol, and dissolved in DEPC-treated deionized water. Total RNA samples were purified further with the Qiagen RNeasy kit, including in-column treatment with RNase-free DNase I as suggested by the manufacturer. The integrity of the RNA samples was checked with an Agilent 2100 Bioanalyzer. RNA samples were collected from three different biological samples prepared in triplicate each time. Gene transcription was examined with the Qiagen real-time one-step QuantiTect SYBR green quantitative reverse transcription (qRT)-PCR system by following the manufacturer's recommendations. Primers BlsAR.rt/7 and BlsAF.rt/8 (see Fig. 2 and Table 2) amplified an internal 180-bp blsA region, while primers RecAF.rt and RecAR.rt (Table 2) amplified an internal 148-bp recA fragment, which was used as an internal control for constitutively expressed genes. A Bio-Rad iCycler was used under the following conditions: an RT cycle of 50°C for 30 min and 95°C for 15 min, followed by 35 amplification cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. These experiments were done in triplicate using three different biological samples. The blsA transcript levels of each sample were normalized to the recA cDNA levels of each RNA sample. Samples containing no reverse transcriptase or template RNA were included as negative controls.
RESULTS
Cell motility is inhibited under blue light at 24°C.
The initial observation of the effect of light on the physiology of A. baumannii ATCC 17978 was made while we were testing the motility of this strain at different temperatures. Switching because of equipment problems from an environmental chamber constantly illuminated by regular fluorescent tubes to one in which lights were always off resulted in the observation that bacteria moved away from the inoculation site on motility plates when incubated overnight (10 to 12 h) at 24°C in darkness but not in the presence of light. This initial unexpected observation was followed by a more detailed analysis using an array of nine LEDs conducted under more controlled conditions. This analysis showed that while cells grew only around the inoculation point on plates incubated in the presence of blue light, cellular halos covering almost half of the surface formed when the motility plates were incubated overnight at 24°C in darkness (Fig. 1). Bacterial cells covered the entire surface of the plate when the incubation in darkness was extended to 24 h at the same temperature, with no appreciable motility changes on plates incubated under illuminated conditions. While red-light illumination had no inhibitory effects on cell motility, incubation of motility plates in the presence of green light resulted in partial motility inhibition compared to that detected with blue light (see Fig. S1A in the supplemental material). These results are consistent with the fact that the emission spectra of the blue and red LEDs do not overlap, while the emission spectra of the blue and green LEDs are superimposed (see Fig. S1B in the supplemental material). It is important to note that the light-mediated response is not due to the effect of light on cell growth and viability. Cells cultured in LB broth displayed similar growth curves and reached comparable viable counts after incubation for up to 96 h at 24°C in the presence or absence of light (see Fig. S2 in the supplemental material).
FIG. 1.
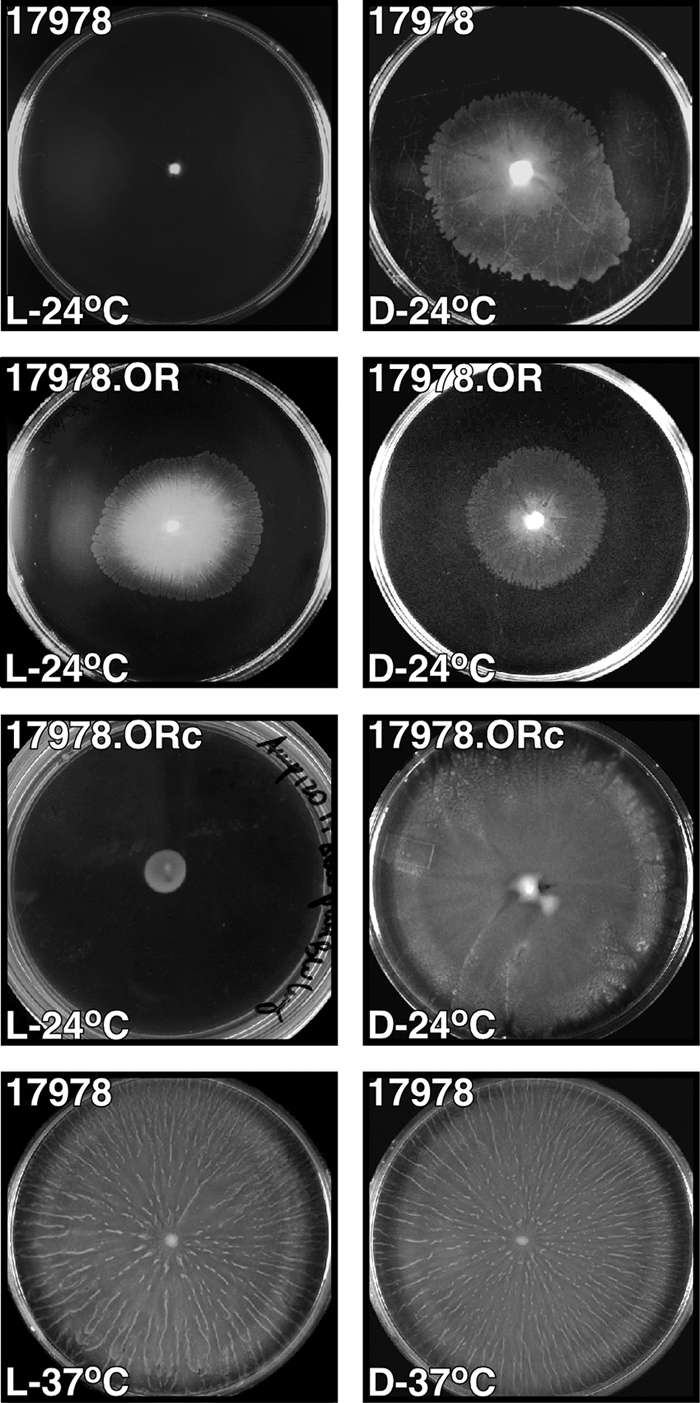
Effects of light and temperature on motility. Cells of the parental strain ATCC 17978 and the ATCC 17978.OR blsA mutant harboring no additional plasmid or transformed with the blsA-complementing plasmid pWHBLSA (ATCC 17978.ORc) were inoculated on the surface of motility plates. Plates were inspected and photographed after incubation overnight (10 to 12 h) in darkness (D) or in the presence of blue light (L) at 24°C or 37°C.
Identification of a chromosomal locus coding for a BLUF-domain containing protein that functions as a photoreceptor.
The results described above suggest that a light-sensing effector must be operating in A. baumannii ATCC 17978. Accordingly, our in silico analysis of the genome of this strain showed that the hypothetical A1S_2225 gene codes for a predicted 18.6-kDa protein harboring a BLUF photosensor domain (pfam04940) in its N-terminal region. This domain harbors two α helices (Fig. 2 B), a structure that is shared with other BLUF domain-containing bacterial proteins significantly related (E values higher than 1 × e−5) to the predicted A1S_2225 product. In contrast, the C-terminal region of the A1S_2225 product, which also harbors two predicted α helices, showed no significant similarity to sequences deposited in GenBank.
FIG. 2.
Genetic organization of the blsA locus and predicted secondary structure of its translational product. (A) Genetic map of the chromosomal region harboring blsA. The horizontal arrows indicate the locations and directions of predicted genes. The numbers and vertical black bar in the upper portion represent base pairs and the location of the NsiI site used for insertion mutagenesis, respectively. The black rectangles, horizontal lines, and numbers indicate the locations of the primers listed in Table 2 that were used to PCR amplify the different chromosomal regions involved in this work. (B) Predicted amino acid sequence, location of the BLUF domain (black horizontal bar), and secondary structure of BlsA: black horizontal line, coil; hatched arrows, β strand; waved line, α helix. The vertical black arrow indicates the site of insertion of the Kmr DNA cassette.
Ni affinity column chromatography of an A1S_2225 overproduced derivative His6 tagged at the amino-terminal end resulted in the isolation of a single 20-kDa protein eluted with 80 mM imidazole (Fig. 3). This purified protein fraction was treated with the TEV protease to obtain the untagged recombinant derivative of A1S_2225 that was used for spectral analysis. The visible absorbance spectrum of dark-adapted A1S_2225 recombinant protein showed the characteristic pattern of flavin-binding proteins, including two major absorbance peaks at 360 nm and 459 nm, with the latter band showing a defined vibrionic structure with shoulders at 426 nm and 480 nm (Fig. 4A). These prominent shoulders on the lowest-energy band suggest that flavin is located in a nonpolar environment. Spectral analysis of dark-adapted versus illuminated protein samples exposed to a blue LED array showed significant changes, with a distinct 12-nm red shift of the 459-nm peak (Fig. 4A; see Fig. S3 in the supplemental material), which is characteristic of BLUF photoreceptors. We then measured the kinetics of recovery to the equilibrium dark state of the activated protein. A sample of the purified protein was illuminated with blue light, and the recovery was followed both in a discontinuous way by recording the full spectra of the sample kept in the dark at different times and by continuously measuring the absorbance at 510 nm (Fig. 4B). Each decay was fitted to an exponential decay function giving similar rate constants: 180 s and 210 s for the continuous and discrete modes, respectively. These values place the A1S_2225 derivative between the slow-recovery AppA (with a recovery time of ca. 1,000 s) and the fast-recovery BlrB and F2 (with recovery times ranging from 2 s to 40 s) BLUF-containing proteins (55).
FIG. 3.
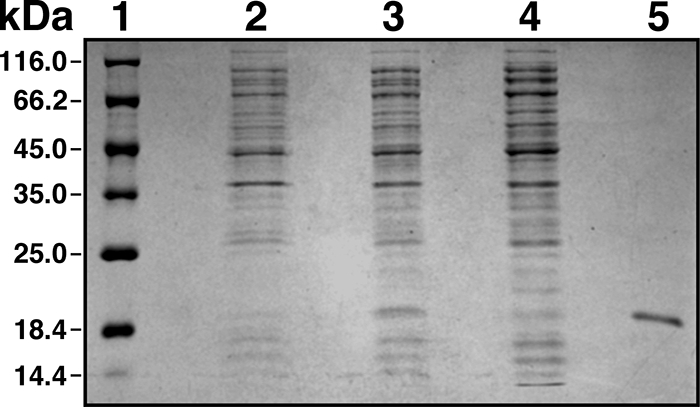
Electrophoretic analysis of bacterial proteins. Proteins present in E. coli BL21 lysates and chromatographic fractions were size fractionated on 15% polyacrylamide gels under denaturing conditions and visualized by staining with Coomassie blue. Lanes: 1, molecular mass markers; 2, cell lysate from uninduced cells; 3, cell lysate from induced cells; 4, NiNTA-agarose chromatographic flowthrough fraction; 5, NiNTA-agarose chromatographic fraction isolated with elution buffer.
FIG. 4.
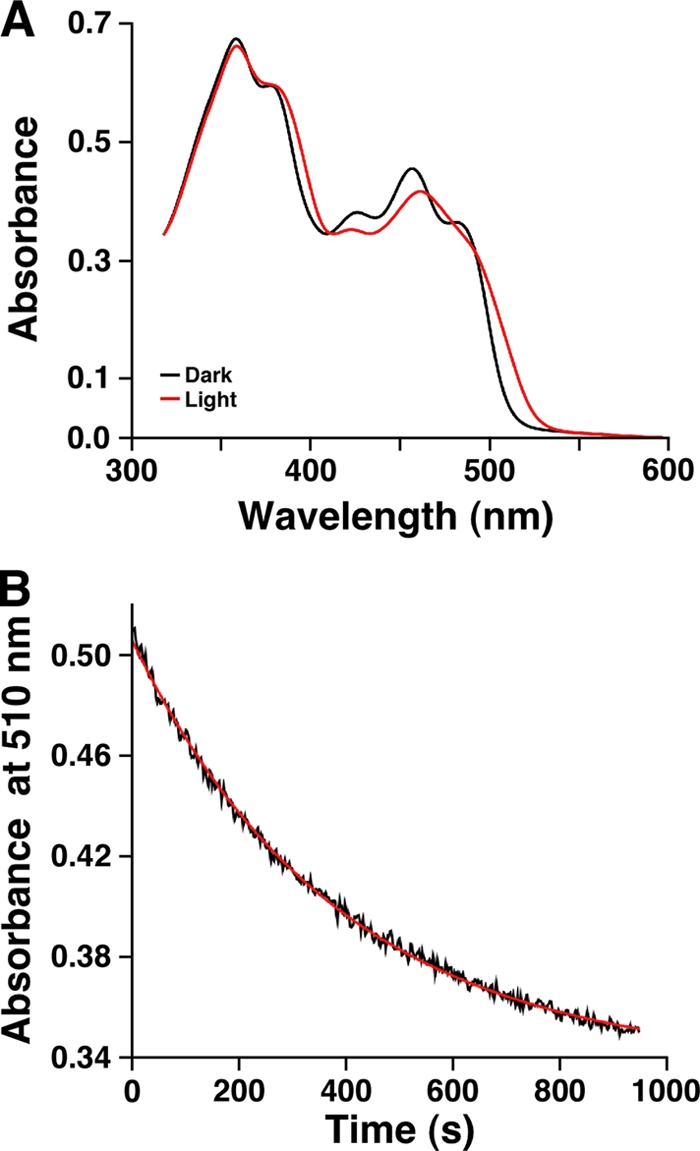
Spectral analysis of recombinant BlsA. (A) UV-visible light spectra of dark and light states of BlsA recombinant protein. For simplicity, the spectra of the dark-adapted sample and a sample illuminated for 220 s are shown. The spectra collected after 10 s, 40 s, 100 s, and 220 s of illumination, together with the spectrum of the dark-adapted sample, are displayed in Fig. S3 in the supplemental material. (B) Kinetics of recovery to the initial receptor state in the dark of purified His-BlsA. The decrease in absorbance at 510 nm is shown in black. The red line shows a fit of the data to an exponential decay function.
The BLUF domain-containing product of A1S_2225 is involved in photoresponse.
The experimental data described so far suggest that the product of the A. baumannii ATCC 17978 A1S_2225 gene is an active photoreceptor protein. Such a possibility is supported by the observation (Fig. 1) that the A1S_2225::aph ATCC 17978.OR isogenic mutant fails to respond to light by forming similar halos on motility plates under dark and illuminated conditions, a response that stands in contrasts to that of the ATCC 17978 parental strain. The photoreceptor function of the A1S_2225 gene product was further confirmed by the observation that electroporation of pWHBLSA, a derivative of the shuttle vector pWH1266 harboring a copy of the A1S_2225 wild-type allele, into ATCC 17978.OR restored the parental photoinhibition phenotype in the complemented isogenic derivative ATCC 17978.ORc (Fig. 1). In contrast, the light response phenotype of the mutant ATCC 17978.OR was not changed upon transformation with the empty pWH1266 vector (data not shown). These results, together with the detection of the predicted A1S_2225 insertion derivative by PCR and Southern blotting, using the aph and A1S_2225 genes as probes, indicate that the phenotype of the ATCC 17978.OR mutant is due to the inactivation of A1S_2225. It is also important to note that the parental strain, the ATCC 17978.OR mutant, and the ATCC 17978.ORc complemented derivative did not display significant differences in growth rate when cultured in LB broth at 24°C or 37°C (data not shown). Furthermore, restriction analysis proved that the pWHBLSA complementing plasmid was stably maintained as an independent replicon without detectable rearrangements in the ATCC 17978.ORc complemented strain. Taken together, all of these observations indicate that the product of the A1S_2225 gene controls the motility of A. baumannii ATCC 17978 at 24°C by sensing blue light. Therefore, this coding region was named blue-light-sensing A (blsA).
Blue light modulates biofilm and pellicle formation through blsA.
Since motility affects biofilm formation by bacteria such as Pseudomonas aeruginosa (38), we determined whether blue light also affects biofilm formation on an abiotic surface such as glass. The incubation of A. baumannii ATCC 17978 for 4 days in LB at 24°C without shaking under blue light produced little or no biofilm on glass tube walls (Fig. 5A). However, when incubated under the same conditions in darkness, this strain formed biofilms on the tube walls at the liquid-air interface, as well as surface pellicles, a multicellular structure that is a type of biofilm formed by other bacterial pathogens such as P. aeruginosa (18). In contrast, the incubation of the ATCC 17978.OR blsA mutant resulted in the formation of similar amounts of pellicles and biofilms on tube walls in cultures incubated in either darkness or blue light (Fig. 5B). The genetic complementation of the ATCC 17978.OR blsA mutant with pWHBLSA restored the differential production of pellicles and biofilms on tube walls in response to illumination as observed with the parental strain. However, this response was not detected when ATCC 17978.OR mutant cells harbored the empty pWH1266 plasmid vector (Fig. 5, panels C and D, respectively). These macroscopic observations were confirmed by spectrophotometrically measuring the biomass of total biofilms (pellicles plus cells attached to the tube walls) formed in each sample after the culture medium was gently removed and the cells were suspended in PBS. This quantitative approach showed that the absence of light resulted in a 4-fold increase in biofilm formation by the A. baumannii ATCC 17978 parental strain compared with that of duplicate samples incubated at 24°C in the presence of blue light (Fig. 5E), a response similar to that detected with the ATCC 17978.ORc mutant complemented with a plasmid copy of the parental blsA allele. In contrast, no noticeable regulation of biofilm formation in response to light was observed with the ATCC 17978.OR blsA insertion mutant and the derivative ATCC 17978.ORp harboring the empty pWH1266 cloning vector (Fig. 5E). It is important to note that these differential biofilm responses could not be attributed to the effect of light on cell viability, considering that there were no significant differences between the growth rates and colony counts of samples cultured in the presence or absence of light at 24°C (see Fig. S2A and B in the supplemental material). Taken together, these results show that the formation of pellicles on the surface of the culture medium and biofilms on the tube walls by cells incubated at 24°C is another cell property that is influenced by blue light through a regulatory process that involves the expression of a functional blsA gene.
FIG. 5.
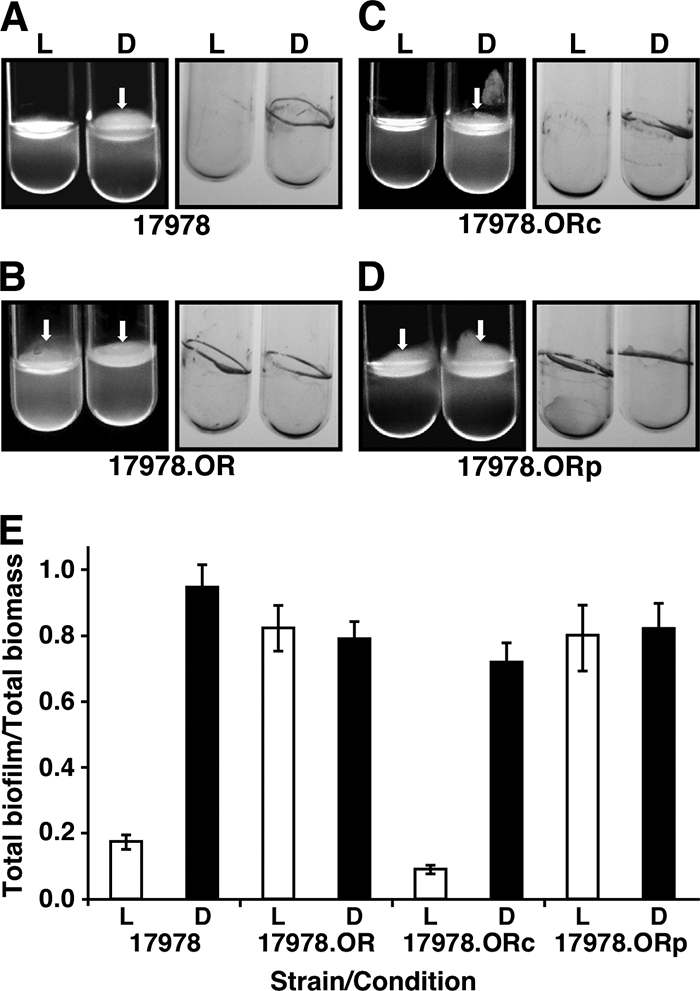
Effects of light on biofilm formation. The biofilms formed by parental strain ATCC 17978 (A) and the ATCC 17978.OR blsA mutant harboring no additional plasmid (B) or transformed with the blsA-complementing derivative pWHBLSA (C) or the shuttle vector pWH1266 (D) were recorded after static incubation for 96 h at 24°C by direct visual inspection and staining with crystal violet. The white arrows point to the pellicles formed on the liquid surface. (E) Quantification of total biofilms (pellicles plus cells attached to tube walls) of duplicate samples shown in the cognate panels. Error bars show the standard error of the mean.
blsA is involved in the killing of C. albicans tup1 filaments.
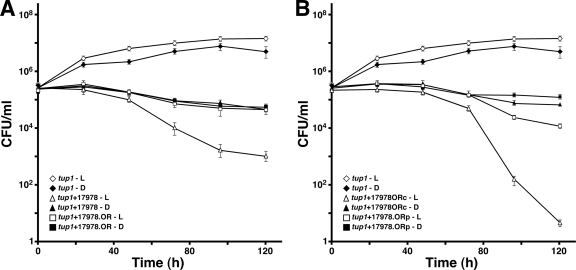
We tested the effect of blue light on the ability of A. baumannii to interact with eukaryotic cells, given our recent report describing the killing of C. albicans tup1 filaments upon their incubation with this bacterial pathogen, a response that parallels the apoptosis of human alveolar epithelial cells and serves as a model to study bacterial virulence (19). Coincubation of tup1 mutant filaments with A. baumannii ATCC 17978 cells resulted in a time-dependent increase of fungal death, which represented a 14,000-fold decrease in filament survival after incubation for 120 h at 24°C in the presence of blue light (compare open diamonds and triangles in Fig. 6A), while a 116-fold decrease was observed when duplicate samples were incubated in darkness (compare closed diamonds and triangles in Fig. 6A). This 121-fold reduction in fungal killing is due to the effect of light on the A. baumannii ATCC 17978 virulence phenotype rather than to a drastic detrimental illumination effect on the viability of the tup1 mutant filaments; only a 2.9-fold growth reduction was detected between fungal filaments incubated in sterile medium in the presence of blue light and those incubated in darkness (compare open and closed diamonds in Fig. 6A). The fact that the number of tup1 mutant filaments was not significantly affected when they were coincubated with ATCC 17967.OR blsA mutant cells in either the presence or the absence of light and the observation that the filament death rate under these conditions was similar to that obtained with filaments coincubated with parental bacterial cells in darkness indicate that blsA also plays a role in the virulence of A. baumannii when tested using the C. albicans virulence model. This possibility was further confirmed by the fact that the ATCC 17978.ORc mutant, complemented with the blsA parental allele, and the ATCC 17978.ORp derivative, complemented with the empty shuttle plasmid pWH1266, displayed a behavior similar to that of the ATCC 17978 parental strain and the ATCC 17978.OR blsA mutant, respectively (Fig. 6B). Notably, the complementation experiments showed that the ATCC 17978.ORc derivative displays a 2.9 × 106-fold increase in virulence compared with that displayed by the parental strain when tup1 filaments and ATCC 17978 bacterial cells are coincubated in the presence of light (Fig. 6B). This enhanced-virulence phenotype and the enhanced-motility phenotype of ATCC 17978.ORc (Fig. 1) are most likely due to the higher copy number of the blsA allele in this plasmid-complemented derivative of the ATCC 17978.OR blsA mutant.
FIG. 6.
Killing of tup1 mutant C. albicans by A. baumannii cells. (A) Fungal filaments were incubated in sterile medium or medium inoculated with bacterial cells from the ATCC 17978 parental strain or the BlsA-deficient derivative ATCC 17978.OR either in the presence of blue light (L) or in darkness (D). (B) Experiment similar to that shown in panel A but using the ATCC 17978.ORc and ATCC 17978.ORp derivatives harboring the complementing plasmid pWHBLSA and the empty vector pWH1266, respectively. Error bars show the standard error of the mean.
Presence of blsA and light response in A. baumannii clinical isolates.
In silico analysis showed that the ATCC 17978 BlsA predicted amino acid sequence is significantly related (E values higher than 1 × e−5) to a large number of bacterial proteins, with those found in the genomes of the A. baumannii ACICU (ACICU_02430), AYE (ABAYE1304), AB0057 (AB57_2585), AB307-0294 (ABBFA_001217) (1, 27, 46), and ATCC 19606T (ZP_05826716.1) strains being the top matches. The top matching protein sequences also include the products of predicted genes found in other members of the Acinetobacter genus, such as the environmental strains A. baylyi ADP1 (4) and A. radioresistens (ZP_05360709). Most of these predicted proteins were annotated as either conserved hypothetical proteins or BLUF domain-containing proteins. Our in silico analysis and resequencing data also showed that when two sequencing errors that mapped outside blsA were corrected by adding a T at positions 2,590,157 and 2,591,288 of the originally reported A. baumannii ATCC 17978 genome sequence (GenBank accession number NC_009085), blsA is flanked by two predicted coding regions (Fig. 2A). The downstream open reading frame is transcribed in the opposite direction and codes for a hypothetical protein, while the one located upstream of blsA and transcribed in the same direction codes for a predicted membrane protein that belongs to the bacterial oligonucleotide/oligosaccharide-binding (OB) fold protein family. Members of this protein family are putative periplasmic proteins that harbor a predicted OB fold structure, which can bind proteins, small molecules, or other ligands recognized by the OB fold structure (21). Interestingly, the gene coding for this type of protein is found in mobile genetic elements and although their function is still unknown, their presence may be associated with bacterial virulence (21). The gene arrangement shown in Fig. 2A is also present in the genome of the aforementioned A. baumannii clinical strains, as well as in the genome of the A. baylyi ADP1 environmental strain. Furthermore, PCR analysis using primers BlsA.R/1 and BlsA.F/2 (Table 2 and Fig. 2A) and total DNA isolated from the A. baumannii strains listed in Table 1, for which there is no genomic information available and which are different representatives of EU clones I, II, and III (Table 1), resulted in the production of a single amplicon which matched the size of that produced when ATCC 17978 total DNA was used as the template (data not shown). Thus, the genetic arrangement shown in Fig. 2A is not a feature unique to the A. baumannii ATCC 17978 clinical isolate.
The motility test proved that isolates AYE and RUH 134 have a light response similar to that of ATCC 17978, while isolates LUH 5875, LUH 13000, and RUH 875 displayed an intermediate response between those of ATCC 17978 and isolates ATCC 19606T, LUH 7672, and LUH 8809. The latter three strains grew only around the inoculation site when incubated in the presence or absence of light. The C. albicans infection assays revealed that strains ATCC 17978, LUH 7672, and LUH 5875 kill the fungal filaments at similar rates. On the other hand, the other strains tested showed various degrees of killing under illumination, ranging from 2.5-fold (strain RUH 134) and 4-fold (strains ATCC 19606T, AYE, and LUH 13000) reductions to a 12-fold reduction (strains LUH 8809 and RUH 875) of the response displayed by ATCC 17978. Whether these variations in motility and killing activity among the strains tested are due to differences in light sensing or the expression of functions associated with these two light-mediated responses is a possibility that must be examined in more detail before drawing final conclusions. Nevertheless, these results indicate that strains other than ATCC 17978 are also able to mount differential motility and killing responses upon illumination. The effect of light on biofilm and pellicle formation by the A. baumannii strains used in this study could not be evaluated because of the wide variations observed among them, even under a single culture condition. Such a response is not surprising, considering the significant variations in biofilm phenotypes displayed by different A. baumannii clinical isolates (10, 36, 53).
Effect of temperature on bacterial response to blue light.
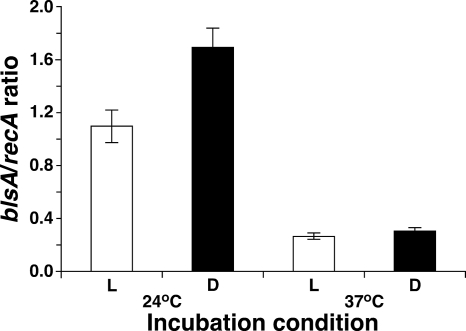
Since A. baumannii is a pathogen that produces serious infections in humans, whose normal temperature is close to 37°C, we tested the effect of this temperature on the A. baumannii ATCC 17978 light-inhibited motility response. The entire surface of each motility plate was covered with a fuzzy layer of cells and striations of cell aggregates radiating from the inoculation site when the plate was incubated at 37°C in the presence or absence of light (Fig. 1, bottom panels). Microscopy of Gram-stained cells lifted from motility plates incubated at 24°C or 37°C showed no detectable differences in cell morphology (data not shown) that could account for this observation. Incubation at 37°C also affected the differential production of biofilms in response to light. The amount of biofilm formed by strain ATCC 17978 under darkness and illumination represented 20% and 25% of the total biomass of each sample, respectively, values that were similar to those produced by the ATCC 17978.OR blsA mutant and the ATCC 17978.ORc and ATCC 17978.ORp complemented derivatives (see Fig. S4 in the supplemental material). Furthermore, these values are comparable to those detected when A. baumannii ATCC 17978 cells were incubated at 24°C in the presence of light (Fig. 5E). As shown by experiments done at 24°C, cell growth and viability were not significantly affected by incubation at 37°C in the presence or absence of light (see Fig. S2 in the supplemental material). These results indicate that temperature could play a role in light response probably due to the temperature-dependent differential expression of blsA. Accordingly, incubation of A. baumannii ATCC 17978 cells at 37°C results in a 5-fold reduction of blsA mRNA levels compared with those of cells incubated at 24°C, independently of whether the cells were incubated in the presence or absence of light (Fig. 7). While the blsA transcript levels were comparable when cells were incubated at 37°C in darkness or in the presence of light, the lack of illumination resulted in a 1.5-fold increase when the cells were incubated at 24°C. Taken together, these results indicate that blsA could control cellular functions and possibly its own production via a temperature response never examined before in this bacterial pathogen.
FIG. 7.
Effects of light and temperature on blsA transcript levels. RNA from A. baumannii ATCC 17978 cells grown overnight on motility plates at 24°C and 37°C in the presence of blue light (L) or in darkness (D) was used as the template for qRT-PCR using blsA-specific primers. Transcription of recA was used as a constitutively expressed internal control. Error bars show the standard error of the mean.
DISCUSSION
It is apparent from our spectral, genetic, and functional data that A. baumannii ATCC 17978 senses and responds to blue light through a process that depends on the expression of a functional bslA gene, which codes for the production of a BLUF-containing photoreceptor. This observation is in agreement with in silico studies predicting the presence of a single gene coding for this type of photoreceptor in fully sequenced and annotated A. baumannii genomes (30, 49), our analysis of Acinetobacter genomic data, and the light-mediated response displayed by the A. baumannii AYE strain, the chromosome of which harbors a blsA homolog highly related to that present in ATCC 17978. The lack of motility displayed by the ATCC 19606T strain, which also has a blsA homolog, could be explained by the fact that ATCC 17978 cells produce pili that do not contain the CsuA/B putative pilin subunit and are structurally different from those detected on the surface of ATCC 19606T cells (C. McQueary and L. A. Actis, unpublished data).
Although photosensing seems to be a widespread phenotype among members of this genus that includes environmental and pathogenic species, there are variations in the number of predicted genes coding for photoreceptors among members of the Acinetobacter genus. For example, the A. baylyi ADP1 genome includes four genes predicted to encode BLUF domain-containing proteins. Also in agreement with previous studies (49) is the observation that BlsA belongs to the most common bacterial BLUF photoreceptors, small proteins containing a flavin-binding photosensing core lacking an effector or output domain(s). In spite of the apparent lack of an output domain(s), it is obvious from our data that BlsA serves as a regulatory factor controlling at least three different cell behaviors. Overall, our findings are compatible with the multistep regulatory process mediated by BlsA, which starts with light sensing and ends with the differential expression of diverse cellular functions, including those involved in motility and the interaction of bacteria with biotic and abiotic surfaces. BlsA could accomplish this role by interacting with unknown downstream signaling proteins with which it could form stable or transient regulatory complexes. In contrast to other systems in which the genes coding for the photosensing and signal-transducing components are located next to each other and are translationally coupled, our genomic analysis indicates that the A. baumannii photosensing system may involve transducers coded for by genes that do not map near blsA, as has been described in several unrelated bacteria (22).
The fact that the motility, virulence, and biofilm formation of ATCC 17978 cells are affected by light and the expression of blsA suggests that this signal and photoreceptor-coding gene, respectively, have a global effect on the physiology of A. baumannii. This hypothesis is supported by the spectral changes displayed by the BslA recombinant derivative upon blue light illumination and the dark-state recovery times, which are within the values of bona fide BLUF-containing bacterial photoreceptors that regulate different cellular functions (49), as well as the pleiotropic effects of the blsA insertion inactivation. Interestingly, these three cellular responses are functionally interrelated although different sets of genes code for their individual components. The negative effect of blue light on A. baumannii biofilm formation is similar to that of Idiomarina loihiensis, a deep sea bacterium that produces a single PYP photoreceptor protein (50). On the other hand, the A. baumannii blsA-mediated light response contrasts with the positive role light plays in biofilm formation by E. coli via the BLUF-EAL YcgF photoreceptor (45) and the attachment of Caulobacter crescentus to glass surfaces mediated by the LovK/LovR two-component light-dependent regulatory system (34). These differences could reflect the diverse sensing and regulatory pathways bacteria use to interact with surfaces (39) rather than differences in the mechanisms by which bacteria sense and respond to light.
The motility of A. baumannii ATCC 17978 cells on semisolid agarose plates is also reduced by blue light through a blsA-dependent process, a response that may explain the lack of biofilm formation under illumination. It is possible that, under darkness, genes coding for functions associated with biofilm formation, such as cell motility and the production of pili and exopolymeric substances, are relieved from blsA-mediated repression, favoring the formation of multicellular structures on solid and liquid surfaces. This possibility is supported by the observation that pilus production and motility are needed for full biofilm development by bacteria such as P. aeruginosa (5). It is also possible that light-mediated responses are part of complex regulatory processes, such as that mediated by YcgF in E. coli (45), which could control gene expression and cellular behavior by sensing interrelated signals (39). Temperature should be included among these signals affecting A. baumannii light-dependent responses since cell motility and biofilm formation at 37°C are independent of illumination. This temperature-dependent response to blue light, also detected in E. coli, may allow bacteria to sense their environmental location (45). This is a reasonable potential response by A. baumannii, considering its ability to grow at different temperatures and persist in hospital settings outside the human host (32, 43). In contrast to motility and biofilm formation, light and blsA play a positive role in the virulence of A. baumannii ATCC 17978 when tested using the killing of C. albicans tup1 mutant filaments (24), a model that in our studies parallels the observations obtained with A549 human alveolar epithelial cell monolayers (19). Interestingly, the role of blsA is reflected by not only a reduced virulence because of its inactivation but also an enhanced virulence phenotype as a consequence of a gene dosage effect when present as a higher-copy-number plasmid-based trait as it is in the case of the complemented derivative ATCC 17978.ORc. A similar effect was observed in B. subtilis, where overproduction of YtvA results in an enhanced light-mediated stress response (3). The potential virulence role of blsA is further supported by the observation that the expression of genes coding for LOV-HK photoreceptors is either constitutive or enhanced under conditions inducing the virulence of the plant pathogen P. syringae pathovar syringae strain DC3000, as well as being required for B. abortus replication in murine macrophages (40). Whether BlsA also plays a role in the pathogenesis of the infections A. baumannii causes in humans is not clear at the moment in view of the lack of understanding of the role of photoreceptors in bacterial virulence. However, it is possible to speculate that photoreceptors may not play a role in systemic infections considering the temperature and the lack of blue illumination of internal human tissues and organs targeted by this pathogen. On the other hand, the function of these receptors could be important in the pathogenesis of surface-exposed wound infections considering the potential exposure of bacteria to light and the relatively lower temperatures recorded in these types of lesions (31). It is also possible that the production of BlsA is responsible for the ability of A. baumannii to persist in medical settings, where it interacts with biotic and abiotic substrata under conditions that would require a gene expression pattern different from that expressed during the infection of the human host.
In summary, our study describes the ability of A. baumannii to sense and respond to blue light, a finding that provides a new and unexpected insight into the physiology of this bacterium. The motility response we observed during our work is also interesting considering that the Acinetobacter genus name (akinetobacter, nonmotile rod) reflects the nonmotile phenotype initially assigned to members of this bacterial group. This is perhaps one of the main reasons why this response, which plays a key role in bacterial cell biology and virulence, has been overlooked in most of the members of this genus.
Supplementary Material
Acknowledgments
Funds from the Agencia Nacional de Promoción Científica y Tecnológica and CONICET, Argentina (to M.A.M. and A.M.V.); U.S. Public Health Service AI070174 and NSF 0420479 grants (to L.A.A.); and Miami University supported this work.
We are grateful to J. Hawes, coordinator of the Miami University Center of Bioinformatics and Functional Genomics, for his support and assistance with automated DNA sequencing, nucleotide sequence analysis, and qRT-PCR analysis of gene expression. We thank L. Dijkshoorn (Department of Infectious Diseases, Leiden University Medical Centre, Leiden, Netherlands) for providing the A. baumannii clone I, II, and III strains used in this work, and Eligio Morandi (Facultad de Ciencias Agrarias, Universidad Nacional de Rosario, Rosario, Argentina) for his assistance in the spectral analysis of the light produced by the LEDs used in this work. We are grateful to R. Morgan-Kiss (Department of Microbiology, Miami University, Oxford, OH) for her comments and suggestions.
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 3.Avila-Pérez, M., K. J. Hellingwerf, and R. Kort. 2006. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188:6411-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barken, K. B., S. J. Pamp, L. Yang, M. Gjermansen, J. J. Bertrand, M. Klausen, M. Givskov, C. B. Whitchurch, J. N. Engel, and T. Tolker-Nielsen. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331-2343. [DOI] [PubMed] [Google Scholar]
- 6.Borer, A., J. Gilad, R. Smolyakov, S. Eskira, N. Peled, N. Porat, E. Hyam, R. Trefler, K. Riesenberg, and F. Schlaeffer. 2005. Cell phones and Acinetobacter transmission. Emerg. Infect. Dis. 11:1160-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72:249-252. [DOI] [PubMed] [Google Scholar]
- 9.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 10.Cevahir, N., M. Demir, I. Kaleli, M. Gurbuz, and S. Tikvesli. 2008. Evaluation of biofilm production, gelatinase activity, and mannose-resistant hemagglutination in Acinetobacter baumannii strains. J. Microbiol. Immunol. Infect. 41:513-518. [PubMed] [Google Scholar]
- 11.Charnot-Katsikas, A., A. H. Dorafshar, J. K. Aycock, M. Z. David, S. G. Weber, and K. M. Frank. 2009. Two cases of necrotizing fasciitis due to Acinetobacter baumannii. J. Clin. Microbiol. 47:258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel, C., S. Haentjens, M. C. Bissinger, and R. J. Courcol. 1999. Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol. Lett. 170:199-209. [DOI] [PubMed] [Google Scholar]
- 13.Dijkshoorn, L., H. Aucken, P. Gerner-Smidt, P. Janssen, M. E. Kaufmann, J. Garaizar, J. Ursing, and T. L. Pitt. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 15.Dorsey, C. W., M. E. Tolmasky, J. H. Crosa, and L. A. Actis. 2003. Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology 149:1227-1238. [DOI] [PubMed] [Google Scholar]
- 16.Dorsey, C. W., A. P. Tomaras, and L. A. Actis. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68:6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 19.Gaddy, J. A., A. P. Tomaras, and L. A. Actis. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77:3150-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaidenko, T. A., T. J. Kim, A. L. Weigel, M. S. Brody, and C. W. Price. 2006. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginalski, K., L. Kinch, L. Rychlewski, and N. V. Grishin. 2004. BOF: a novel family of bacterial OB-fold proteins. FEBS Lett. 567:297-301. [DOI] [PubMed] [Google Scholar]
- 22.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497-500. [DOI] [PubMed] [Google Scholar]
- 23.Han, Y., M. H. Meyer, M. Keusgen, and G. Klug. 2007. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol. Microbiol. 64:1090-1104. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 25.Houben, K., D. Marion, N. Tarbouriech, R. W. Ruigrok, and L. Blanchard. 2007. Interaction of the C-terminal domains of Sendai virus N and P proteins: comparison of polymerase-nucleocapsid interactions within the paramyxovirus family. J. Virol. 81:6807-6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunger, M., R. Schmucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45-51. [DOI] [PubMed] [Google Scholar]
- 27.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Leong, S. A., G. S. Ditta, and D. R. Helinski. 1982. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for δ-aminolevulininc acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257:8724-8730. [PubMed] [Google Scholar]
- 30.Losi, A., and W. Gartner. 2008. Bacterial bilin- and flavin-binding photoreceptors. Photochem. Photobiol. Sci. 7:1168-1178. [DOI] [PubMed] [Google Scholar]
- 31.McGuiness, W., E. Vella, and D. Harrison. 2004. Influence of dressing changes on wound temperature. J. Wound Care 13:383-385. [DOI] [PubMed] [Google Scholar]
- 32.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell, E. B., and S. Crosson. 2008. Photoregulation in prokaryotes. Curr. Opin. Microbiol. 11:168-178. [DOI] [PubMed] [Google Scholar]
- 34.Purcell, E. B., D. Siegal-Gaskins, D. C. Rawling, A. Fiebig, and S. Crosson. 2007. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. U. S. A. 104:18241-18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Baño, J., S. Marti, S. Soto, F. Fernandez-Cuenca, J. M. Cisneros, J. Pachon, A. Pascual, L. Martinez-Martinez, C. McQueary, L. A. Actis, and J. Vila. 2008. Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 14:276-278. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 39.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 40.Swartz, T. E., T. S. Tseng, M. A. Frederickson, G. Paris, D. J. Comerci, G. Rajashekara, J. G. Kim, M. B. Mudgett, G. A. Splitter, R. A. Ugalde, F. A. Goldbaum, W. R. Briggs, and R. A. Bogomolni. 2007. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 317:1090-1093. [DOI] [PubMed] [Google Scholar]
- 41.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473-3484. [DOI] [PubMed] [Google Scholar]
- 42.Tomaras, A. P., M. J. Flagler, C. W. Dorsey, J. A. Gaddy, and L. A. Actis. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398-3409. [DOI] [PubMed] [Google Scholar]
- 43.Towner, K. J. 2009. Acinetobacter: an old friend, but a new enemy. J. Hosp. Infect. 73:355-363. [DOI] [PubMed] [Google Scholar]
- 44.Towner, K. J., E. Bergogne-Berezin, and C. A. Fewson. 1991. Acinetobacter: portrait of a genus, p. 1-24. In K. J. Towner, E. Bergogne-Berezin, and C. A. Fewson (ed.), The biology of Acinetobacter. Plenum Press, New York, NY.
- 45.Tschowri, N., S. Busse, and R. Hengge. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23:522-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J. M. Claverie, D. Raoult, C. Medigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Broek, P. J., J. Arends, A. T. Bernards, E. De Brauwer, E. M. Mascini, T. J. van der Reijden, L. Spanjaard, E. A. Thewessen, A. van der Zee, J. H. van Zeijl, and L. Dijkshoorn. 2006. Epidemiology of multiple Acinetobacter outbreaks in The Netherlands during the period 1999-2001. Clin. Microbiol. Infect. 12:837-843. [DOI] [PubMed] [Google Scholar]
- 48.van den Broek, P. J., T. J. van der Reijden, E. van Strijen, A. V. Helmig-Schurter, A. T. Bernards, and L. Dijkshoorn. 2009. Endemic and epidemic Acinetobacter species in a university hospital: an 8-year survey. J. Clin. Microbiol. 47:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Horst, M. A., J. Key, and K. J. Hellingwerf. 2007. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol. 15:554-562. [DOI] [PubMed] [Google Scholar]
- 50.van der Horst, M. A., T. P. Stalcup, S. Kaledhonkar, M. Kumauchi, M. Hara, A. Xie, K. J. Hellingwerf, and W. D. Hoff. 2009. Locked chromophore analogs reveal that photoactive yellow protein regulates biofilm formation in the deep sea bacterium Idiomarina loihiensis. J. Am. Chem. Soc. 131:17443-17451. [DOI] [PubMed] [Google Scholar]
- 51.van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 52.Villegas, M. V., and A. I. Hartstein. 2003. Acinetobacter outbreaks, 1977-2000. Infect. Control Hosp. Epidemiol. 24:284-295. [DOI] [PubMed] [Google Scholar]
- 53.Wroblewska, M. M., A. Sawicka-Grzelak, H. Marchel, M. Luczak, and A. Sivan. 2008. Biofilm production by clinical strains of Acinetobacter baumannii isolated from patients hospitalized in two tertiary care hospitals. FEMS Immunol. Med. Microbiol. 53:140-144. [DOI] [PubMed] [Google Scholar]
- 54.Zimbler, D. L., W. F. Penwell, J. A. Gaddy, S. M. Menke, A. P. Tomaras, P. L. Connerly, and L. A. Actis. 2009. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22:23-32. [DOI] [PubMed] [Google Scholar]
- 55.Zirak, P., A. Penzkofer, T. Schiereis, P. Hegemann, A. Jung, and I. Schlichting. 2006. Photodynamics of the small BLUF protein BlrB from Rhodobacter sphaeroides. J. Photochem. Photobiol. B 83:180-194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.