Abstract
Mycobacterium tuberculosis is known to reside latently in a significant fraction of the human population. Although the bacterium possesses an aerobic mode of metabolism, it adapts to persistence under hypoxic conditions such as those encountered in granulomas. While in mammalian systems hypoxia is a recognized DNA-damaging stress, aspects of DNA repair in mycobacteria under such conditions have not been studied. We subjected Mycobacterium smegmatis, a model organism, to the Wayne's protocol of hypoxia. Analysis of the mRNA of a key DNA repair enzyme, uracil DNA glycosylase (Ung), by real-time reverse transcriptase PCR (RT-PCR) revealed its downregulation during hypoxia. However, within an hour of recovery of the culture under normal oxygen levels, the Ung mRNA was restored. Analysis of Ung by immunoblotting and enzyme assays supported the RNA analysis results. To understand its physiological significance, we misexpressed Ung in M. smegmatis by using a hypoxia-responsive promoter of narK2 from M. tuberculosis. Although the misexpression of Ung during hypoxia decreased C-to-T mutations, it compromised bacterial survival upon recovery at normal oxygen levels. RT-PCR analysis of other base excision repair gene transcripts (UdgB and Fpg) suggested that these DNA repair functions also share with Ung the phenomenon of downregulation during hypoxia and recovery with return to normal oxygen conditions. We discuss the potential utility of this phenomenon in developing attenuated strains of mycobacteria.
The genus Mycobacterium includes some of the most important human pathogens, such as M. tuberculosis and M. leprae, the causative agents of tuberculosis and leprosy, respectively. M. tuberculosis is known to reside latently in a significant fraction of the human population (6). The bacterium possesses an aerobic mode of metabolism. However, it undergoes a change in gene expression (21) to enable it to adapt to microaerophilic conditions, such as those found in granulomas (37). Although animal model systems are available to establish granulomas harboring the pathogen (13), the availability of sufficient bacteria from such models to allow microbiological/biochemical studies has remained challenging.
To circumvent this difficulty, Wayne and coworkers developed an in vitro hypoxia model of dormancy for M. tuberculosis for growth under sealed and slow stirring conditions (39), in which the bacterium gradually shifts from the log phase of growth to a distinct nonreplicative persistence stage characterized by a state of prolonged viability but very low metabolic activity. It has been argued that this process of in vitro hypoxia leading to bacterial persistence shares similarities with the actual infection model, in which bacteria remain within the microaerophilic environment of granulomas (40). The conditions for hypoxic growth of M. smegmatis have also been established (5), and it has been observed that under sealed and slow stirring conditions, initially there is a rapid increase in the viable count due to aerobic growth. This growth is then followed by a brief phase of slower growth under the microaerophilic conditions (generated upon consumption of the available oxygen). At the end of this growth, bacteria are in a phase of an extended life span characterized by a slow decline in the viable count.
Biochemical and genetic analyses of bacteria subjected to this in vitro hypoxia model have yielded a wealth of knowledge about the metabolic status of the dormant bacterium. It has been shown that there is considerable synthesis of RNA but not DNA in M. tuberculosis when subjected to hypoxia (39). Also, during this stage, bacteria are highly susceptible to the action of metronidazole, a prodrug, which is activated under the reducing environmental conditions and inflicts DNA damage (7, 12). It has been shown that M. smegmatis, a nonpathogenic and fast-growing mycobacterium, when subjected to the Wayne's model of hypoxia shows physiological changes similar to those observed in M. tuberculosis in terms of biochemical properties and metronidazole sensitivity (19). Further, a comparison of the genome sequences of M. tuberculosis and M. smegmatis showed that the latter retains most of the genes involved in adaptation to hypoxia (35). Given the tractability of genetically manipulating M. smegmatis, it has been found to serve as a suitable model to study hypoxia-induced changes in mycobacteria (19). The hypoxia conditions are known to result in generation of DNA damage, such as base modifications, abasic sites, and single-stranded DNA breaks, in mammalian systems (10, 20). However, aspects of DNA damage (and repair) in mycobacteria under such conditions have not been studied.
Mycobacterial genomes, owing to their G+C-rich nature, are naturally at high risk of cytosine deamination, which leads to the occurrence of uracils in DNA. Such a conversion of cytosines to uracils could be enhanced by the habitat of the pathogen in host macrophages, which respond by generating reactive oxygen and nitrogen intermediates, compounds which are known to enhance cytosine deamination (8, 41). In fact, it has been shown that uracil DNA glycosylase (Ung) provides a crucial DNA repair activity in mycobacteria (28, 36). Furthermore, we have reported that M. smegmatis strains deficient in nucleotide excision repair or uracil excision repair have a compromised fitness under hypoxia (17). In this study, using the M. smegmatis model, we have addressed the temporal aspects of Ung-mediated repair during in vitro-generated hypoxia.
MATERIALS AND METHODS
Strains, DNA oligomers, and plasmids.
The details of M. smegmatis mc2155 (31) derivatives and DNA oligomers and plasmids are given in Tables 1 and 2. M. smegmatis strains were grown in Dubos broth base with 0.2% (vol/vol) Tween 80 and supplemented with 10% albumin-dextrose complex (ADC; BBL). For growth on a solid surface, 1.5% agar was included in Luria-Bertani (LB) medium containing 0.05% Tween 80. When specified, 7H10 medium (Difco) containing 0.5% (vol/vol) glycerol was used. Media were supplemented with hygromycin and kanamycin at 50 μg ml−1, when needed.
TABLE 1.
DNA oligomers and plasmids
| Oligomer or plasmid | Sequence (5′ to 3′) or description | Reference or source |
|---|---|---|
| Oligomers | ||
| Msm-hspXRT_Fp | CGGCGTGTTGACCATCAAG | This study |
| Msm-hspXRT_Rp | GATCGTGAGGATGCCTTTGTC | |
| Msm-udgBRT_Fp | GGCACAGTGGACGGGTGT | |
| Msm-udgBRT_Rp | GAGATCATCGCGTCGAGGTC | |
| Msm-16SRT_Fp | AAGCGCAAGTGACGGTATGTG | |
| Msm-16SRT_Rp | AAGCTGTGAGTTTTCACGAACAAC | |
| Msm-ungRT_Fp | GCAGCCTGTCCAACATCTTCAC | |
| Msm-ungRT_Rp | CGGCCTCCCATCCTTTACC | |
| Msm-mutMRT_Fp | CACGTCGTTCGATTCGTTGTAC | |
| Msm-mutMRT_Rp | GCTGACATCTCGGGCAGTAGA | |
| Msm-uvrBRT_Fp | GATGGCGCCCAACAAGAC | |
| Msm-uvrBRT_Rp | CGTTGATCGAGCTGTCCTTCTC | |
| narK2_Fp | GATATCGTGATCCGACTGGTCGCC | |
| narK2_Rp | CCATGGCGAACTCCGCGCCCC | |
| PTUa | 5′-A∼T∼ATACCGCGGUCGGCCGATCAAGCTTA∼T∼T-3′ | 16 |
| Plasmids | ||
| pJet1.2 | E. coli vector for cloning PCR products | MBI Fermentas |
| pRARE | E. coli plasmid (p15a origin of replication, Cmr), encodes tRNA genes (proL, leuW, metT, argW, thrT, glyT, tyrU, thrU, argU, and ileX) | Novagen |
| pETMsmUng | pET 11D E. coli expression construct harboring ung ORF from M. smegmatis SN2 between NcoI and EcoRI sites (renamed from pETMsmUDG) | 2 |
| pRSETbMsmUng | pRSET E. coli expression construct harboring M. smegmatis ung between NcoI and HindIII sites along with a 40-amino-acid N-terminal presequence | This study |
| pRSET-11-MsmUng | Derivative of pRSETbMsmUng in which the N-terminal 40-amino-acid presequence has been reduced to 11 amino acids | |
| pTrcHisMsmUng | E. coli expression construct with M. smegmatis ung along with 11-amino-acid N-terminal tag from pRSET-11-MsmUng cloned between NdeI and HindIII sites | |
| pTrcMsmUng | pTrc99c harboring M. smegmatis ung ORF between NcoI and HindIII sites | 1 |
| pJet1.2narK2 | pJet vector containing ∼300 bp of narK2 promoter within EcoRV and NcoI sites | This study |
| pTrcnarK2ung | pTrcMsmUng vector with narK2 promoter cloned upstream of ung ORF between EcoRV and NcoI sites | This study |
| pMV361 (Hygr) | Integrative vector in mycobacteria containing L5 att region with E. coli origin of replication and Hygr marker | 33 |
| pMV361Δhsp | Derivative of pMV361 vector with hsp60 promoter truncated by BsrDI digestion, end filled, and ligated | This study |
| pMVnarK2ung | pMV361Δhsp in which narK2ung from pTrcnarK2ung has been cloned between PvuII and HindIII sites | This study |
The ∼ symbols indicate phosphorothioate modifications in the oligonucleotide.
TABLE 2.
List of M. smegmatis strains used in the study
| M. smegmatis strain | Relevant characteristic(s) | Reference |
|---|---|---|
| SN2 | Laboratory strain | 23 |
| mc2155 or wild type | High-efficiency transformation strain | 31 |
| WT(L5 att::pMV361) | mc2155 harboring an integrative vector, pMV361 (Hygr), at the L5 att site of the chromosome | This study |
| ung::kan strain | mc2155 with ung gene disrupted with kan cassette | 36 |
| ung::kan(L5 att::narK2ung) strain | ung::kan strain harboring the integrative vector, pMVnarK2ung (Hygr), at the L5 att site of the chromosome | This study |
| WT(L5 att::narK2ung) | mc2155 strain harboring pMVnarK2ung at the L5 att site of the chromosome |
In vitro dormancy setup.
Isolated colonies of M. smegmatis strains obtained on 7H10 plates containing appropriate antibiotics were grown to an optical density at 600 nm (OD600) of ∼0.6 (∼1.5× 108 CFU/ml) in triplicates, and 0.2-ml aliquots of these cultures were inoculated into 30-ml screw-cap tubes with a headspace ratio of 0.5 (20 ml medium and 10 ml air space). The cultures were stirred at 180 rpm using a multipoint magnetic stirrer at 37°C for 10 days (17). The viability of cultures was determined by serial dilution plating at different times. For RNA isolation, hypoxia cultures (400 ml) were set up in screw-cap 600-ml-capacity conical flasks and subjected to slow stirring conditions as mentioned above. Following 10 days of incubation (hypoxia phase), 200 ml of culture was harvested immediately and processed. The remaining 200 ml was aerated and incubated at 37°C for 1 h under vigorous shaking conditions (recovery phase) prior to RNA isolation. A hypoxic culture of M. tuberculosis H37Ra was established by inoculating 4 ml of log-phase cells (OD600, ∼0.6) into 400 ml of Dubos medium containing ADC supplement in a screw-cap 600-ml-capacity conical flask. Methylene blue was added to the medium at a final concentration of 1.5 μg ml−1 to monitor oxygen depletion. The flasks were subjected to slow stirring (140 rpm using a multipoint magnetic stirrer) and maintained at 37°C for 14 days. The fading of methylene blue was observed on the sixth day, and decolorization was observed on the 12th day after inoculation.
Preparation of cell extracts.
M. smegmatis cells from hypoxic cultures or recovery cultures (1 h under normal oxygen levels after release from hypoxia) grown at 37°C were harvested by centrifugation for 5 min at 10,000 rpm using an AF5004CA rotor (Kubota) at 4°C. The hypoxia culture (400 ml) of M. tuberculosis H37Ra was harvested at 4°C by centrifugation for 10 min at 7,500 rpm in a Sorvall RCB plus GSA rotor. The bacterial pellets were suspended in TME (25 mM Tris-HCl [pH 8.0], 1 mM Na2EDTA, and 2 mM β-mercaptoethanol), repelleted by centrifugation, and stored at −20°C. The pellet was taken up in 0.3 ml of TME and ultrasonicated using a Vibra Cell (Sonics) five times for 30 s each (50% duty cycle). The lysates were centrifuged using a J70759 rotor (Kubota) at 14,000 rpm for 30 min at 4°C, and the supernatant was stored as cell lysates.
Ung assays.
Reaction mixtures (15 μl) were set up with 0.5 pmol phosphorothioate and uracil (PTU) oligonucleotides (32P-5′-end labeled; ∼5,000 cpm) and cell extract (1 μg total protein) in 1× Ung buffer (50 mM Tris-HCl [pH 8.0], 1 mM Na2EDTA, 1 mM dithiothreitol, and 25 μg ml−1 bovine serum albumin), incubated at 37°C for a time course analysis (0, 5, and 15 min), stopped by addition of 5 μl 0.2 N NaOH and heating at 90°C for 15 min, and analyzed by electrophoresis on 8 M urea-15% polyacrylamide gels (27).
Generation of overexpression construct, purification of M. smegmatis Ung, production of anti-Ung antibodies, and immunoblot analysis.
The M. smegmatis ung gene sequence was retrieved from the TIGR database, and the M. smegmatis ung open reading frame (ORF) was PCR amplified using forward and reverse primers (5′-AGGGATCCCATGGCCGCACGGC-3′ and 5′-GGAATTCCTACTAGGGCAACTTCC-3′, respectively; restriction sites are shown in italics), M. smegmatis SN2 genomic DNA, and Pfu DNA polymerase. The amplicon (∼700 bp) was digested with NcoI and EcoRI and cloned into similarly digested pET11d to generate pETMsmUng and verified by DNA sequencing of the complete ORF (2). The NcoI-HindIII fragment from pETMsmUng was subcloned into similarly digested pRSETb to generate pRSETbMsmUng containing a 40-amino-acid presequence. This construct was digested with NcoI and NheI, blunt ended by filling in using Klenow fragment of DNA polymerase I, and ligated to generate pRSET-11-MsmUng containing an 11-amino-acid presequence (MHHHHHHGMAS). Subsequently, the NdeI-HindIII fragment from this expression plasmid was subcloned into pTrc99CNdeI (30) between the NdeI and HindIII sites to generate pTrcHisMsmUng.
To purify M. smegmatis Ung, fresh transformants of Escherichia coli BW310 (ung deficient) harboring pRARE and pTrcHisMsmUng plasmids were grown in 1.5 liter LB containing ampicillin and chloramphenicol to an OD600 of ∼0.6. M. smegmatis Ung expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 4 h at 37°C, and the cells were harvested by centrifugation at 8,000 rpm for 10 min using a JA14 rotor (Beckman) at 4°C. The pellet was suspended in buffer A (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 10% [vol/vol] glycerol) and lysed by ultrasonication in a Vibra Cell (Sonics). The lysate was centrifuged at 12,000 rpm at 4°C using a JA14 rotor (Beckman) for 40 min. The supernatant was loaded onto a 5-ml Ni-nitrilotriacetic acid (NTA) column (Amersham) preequilibrated with buffer A. The column was washed with buffer A, and the protein was eluted with a gradient of 0 to 500 mM imidazole in buffer A. Fractions enriched for M. smegmatis Ung were pooled and dialyzed against buffer A. The dialyzed protein containing 20 mM imidazole was loaded again onto the Ni-NTA column and eluted with 500 mM imidazole. Fractions containing M. smegmatis Ung at near-homogeneity were pooled, dialyzed against 50% (vol/vol) glycerol containing 500 mM NaCl and 10 mM Tris-HCl (pH 7.5), and stored at −20°C.
Antibodies against M. smegmatis Ung were raised in a New Zealand rabbit (11) by using Freund's incomplete adjuvant. For immunoblot analysis, equal amounts (∼30 μg) of total cell proteins from different stages of growth were separated on a sodium dodecyl sulfate (SDS)-15% polyacrylamide gel and transferred onto a polyvinylidene difluoride (PVDF) membrane using transfer buffer (25 mM Tris-HCl [pH 8.8], 200 mM glycine, and 0.1% [wt/vol] SDS containing 15% [vol/vol] methanol) in a semidry transfer apparatus (Bio-Rad) for 1 h at 15 V. The membrane was treated with blocking buffer containing 5% (wt/vol) skimmed milk powder in Tris-buffered saline-Tween (TBST; 20 mM Tris-HCl [pH 7.5], 0.9% [wt/vol] NaCl, and 0.2% [vol/vol] Tween 20) overnight at 4°C. The serum (10 μl) containing M. smegmatis Ung antibodies was incubated with ∼60 μg of cell extract from the M. smegmatis ung::kan strain for 30 min at room temperature and diluted to 15 ml in the blocking buffer to probe the blot for 2 h at room temperature. The membrane was washed thrice with TBST for 15 min each and probed with anti-rabbit IgG antibody conjugated with horseradish peroxidase (HRP) at a dilution of 1:5,000 in the blocking buffer for 2 h. The membrane was washed thrice, as above, and developed using the ECL Plus Western blotting detection system (GE Healthcare) as per the manufacturer's protocol.
RNA isolation from M. smegmatis.
Bacteria were harvested from triplicate 200-ml cultures (mid-log, hypoxia, or recovery phase) at 4°C for 5 min at 10,000 rpm using an AF5004CA rotor (Kubota) and taken up in 100 μl of RNAlater solution (Sigma). Lysis solution (400 μl) containing 30 mM Tris-HCl (pH 7.4), 5 mM Na2EDTA, 100 mM NaCl, 1% (vol/vol) SDS, 100 mM β-mercaptoethanol, and 5 mM vanadyl ribonucleoside complex plus 1 ml Tri reagent (Sigma) was added to the cell suspension and ultrasonicated five times for 30 s each at a 50% duty cycle under ice using a Vibra Cell apparatus (Sonics). RNA was extracted according to the manufacturer's protocol.
cDNA synthesis and quantitative real-time RT-PCR analysis.
The total RNA preparation (∼8 to 10 μg) was treated with RNase-free DNase I (8 U; MBI Fermentas) for 60 min. The absence of DNA was confirmed by the absence of PCR products upon the use of DNase I-treated RNA samples as templates. Subsequently, the RNA (∼1 μg) was used for cDNA synthesis by using Moloney murine leukemia virus (MMLV) reverse transcriptase (RT; MBI Fermentas). Oligonucleotides were designed to amplify ∼200 bp of the various ORFs (ung, udgB, fpg [mutM], uvrB, and 16S rRNA sequences) (Table 1). The mix containing RNA and 25 pmol of reverse primer for each gene was heated to 65°C for 5 min and snap-chilled on ice. The primer was annealed to the template at 60°C for 5 min, chilled, and mixed with a cocktail containing 1 mM deoxynucleoside triphosphates (dNTPs), RNAguard, and 40 U MMLV reverse transcriptase to a total volume of 20 μl. The primers annealed to RNA template were extended at 45°C for 1 h and heated at 70°C for 10 min to terminate the reaction.
Aliquots of cDNAs (25 ng) were used as templates for quantitative real-time PCR with SYBR green PCR mix (Finnzymes, Finland) containing a 1× final concentration of real-time master mix and 10 pmol each of the forward and reverse primers in an ABI Prism 7900HT machine (Applied Biosystems). Amplification of 16S rRNA cDNA served as an internal control. Analyses were carried out for triplicate samples of RNA prepared from mid-log-phase, 10-day hypoxia, and recovery-phase cells. Changes in the expression levels were calculated by the comparative threshold cycle (2−ΔΔCT) method (29). The differences in the expression patterns were analyzed for significance by using Student's paired t test.
Generation of pMVnarK2ung and its introduction into M. smegmatis.
The DNA sequence (∼300 bp) upstream of the narK2 gene (Rv1737c) of M. tuberculosis was retrieved from Tuberculist (http://genolist.pasteur.fr/TubercuList), and the oligomeric DNA primers narK2_Fp and narK2_Rp were designed to contain EcoRV and NcoI restriction sites, respectively. The promoter region of narK2 was amplified in a PCR mixture (50 μl) containing ∼300 ng of M. tuberculosis H37 Rv DNA, 200 μM dNTPs, 20 pmol each of narK2_Fp and narK2_Rp primers, and 1 U Dynazyme EXT enzyme (Finnzymes, Finland) in 1× Dynazyme EXT buffer. PCR conditions included an initial heating of 94°C for 4 min, followed by 30 cycles of heating at 94°C for 1 min, 60°C for 30 s, and 72°C for 45 s, and a final extension for 10 min at 72°C. The amplicon was gel purified and cloned into pJet 1.2 (MBI Fermentas). The narK2 promoter DNA was released by EcoRV and NcoI digestion and cloned into similarly digested pTrcMsmUng (1). The clones were verified by DNA sequencing (Macrogen, South Korea). An ∼1.0-kb fragment consisting of narK2 promoter and the ung open reading frame was released by EcoRV and HindIII digestion and cloned into PvuII- and HindIII-digested pMV361Δhsp to generate pMVnarK2ung. pMVnarK2ung was electroporated into M. smegmatis to integrate at the L5 att site in the genome.
Analysis of the mutation spectrum.
Bacterial cultures (20 ml, in quintuplet) were subjected to 10 days of hypoxia and harvested at 7,500 rpm in an AF5004CA rotor (Kubota) at 4°C for 10 min, spread on rifampin plates, and incubated for 4 days at 37°C. The isolated rifampin-resistant (Rifr) colonies were used in a colony PCR to amplify the rifampin resistance-determining region (RRDR) of rpoB and sequenced (18).
RESULTS
Growth of M. smegmatis under hypoxia.
Culturing of M. smegmatis under hypoxia has been reported previously (5, 19). A similar hypoxia setup in our study showed that the maximal increase in the viable count of bacteria occurred within 3.5 days. This period was then followed by a phase of smaller increases in the viable counts, and thereafter essentially no significant changes occurred in the viable counts of the bacteria for up to 10 days. The 10-day hypoxia culture corresponded to a phase defined by the extended life span of the bacterium under hypoxic growth. In addition, as reported earlier (3, 19), we noted an upregulation of hspX (MSMEG_2031c) mRNA in the 10-day hypoxia culture, which validated the hypoxic culture conditions of this study (data not shown).
Ung activity during hypoxic growth.
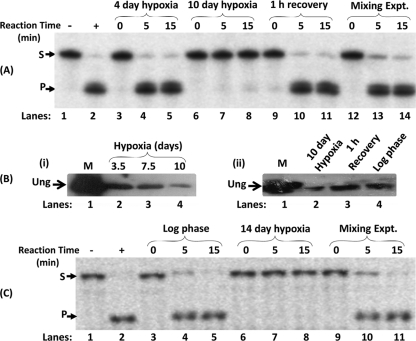
Previous studies (13, 17) implicated a crucial role for DNA repair in bacterial survival under hypoxia. However, a time course analysis of Ung activity in the cell extracts prepared from different stages of hypoxia showed a decrease in Ung activity with the progression of hypoxia to 10 days (Fig. 1A; compare the intensities of the product bands in lanes 7 and 8 with those in lanes 4 and 5). Interestingly, the Ung activity was restored during the reactivation phase of growth (1 h at 37°C) from hypoxia to normal oxygen levels (lanes 10 and 11). As a control, when equal amounts of extracts from the 10-day hypoxia and the recovery-phase cells were mixed, Ung activity was observed (lanes 13 and 14), ruling out the possibility that the lack of Ung activity in the 10-day hypoxia cell extracts was due to the presence of some inhibitors. Furthermore, as shown in Fig. 1B (panel i), immunoblot analysis of the similarly prepared extracts revealed that there was a notable decrease in Ung during 10-day hypoxia (compare lane 4 with lanes 2 and 3), and the Ung level was restored to the log-phase level during recovery with normal oxygen levels (Fig. 1B, panel ii, compare lane 3 with lanes 2 and 4, respectively). These data show that the Ung level decreases during hypoxia, and the same is then restored during recovery under normal oxygen conditions. To analyze if such a phenomenon occurs in other mycobacteria, we used M. tuberculosis H37Ra as a model. The hypoxia growth conditions (see Materials and Methods) were those established earlier (39). As shown in Fig. 1C, the phenomenon of downregulation of Ung during hypoxia is conserved in M. tuberculosis (compare lanes 4 and 5 with lanes 7 and 8).
FIG. 1.
(A) Uracil excision activity assay results for different stages of M. smegmatis growth. Cell extracts (1 μg each) from different stages of hypoxia and recovery phases were used for a time course analysis of uracil excision. The activity assays were performed for 0, 5, and 15 min at 37°C using a 32P-5′-end-labeled 30-mer DNA oligomer (PTU) containing uracil at the 12th position. In vitro mixing (lanes 12 to 14) refers to mixing of equal amounts of extracts from the 10-day hypoxia and recovery-phase cell extracts. M. smegmatis Ung (100 ng) was used as a positive control (lane 2). Following the reaction, the samples were further processed (see Materials and Methods) and analyzed on 8 M urea-15% polyacrylamide gels. S and P refer to substrate and product, respectively. (B) Immunoblot analysis of M. smegmatis cell extracts. (i) Equal amounts of cell extracts (∼30 μg) from different stages of hypoxia (3.5, 7.5, and 10 days) were separated on an SDS-15% polyacrylamide gel along with 200 ng of pure M. smegmatis Ung protein as marker (lane 1; M). The protein samples were transferred onto a PVDF membrane, probed with anti-M. smegmatis Ung antiserum, and developed using goat anti-rabbit IgG conjugated with HRP and using an enhanced chemiluminescence reagent. (ii) Equal amounts of cell extracts (∼30 μg) from 10-day hypoxia, 1-h recovery, and mid-log-phase cultures were separated on an SDS-15% polyacrylamide gel along with 200 ng of pure M. smegmatis Ung protein as marker (lane 1; M) and processed as described above. (C) Uracil excision activity assay results from different stages of M. tuberculosis growth. Cell extracts (1 μg each) from log-phase and 14-day hypoxia cultures were used for a time course analysis of uracil excision as described for panel A.
Generation of a hypoxia-inducible Ung expression construct.
The promoter element narK2, a hypoxia-inducible gene in M. tuberculosis, has been exploited for dormancy-specific gene expression in Mycobacterium bovis BCG (25). To explore the utility of this promoter in M. smegmatis, we generated pMVnarK2ung (which integrates into the L5 att site in the genome) by cloning the M. smegmatis ung open reading frame downstream of the narK2 promoter (Fig. 2A), and we introduced it into an ung-deficient strain of M. smegmatis (ung::kan) to avoid background from the native ung gene (during expression analysis). As shown in Fig. 2B, the strain showed no detectable Ung activity during the log phase of growth (lanes 4 and 5). However, when the culture was subjected to growth under hypoxia (10 days), Ung activity could be detected (lanes 7 and 8), suggesting that the narK2 promoter directed hypoxia-specific gene expression in M. smegmatis.
FIG. 2.
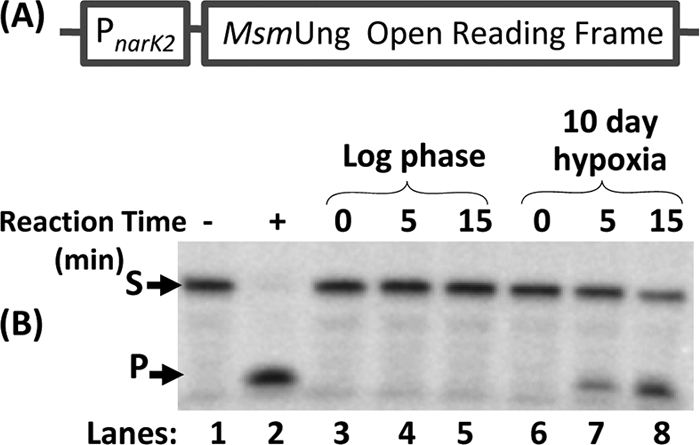
Expression analysis of the narK2 promoter. (A) Schematic representation of the pMVnarK2ung construct, depicting the promoter of narK2 from M. tuberculosis H37Rv cloned upstream of M. smegmatis Ung. (B) The mycobacterial vector pMVnarK2ung was introduced into M. smegmatis ung::kan by electroporation. The strain was grown to mid-log phase and subjected to 10 days of hypoxia. Cell extracts (1 μg each) from mid-log (lanes 3 to 5) and hypoxia (lanes 6 to 8) stages were used in a time course assay for uracil excision using 5′-32P-end labeled PTU. Purified M. smegmatis Ung (100 ng) was used as a positive control (lane 2). S and P refer to substrate and product, respectively.
Misexpression of Ung in M. smegmatis during hypoxia.
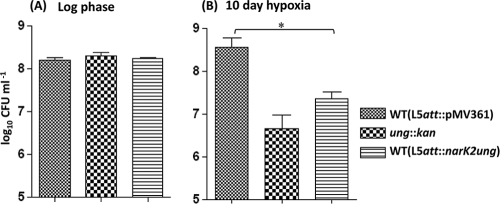
Having verified that the narK2 promoter functions specifically during hypoxia, we introduced the pMVnarK2ung construct into wild-type M. smegmatis and subjected the strain to hypoxic conditions of growth. The survival of this strain was then compared with the survival of the parent strain harboring the vector alone at the L5 att site (L5att::pMV361), as well as an ung-deficient (ung::kan) M. smegmatis strain. As shown in Fig. 3, while the viable counts of all three strains were comparable in the mid-log phase (Fig. 3A), upon subjecting the strains to 10-day hypoxia, the viable counts of M. smegmatis harboring narK2ung or that was deficient in Ung decreased significantly (Fig. 3B). The decrease in the viability of the ung-deficient (ung::kan) strain, during hypoxia, was also reported previously (17).
FIG. 3.
Viable counts of M. smegmatis strains in mid-log and 10-day hypoxia cultures. M. smegmatis mc2155 harboring pMV361 alone or pMVnarK2ung, where ung is driven by the narK2 promoter (a hypoxia-specific promoter), and the ung::kan strain were grown to mid-log phase in triplicate in Dubos medium with ADC supplement and subjected to hypoxic culture (10-day hypoxia). Viabilities of the strains grown to mid-log phase (A) and after subjecting them to 10-day hypoxia (B) were determined by the serial dilution plating method. The differences in viable counts were analyzed for statistical significance by performing Student's t test using GraphPad Prism 4.03 software. *, P < 0.05.
Mutational spectrum during hypoxia.
It was reported earlier that cells subjected to hypoxia display increased mutations (14, 20, 42). To determine the nature of mutations during hypoxia, we sequenced RRDR amplicons from the rifampin-resistant isolates. Sequencing results (Table 3) showed that the major spectrum of mutations in the wild-type strain was represented by A-to-G and C-to-T transitions (43% and 45%, respectively). However, in the strain expressing Ung (under the narK2 promoter), the spectrum was more biased toward A-to-G transitions (72%; 34 out of 47 samples), and the C-to-T mutations decreased to nearly half the value observed in the parent strain. Interestingly, under the same conditions, the M. smegmatis (ung::kan) strain revealed a major shift in the mutational spectrum toward C-to-T mutations (86%; 24 out of 28 samples).
TABLE 3.
Spectrum of mutations in the RRDR locus of rifampin-resistant isolates of three M. smegmatis strains
| Mutations detected | % of M. smegmatis isolates with indicated mutations (no. with mutations/total)a |
||
|---|---|---|---|
| WT (hyg)b | ung::kanc | pMVnarK2ungd | |
| A→G, T→C | 43 (18/42) | 14 (4/28) | 72 (34/47) |
| C→T, G→A | 45 (19/42) | 86 (24/28) | 21 (10/47) |
| C→G, G→C | 5 (2/42) | None | 2 (1/47) |
| C→A, G→T | 5 (2/42) | None | None |
| A→C, T→G | 2 (1/42) | None | 4 (2/47) |
Bacterial cultures (five replicates for each strain) were subjected to hypoxia and, at the end of the 10-day incubation, plated on medium containing rifampin. The RRDR loci from the rifampin-resistant colonies were sequenced. Values in parentheses are the number of isolates with the indicated mutations/the total number with mutations in the RRDR locus.
A total of 50 samples were sequenced for mutations in the RRDR, of which 8 samples did not have mutations.
A total of 45 samples were sequenced for mutations in the RRDR, of which 17 samples did not have mutations.
A total of 63 samples were sequenced for mutations in the RRDR, of which 16 samples did not have mutations.
Analysis of expression of other DNA repair genes under hypoxia.
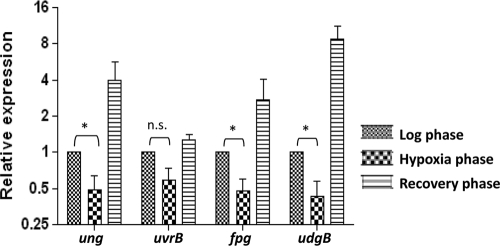
To address if the downregulation of DNA repair functions was specific to Ung, we carried out quantitative real-time PCR analysis for the presence of the mRNAs corresponding to the other base excision and nucleotide excision repair enzymes (ung, udgB, fpg, and uvrB). As shown in Fig. 4, the levels of mRNAs corresponding to ung, fpg, and udgB showed a significant decrease (although to various extents) during 10 days of hypoxia (values shown are relative to the mid-log-phase control). And, as was the case with ung, transcription of both fpg and udgB increased substantially during the recovery of the culture under normal oxygen conditions. Although the transcripts of uvrB also appeared to follow the same trend, the changes were not statistically significant (Fig. 4).
FIG. 4.
Analyses of DNA repair gene mRNAs by quantitative real-time PCR. Total RNA were isolated in triplicate and processed to remove contaminating DNA. The cDNA was synthesized using gene-specific primers. The mRNA levels were determined using quantitative real-time PCR mix containing SYBR green dye. Amplification of 16S rRNA was used as an internal control. The expression level of each gene was determined by the comparative CT method after normalizing with a 16S rRNA control. The mean expression levels ± standard deviations for each gene under different conditions are plotted (using a log2 scale), and the differences in expression were analyzed for statistical significance by performing a paired Student's t test using GraphPad Prism 4.03 software. *, P < 0.05.
DISCUSSION
The landmark study of Wayne and Hayes (39) reported that as bacteria sense depletion of oxygen levels, they differentiate to a nondividing persistent population. Microarray analyses of total RNA preparations have shown significant similarities in the transcriptional responses of bacteria subjected to hypoxia or exposed to nitric oxide (38). However, such analyses using M. tuberculosis RNA from different stages of hypoxia or other stresses have not been able to reveal changes in the expression of DNA repair genes (4, 21). Interestingly, our analysis using a candidate gene approach in the M. smegmatis model, showing downregulation of DNA repair functions during hypoxia, raises the possibility that the DNA repair genes are similarly downregulated during the latent stages of M. tuberculosis infection.
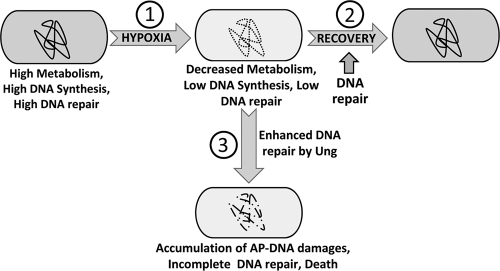
Under aerobic conditions, Ung of M. smegmatis is active throughout the exponential and the stationary phases (23, 24). Under hypoxia, the bacterial metabolism is low, and since bacteria are not dividing, downregulation of DNA repair functions (as observed) may be favored. To understand the physiological significance of downregulation of Ung during hypoxia, we performed two kinds of experiments. In the first case, we investigated the impact of ung gene knockout in M. smegmatis (reference 17 and this report), and we found that Ung is important and its knockout leads to a decrease in the viability of M. smegmatis upon recovery from hypoxia. In the second approach, we engineered misexpression of Ung in M. smegmatis under hypoxic conditions. Such a treatment also resulted in detrimental effects on the survival of bacteria (Fig. 3B). A similar phenotype in both the approaches may, at the first instance, seem counterintuitive. However, it is known that generation of excessive AP sites due to expression of Ung is toxic to cells containing high levels of uracil in the genome (9, 34). It has also been reported that under excessive DNA damage by nitric oxide intermediates, actions of DNA glycosylases lead to toxicity in AP endonuclease-deficient bacteria. Interestingly, this toxic effect could be suppressed by inactivation of the base excision repair enzymes (32). Hence, a likely model to understand the physiological significance of Ung downregulation during hypoxia in M. smegmatis (and possibly in other mycobacteria) is that during hypoxia there is substantial deamination of cytosines to uracils. Because of the general downregulation of most genes and low cellular metabolism, a low level of Ung (meaning fewer events of uracil excision) may be strategic in synchronizing the frequency of uracil excisions with the available activities of the downstream enzymes of the base excision repair pathway and in avoiding accumulation of “unattended” AP sites. As the accumulation of AP sites is toxic (34), it is physiologically sagacious to downregulate base excision repair enzymes in general (e.g., Ung, UdgB, and Fpg, as examined in this study). Under such circumstances, and assuming that hypoxia results in accumulation of uracils in DNA, it is not surprising that misexpression of Ung is detrimental to the viability of the bacterium (Fig. 5).
FIG. 5.
Model of detrimental effects of Ung deficiency and misexpression during hypoxia. Bacterial physiology during the log phase (extreme left) is characterized by high metabolism and efficient DNA synthesis and repair. Under hypoxic stress (step 1), bacterial metabolism declines and is characterized by low DNA synthesis (39) and repair (present study). The ability of hypoxia to inflict DNA damage may lead to persistence of bacteria with damage in the genome (depicted by the dotted line). Release of bacteria from hypoxia to normal oxygen conditions (step 2) leads to reactivation of DNA repair processes (present study), and repair of the accumulated damage (depicted by the bold line) allows bacterial multiplication. Misexpression of Ung (step 3) under hypoxia leads to untimely excision of uracil in DNA, committing the bacteria to the base excision repair pathway even when the downstream repair functions may be deficient. Inability to complete DNA repair leads to accumulation of AP sites and other damage, such as DNA breaks (depicted by the broken line), which are detrimental to bacterial survival. In such a model, Ung deficiency at step 2 would lead to deficient uracil repair, an increase in C-to-T mutations, and detrimental effects on survival. Misexpression of Ung at step 3 would result in excessive uracil release and accumulation of AP sites and other irreparable DNA damage, which too would be detrimental to the survival of the bacteria.
To further understand the mechanism of the detrimental impacts of both the Ung deficiency and Ung misexpression during hypoxia, we carried out mutational spectrum analysis by sequencing the RRDR of rpoB (mutations that confer rifampin resistance to the bacterium). As shown in Table 3, the mutation spectrum analysis of the wild-type strain showed that both A-to-G and C-to-T mutations accounted for the majority of colonies with the Rifr phenotype (43% and 45%, respectively). Interestingly, while the spectrum of Ung-deficient strains was expectedly biased toward C-to-T mutations, that of the Ung-misexpressed strain was biased toward A-to-G mutations. Although the precise mechanisms of how these mutations arise have not been investigated, the differences in the mutational spectra, at least, support that the underlying reasons of the detrimental impacts of Ung deficiency and Ung misexpression, as discussed above, are different.
In E. coli, it is known that increased expression of CpxR (a part of the CpxR/CpxA two-component system) during stationary phase leads to transcriptional downregulation of the ung gene and an increase in C-to-T mutations (22). In mycobacteria, DosR, DosS, and DosT proteins, as a part of the two-component system, have been implicated in sensing oxygen levels. And, as a hypoxia sensor, DosT is known to activate transcription of several genes (15, 26). It would be interesting to investigate if the DNA repair genes are downregulated through a network of this two-component system. Further, our observation that hypoxia-specific misexpression of Ung is detrimental to efficient recovery of bacteria raises an interesting question as to whether such an approach may be useful in further engineering of the attenuated strains of mycobacteria by introducing copies of DNA repair genes under the control of a hypoxia-specific promoter(s). Even if such strains established latent infection in the host, they would be highly compromised for their reactivation. However, in such studies it would also be important to consider the possibility of compensatory promoter mutations which may inactivate them for expression during hypoxia.
Acknowledgments
We thank Kervin Rex for technical help and our other laboratory colleagues for suggestions and critical reading of the manuscript.
This work was supported by funding from the Department of Biotechnology, New Delhi, India. K.K. was supported by a senior research fellowship of the Council of Scientific and Industrial Research, New Delhi, India.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Acharya, N. 2003. Mechanistic studies on uracil DNA glycosylases from Escherichia coli and mycobacteria: interaction with uracil containing DNA, single stranded DNA binding proteins (SSBs) and an inhibitor protein (Ugi). Ph.D. thesis. Indian Institute of Science, Bangalore, India.
- 2.Acharya, N., and U. Varshney. 2002. Biochemical properties of single stranded DNA binding protein from Mycobacterium smegmatis, a fast growing mycobacterium and its physical and functional interaction with uracil DNA glycosylases. J. Mol. Biol. 318:1251-1264. [DOI] [PubMed] [Google Scholar]
- 3.Bagchi, G., Mayuri, and J. S. Tyagi. 2003. Hypoxia-responsive expression of Mycobacterium tuberculosis Rv3134c and devR promoters in Mycobacterium smegmatis. Microbiology 149:2303-2305. [DOI] [PubMed] [Google Scholar]
- 4.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 5.Dick, T., B. H. Lee, and B. Murugasu-Oei. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163:159-164. [DOI] [PubMed] [Google Scholar]
- 6.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, D. I. 1986. Reduction of nitroimidazoles in vitro and DNA damage. Biochem. Pharmacol. 35:53-58. [DOI] [PubMed] [Google Scholar]
- 8.Fang, F. C. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadsden, M. H., E. M. McIntosh, J. C. Game, P. J. Wilson, and R. H. Haynes. 1993. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 12:4425-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grishko, V., M. Solomon, J. F. Breit, D. W. Killilea, S. P. Ledoux, G. L. Wilson, and M. N. Gillespie. 2001. Hypoxia promotes oxidative base modifications in the pulmonary artery endothelial cell VEGF gene. FASEB J. 15:1267-1269. [DOI] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Jackson, D., A. Salem, and G. H. Coombs. 1984. The in-vitro activity of metronidazole against strains of Escherichia coli with impaired DNA repair systems. J. Antimicrob. Chemother. 13:227-236. [DOI] [PubMed] [Google Scholar]
- 13.Kesavan, A. K., M. Brooks, J. Tufariello, J. Chan, and Y. C. Manabe. 2009. Tuberculosis genes expressed during persistence and reactivation in the resistant rabbit model. Tuberculosis (Edinb.) 89:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo, A., R. Safaei, M. Mishima, H. Niedner, X. Lin, and S. B. Howell. 2001. Hypoxia-induced enrichment and mutagenesis of cells that have lost DNA mismatch repair. Cancer Res. 61:7603-7607. [PubMed] [Google Scholar]
- 15.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 104:11568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, P., K. Krishna, R. Srinivasan, P. Ajitkumar, and U. Varshney. 2004. Mycobacterium tuberculosis and Escherichia coli nucleoside diphosphate kinases lack multifunctional activities to process uracil containing DNA. DNA Repair (Amst.) 3:1483-1492. [DOI] [PubMed] [Google Scholar]
- 17.Kurthkoti, K., P. Kumar, R. Jain, and U. Varshney. 2008. Important role of the nucleotide excision repair pathway in Mycobacterium smegmatis in conferring protection against commonly encountered DNA-damaging agents. Microbiology 154:2776-2785. [DOI] [PubMed] [Google Scholar]
- 18.Kurthkoti, K., T. Srinath, P. Kumar, V. S. Malshetty, P. B. Sang, R. Jain, R. Manjunath, and U. Varshney. 2010. A distinct physiological role of MutY in mutation prevention in mycobacteria. Microbiology 156:88-93. [DOI] [PubMed] [Google Scholar]
- 19.Mayuri, G. Bagchi, T. K. Das, and J. S. Tyagi. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol. Lett. 211:231-237. [DOI] [PubMed] [Google Scholar]
- 20.Moller, P., S. Loft, C. Lundby, and N. V. Olsen. 2001. Acute hypoxia and hypoxic exercise induce DNA strand breaks and oxidative DNA damage in humans. FASEB J. 15:1181-1186. [DOI] [PubMed] [Google Scholar]
- 21.Muttucumaru, D. G., G. Roberts, J. Hinds, R. A. Stabler, and T. Parish. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb.) 84:239-246. [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara, H., J. Teramoto, K. Hirao, K. Yamamoto, A. Ishihama, and R. Utsumi. 2004. Negative regulation of DNA repair gene (ung) expression by the CpxR/CpxA two-component system in Escherichia coli K-12 and induction of mutations by increased expression of CpxR. J. Bacteriol. 186:8317-8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purnapatre, K., and U. Varshney. 1998. Uracil DNA glycosylase from Mycobacterium smegmatis and its distinct biochemical properties. Eur. J. Biochem. 256:580-588. [DOI] [PubMed] [Google Scholar]
- 24.Purnapatre, K. 1999. Uracil DNA glycosylase from mycobacteria and Escherichia coli: mechanism of uracil excision from synthetic substrates and differential interaction with uracil DNA glycosylase inhibitor (Ugi) and single stranded DNA binding proteins (SSBs). Ph.D. thesis. Indian Institute of Science, Bangalore, India.
- 25.Rao, S. P., L. Camacho, B. Huat Tan, C. Boon, D. G. Russel, T. Dick, and K. Pethe. 2008. Recombinase-based reporter system and antisense technology to study gene expression and essentiality in hypoxic nonreplicating mycobacteria. FEMS Microbiol. Lett. 284:68-75. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen, T. D., and K. J. Livak. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101-1108. [DOI] [PubMed] [Google Scholar]
- 30.Singh, N. S., R. Ahamad, and U. Varshney. 2008. Recycling of ribosomal complexes stalled at the step of elongation in Escherichia coli. J. Mol. Biol. 380:451-464. [DOI] [PubMed] [Google Scholar]
- 31.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 32.Spek, E. J., L. N. Vuong, T. Matsuguchi, M. G. Marinusand, and B. P. Engelward. 2002. Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases. J. Bacteriol. 184:3501-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, A. F., and B. Weiss. 1982. Role of exonuclease III in the base excision repair of uracil-containing DNA. J. Bacteriol. 151:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi, J. S., and D. Sharma. 2002. Mycobacterium smegmatis and tuberculosis. Trends Microbiol. 10:68-69. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesh, J., P. Kumar, P. S. Krishna, R. Manjunath, and U. Varshney. 2003. Importance of uracil DNA glycosylase in Pseudomonas aeruginosa and Mycobacterium smegmatis, G+C-rich bacteria, in mutation prevention, tolerance to acidified nitrite, and endurance in mouse macrophages. J. Biol. Chem. 278:24350-24358. [DOI] [PubMed] [Google Scholar]
- 37.Via, L. E., P. L. Lin, S. M. Ray, et al. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 41.Wink, D. A., K. S. Kasprzak, C. M. Maragos, R. K. Elespuru, M. Misra, T. M. Dunams, T. A. Cebula, W. H. Koch, A. W. Andrews, J. S. Allen, et al. 1991. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science 254:1001-1003. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, J., L. Narayanan, S. Rockwell, and P. M. Glazer. 2000. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 60:4372-4376. [PubMed] [Google Scholar]