Abstract
Cytochrome oxidases are perfect model substrates for analyzing the assembly of multisubunit complexes because the need for cofactor incorporation adds an additional level of complexity to their assembly. cbb3-type cytochrome c oxidases (cbb3-Cox) consist of the catalytic subunit CcoN, the membrane-bound c-type cytochrome subunits CcoO and CcoP, and the CcoQ subunit, which is required for cbb3-Cox stability. Biogenesis of cbb3-Cox proceeds via CcoQP and CcoNO subcomplexes, which assemble into the active cbb3-Cox. Most bacteria expressing cbb3-Cox also contain the ccoGHIS genes, which encode putative cbb3-Cox assembly factors. Their exact function, however, has remained unknown. Here we analyzed the role of CcoH in cbb3-Cox assembly and showed that CcoH is a single spanning-membrane protein with an N-terminus-out-C-terminus-in (Nout-Cin) topology. In its absence, neither the fully assembled cbb3-Cox nor the CcoQP or CcoNO subcomplex was detectable. By chemical cross-linking, we demonstrated that CcoH binds primarily via its transmembrane domain to the CcoP subunit of cbb3-Cox. A second hydrophobic stretch, which is located at the C terminus of CcoH, appears not to be required for contacting CcoP, but deleting it prevents the formation of the active cbb3-Cox. This suggests that the second hydrophobic domain is required for merging the CcoNO and CcoPQ subcomplexes into the active cbb3-Cox. Surprisingly, CcoH does not seem to interact only transiently with the cbb3-Cox but appears to stay tightly associated with the active, fully assembled complex. Thus, CcoH behaves more like a bona fide subunit of the cbb3-Cox than an assembly factor per se.
The heme-copper oxidases comprise a family of functionally related enzyme complexes, which terminate respiratory chains in both prokaryotic and eukaryotic cells (38). In addition to their physiological importance for all oxygen-dependent cells, these enzyme complexes have been used as model complexes for studying the assembly of cofactor-containing multisubunit protein complexes (18). All members of the heme-copper oxidase family are characterized by the presence of a membrane integral subunit I, which binds a low-spin heme and a characteristic high-spin heme-CuB binuclear center. There is, however, a remarkable diversity in respect to subunit composition, electron donor, and oxygen affinity. Due to this diversity, members of the heme-copper oxidase family have been classified into three major subfamilies (33). Subfamily A heme-copper oxidases are the most abundant and are present in mitochondria and in many bacterial species. They use two proton transfer pathways (21), while subfamily B heme-copper oxidases appear to use only one proton channel (5, 17). The bacterial cbb3-type cytochrome (cyt) c oxidases (cbb3-Cox) constitute the prototype for the C subfamily and are the second-most-abundant oxidases (8, 17). They are encoded by the ccoNOQP operon (fixNOQP in rhizobia) and have so far been found only in eubacterial species. ccoN encodes the catalytic subunit I, which harbors the conserved histidines required for ligating the heme b and the heme b3/CuB binuclear centers (11, 12, 37). Different from the aa3-type cytochrome c oxidases (aa3-Cox), cbb3-Cox lack a second Cu center (CuA) in subunit II and instead contain two membrane-bound c-type cytochrome subunits, the monoheme subunit CcoO and the diheme subunit CcoP. As in many bacterial cytochrome oxidases, a fourth subunit (called CcoQ) is present in most cbb3-Cox. This subunit does not seem to contain any cofactor and is not essential for function. Nevertheless, in the absence of CcoQ the amount of the functional cbb3-Cox complex is reduced in Rhodobacter capsulatus (34).
The first X-ray structure of a cbb3-Cox from Pseudomonas stutzeri has recently been solved at 3.2 Å (3). The architecture of the catalytic CcoN subunit displays a remarkable similarity to the known structures of cytochrome oxidases of the A and B families (21, 36, 47, 54) despite the rather low level of sequence conservation. Differences are detectable in the heme b and heme b/CuB environments, which probably contribute to the high level of oxygen affinity of the cbb3-Cox. Based on the orientation of the CcoP and CcoO subunits relative to the CcoN subunit, electrons are most likely transferred from a donor cytochrome to the partly solvent exposed outer heme group of CcoP and subsequently via the inner heme group of CcoP to CcoO. From CcoO, the electron is then shuttled via heme b to the heme b3/CuB binuclear center, where oxygen reduction takes place.
The structural diversity of the heme-copper oxidase family is also reflected by the requirement for different assembly factors. For aa3-Cox, more than 30 proteins are required for their assembly in eukaryotes (4, 23, 46). Of those, only a few are also required for the assembly of the simpler bacterial aa3-Cox. Examples are Surf1, which is required for heme a insertion into subunit I (2, 45), and CoxG, a Cox11 homologue required for Cu delivery to the CuB site (13, 19). Neither Surf1 nor CoxG appears to be required for cbb3-Cox assembly (1, 2).There are also indications, however, for a dual function of assembly factors in both aa3-Cox and cbb3-Cox assembly. SenC, a homologue of the mitochondrial ScoI, is involved in CuA center assembly in aa3-type cytochrome oxidases (20) but is also required for cbb3-Cox activity in R. capsulatus (49) and Pseudomonas aeruginosa (9) but not in Bradyrhizobium japonicum (1). Since cbb3-Cox lack a CuA center, the exact function of SenC/ScoI for cbb3-Cox activity remains to be determined.
The active R. capsulatus cbb3-Cox migrates on blue native (BN) gels as a 230-kDa complex that is assembled via a 210-kDa CcoNO subcomplex to which an approximately 50-kDa CcoQP complex is recruited (26, 34). This multistep assembly depends on the proteins encoded by the ccoGHIS gene cluster (25, 26, 35), which is located immediately downstream of the ccoNOQP operon in many bacteria (8, 22, 24). The ccoG gene encodes a putative polyferrodoxin and is the most conserved of the four genes (8). Nevertheless, its deletion in R. capsulatus does not abolish the cbb3-Cox activity (25). A different phenotype is observed with a ccoI deletion; here no cbb3-Cox activity is detectable, and the cbb3-Cox subunits fail to form a stable complex in the membrane (26). The ccoI gene product shows all features of a P-type ATPase, and it has been suggested that it is involved in copper delivery to cbb3-Cox (25, 35), although this suggestion still awaits experimental verification. Whether CcoI is a dedicated Cu delivery system for cbb3-Cox is also unknown; it has recently been suggested that the CcoI homologue in Rubrivivax gelatinosus might also be involved in providing Cu to other Cu-containing enzymes like NosZ (14). The ccoS gene encodes a small, putative membrane protein, which appears to be specifically involved in cofactor insertion. In the absence of CcoS, cbb3-Cox assembles without the heme b and the heme b3/CuB binuclear centers (25, 26). Whether CcoS is able to bind heme or Cu directly is currently unknown, but its primary sequence does not show any motifs that would point in this direction. Finally, CcoH also encodes a small putative membrane protein without any defined motif or any homology to proteins of known function. Deleting ccoH prevents the stable assembly of cbb3-Cox (26), but the exact function of CcoH has remained unclear.
The available data indicate that distinct cellular machineries are involved in catalyzing similar, or even identical, steps during the assembly of aa3-Cox and cbb3-Cox The frequent occurrence of cbb3-Cox in many pathogenic bacteria and the observation that this enzyme is the only terminal oxidase recognized in the genomes of Helicobacter pylori and Neisseria meningitides (30) render the assembly process and the involved assembly factors attractive as targets for antibacterial treatments. In the current study, we analyzed the function of CcoH in cbb3-Cox assembly and stability. Our data demonstrate that CcoH is a single spanning-membrane protein with an N-terminus-out-C-terminus-in (Nout-Cin) topology that specifically contacts the CcoP subunit of cbb3-Cox. Surprisingly, CcoH does not seem to interact only transiently with the cbb3-Cox, but under the tested conditions, it stayed tightly associated with the active, fully assembled complex, almost behaving like a bona fide subunit of the cbb3-Cox rather than an assembly factor per se.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used are listed in Table 1. Escherichia coli strains harboring plasmids were grown in LB medium supplemented with the appropriate antibiotics (100, 50, and 12.5 μg/ml of ampicillin, kanamycin, and tetracycline, respectively). R. capsulatus strains were grown in Sistrom's minimal A medium or in enriched MPYE medium (6, 44) at 35°C in liquid cultures in the dark with the appropriate antibiotics (10 and 2.5 μg/ml of kanamycin and tetracycline, respectively). For semiaerobic growth, 500-ml cultures were grown in the dark in 1,000-ml flasks and were shaken at 110 rpm.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype | Relevant phenotype or applicationb | Source or reference |
|---|---|---|---|
| Strain | |||
| Escherichia coli | |||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 52 | |
| HB101 | F−proA2 hsdS20(rB− mB−) recA13 ara-14 lacY1 galK2 rpsL20 supE44 xyl-5 mtl-1 | 52 | |
| Rhodobacter capsulatus | |||
| MT1131 | crtD121 rifR | Wild-type, NADI+ | 41 |
| MT1131-pCW25 | pRK415-ccoNOQP-ccoGHIS | cbb3-Cox overexpression | 29 |
| GK32 | ΔccoNO::kan | NADI− | 30 |
| M7G | ccoP269 (stopc) | NADI− | 28 |
| CW6 | ΔccoH::kan | NADI− | 30 |
| CW1 | ΔccoGHIS::spe | NADI− | 30 |
| Plasmid | |||
| pRK415-ccoH | ccoH cloned into pRK415 via BamHI/XbaI | In vivo expression of CcoH | 27 |
| pRK415-ccoH-M1 | ccoH cloned into pRK415 without second HD via BamHI/XbaI | In vivo expression of ccoH lacking the second HD | This work |
| pET19b/pET22b | Ampr | Novagen | |
| pET19b-ccoH | ccoH cloned into pET19b via NdeI/BamHI | In vitro synthesis of N-His-CcoH | 27 |
| pET19bccoH-M0 | ccoH cloned into pET19b via NdeI/BamHI (105)TTC→AUG | In vitro synthesis of N-His-CcoH lacking the cytoplasmic loop | This work |
| pET19bccoH-M1 | ccoH cloned into pET19b via NdeI/BamHI (378)TGG→AUG | In vitro synthesis of N-His-CcoH lacking the second HD | This work |
HD, hydrophobic domain; N-His-CcoH, N-terminally His-tagged CcoH.
NADI+ indicates that the strain is positive in the NADI reaction (see Materials and Methods), while NADI− indicates that the strain does not respond within 30 min to the NADI reaction.
R. capsulatus M7G carries a premature stop codon at position 269.
Molecular-genetics techniques.
Standard molecular-genetics techniques were performed as described by Sambrook et al. (40). For in vitro synthesis, the ccoH gene was cloned into pET19b and pET22b (Novagen, Bad Schwalbach, Germany) by using BamHI and NdeI restriction sites to obtain N-terminally or C-terminally His-tagged CcoH derivatives under the control of the T7 promoter. Truncated CcoH versions for in vitro synthesis were generated by incorporating stop codons into the ccoH sequence by using the following primers: 5′-CGCGGAATAAACCTAGTCGGTCCGGGTC-3′ and 5′-CCCCTCAGCCTTGCCCCCGGGGCCTG-3′ for deleting the second hydrophobic segment and 5′-CCCCGGCTAGGTCTTCACC-3′ and 5′-CTTGAAGTGGCGAATTCCTATGTCG-3′ for deleting the entire cytoplasmic domain. For in vivo truncation of CcoH, an additional stop codon and an XbaI restriction side were introduced into CcoH in pET17b-CcoGHI by using primers 5′-GTCCGT CCG GGTCATCTGC-3′ and 5′-TGATCTAGATCCGCGCCCCTC-3′, and the modified ccoH gene was subsequently subcloned into pRK415 using BamHI and XbaI.
Preparation of ICMs and high-speed supernatants.
High-speed supernatants (S-135 extracts) of cell homogenate from wild-type R. capsulatus strain MT1131 and intracytoplasmic membranes (ICMs) from MT1131 and mutant strains were prepared essentially as described previously (55), with the following modification: cells were grown semiaerobically at 35°C. Membrane-free S-135 extracts were obtained from strain MT1131 by French pressing cell suspensions at 8,000 lb/in2 followed by a first centrifugation at 30,000 × g for 30 min. The resulting supernatant (S-30) was centrifuged in 1-ml aliquots in a Beckmann TLA-100 ultracentrifuge at 90,000 rpm for 9 min using a TLA-100.2 rotor. The top 750-μl of the supernatant was used as an S-135 extract for cell-free protein synthesis. Preparation of Helicobacter pylori membranes was done according to the protocol designed for E. coli membranes (7). In brief, after breaking the cells using a French-pressing cell and a first centrifugation as described above, the pellet was resuspended and centrifuged for 2 h at 40,000 rpm in a Beckmann Ti50 rotor. The crude membrane pellet was then resuspended and subjected to three-step sucrose gradient centrifugation as described previously (7). Membranes were finally resuspended in INV buffer (50 mM triethanolamine acetate [pH 8.0]-250 mM sucrose).
In vitro protein synthesis, protease protection assay, and carbonate resistance.
T7 RNA polymerase-dependent in vitro expression of ccoH was achieved using plasmid pET19b-ccoH. Cell-free protein synthesis using [35S]methionine and cysteine with R. capsulatus S-135 extracts was carried out for 30 min at 35°C as described previously (15, 26, 55). For cotranslational integration of in vitro-synthesized proteins into membranes, ICMs were added after 5 min of synthesis, and the reaction mixture was incubated for 25 min at 35°C. For protease treatment, samples were incubated with 0.5 mg of proteinase K/ml for 20 min at 25°C followed by trichloroacetic acid (TCA) precipitation (5% final concentration). The TCA pellet was resuspended in sodium dodecyl sulfate (SDS) loading buffer and loaded onto an 18% SDS-polyacrylamide gel. For analyzing carbonate resistance, freshly prepared Na2CO3 (pH 11) was added to the in vitro reaction mixture (final concentration, 0.18 M), and the samples were incubated on ice for 30 min. After the mixture was centrifuged in a Beckmann TLA-100.3 rotor for 15 min at 70,000 rpm, the pellet thus obtained was directly dissolved in SDS loading buffer. The supernatant was neutralized with glacial acetic acid, TCA precipitated, and centrifuged for 10 min at 20,000 × g, and the pellet obtained was dissolved in SDS loading buffer. Radiolabeled proteins were visualized by phosphorimaging using a Molecular Dynamics phosphorimager and quantified by using ImageQuant software from Molecular Dynamics.
BN-PAGE.
For analyses by BN-polyacrylamide gel electrophoresis (BN-PAGE), ICMs (50 μg of total protein) were resuspended in 10 μl of 2× lysis buffer (50 mM NaCl, 5 mM 6-aminohexanoic acid, and 50 mM imidazole/HCl [pH 7.0]), adjusted to 20 μl with water, and solubilized with n-dodecylmaltoside (DDM; Roche Diagnostics, Mannheim, Germany) at a 1:1 (wt/wt) ICM protein-to-detergent ratio from a 10% dodecylmaltoside stock solution in lysis buffer (final DDM concentration, 1%). After being incubated for 10 min at 25°C, the samples were centrifuged for 15 min at 70,000 rpm in a Beckmann TLA-100.3 rotor. After centrifugation, 15 μl of the supernatant was supplemented with 2 μl of loading buffer (5% Coomassie blue in 500 mM 6-aminohexanoic acid) and 5 μl of 50% glycerol and then loaded onto a 5 to 20% BN-polyacrylamide gel.
Activity assays. (i) Oxygen uptake.
N,N,N,N-tetramethyl-p-phenylenediamine (TMPD) oxidase activity was measured at 28°C in a closed reaction chamber (1-ml volume) with a fiber optic oxygen meter (Fibox 3; PreSens GmbH, Regensburg, Germany). R. capsulatus membranes were dissolved in ICM buffer (50 mM triethanolamine, 1 mM EDTA, 1 mM dithiothreitol [DTT], and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) to a final concentration of approximately 0.1 mg/ml. Oxygen consumption was initiated by the addition of 10 μl of 1 M sodium ascorbate (final concentration, 10 mM) and 5 μl of 24 mM TMPD (final concentration, 0.2 mM). Oxygen consumption was recorded at 28°C using OxyView 3.5.1 software (PreSens GmbH) and was terminated after several minutes of recording by the addition of 0.1 mM NaCN (final concentration). Net TMPD oxidase activity was determined by subtracting the endogenous respiration rate from that induced by ascorbate.
(ii) In-gel heme staining and on-plate or in-gel NADI staining.
To reveal the c-type cytochromes, SDS-Tris-tricine-polyacrylamide gels were treated with 3,3,5,5-tetramethylbenzidine (TMBZ) according to the method of Thomas et al. (51). For activity staining of cbb3-Cox in growing cells via α-naphthol plus N,N-dimethyl-p-phenylenediamine (DMPD) (NADI) reaction, cell cultures were grown on MPYE plates with the appropriate antibiotics at 35°C. Staining was performed by incubating the plates with a 1:1 (vol/vol) mixture of 35 mM α-naphthol dissolved in ethanol and 30 mM NADI in water, which turned blue in the presence of active enzyme. NADI staining of BN-polyacrylamide gels was performed as described previously (26).
Immunodetection methods.
For immunoblot analyses, proteins were electroblotted onto Immobilon-P transfer membranes. Polyclonal antibodies against CcoP, CcoN, and CcoH were used with horseradish peroxidase-conjugated goat anti-rabbit antibodies from Caltag Laboratories (Burlingame, CA) as secondary antibodies, and enhanced chemiluminescence (ECL) reagent (GE Healthcare, Munich, Germany) was used as the detection substrate. Antibodies against the His6 tag were obtained from Roche (Mannheim, Germany). Immunoprecipitations were performed as described previously (26).
RNA isolation and RT-PCR.
Semiaerobic cultures of R. capsulatus in MPYE medium were grown to mid-log phase (optical density at 652 nm [OD652], 0.8 to 1.0). Approximately 1× 109 cells were centrifuged, incubated in TE buffer (10 mM Tris-0.5 mM EDTA [pH 7]) containing 10 mg/ml of lysozyme for 15 min in 37°C and passed 5 times through an 0.8-μm-diameter needle into RNase-free tubes. Isolation of total RNA was achieved by using a GE Healthcare minispin kit according to the suggested protocol. Two micrograms of total RNA was treated with DNase I for 30 min at 25°C in the presence of the RNase inhibitor RNasin. Fifty nanograms of treated RNA was used in reverse transcriptase (RT)-PCRs with a OneStep RT-PCR kit (Qiagen, Hilden, Germany). The primers used for detection of ccoH mRNA were 5′-CGACCAGAACCTCGAGCTGG-3′ and 5′-ACCGCTGACCGGCCGCAAG-3′; the primers used for detection of ccoN mRNA were 5′-CATGTTGCACATCGTCAACAACC-3′ and 5′-CGAAGGTGATCATGCCGTTCC-3′. As a control for detecting genomic DNA contamination, PCRs were performed with the same primer without the reverse transcriptase step using Phusion DNA polymerase (Finnzymes USA). Samples were separated on a 1.2% agarose gel.
RESULTS
CcoH is required for the activity and stability of cbb3-type cytochrome oxidase.
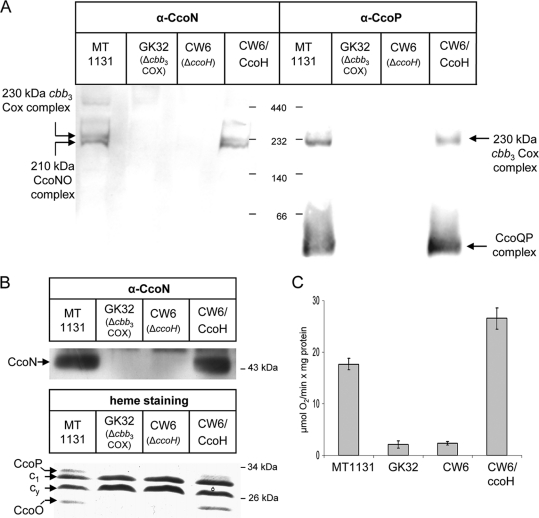
The crucial importance of CcoH for cbb3-Cox assembly and activity has been demonstrated in previous studies (25, 26); however, the exact function of CcoH has remained unclear. In R. capsulatus, the active cbb3-Cox is detectable as a 230-kDa complex in BN-PAGE and is assembled via a 210-kDa intermediate containing at least CcoN and CcoO and an approximately 50-kDa intermediate containing at least CcoP and CcoQ (26, 34). In the ccoH deletion mutant CW6, neither the fully assembled 230-kDa complex nor the 210-kDa and 50-kDa intermediates were detectable (Fig. 1A); thus, CW6 displayed the same phenotype as the cbb3-Cox knockout strain GK32. In SDS-PAGE, the catalytic subunit CcoN of cbb3-Cox was undetectable in membranes of CW6 by immunodetection. The c-type cyt subunits CcoO and CcoP were also undetectable by heme staining, while the absence of CcoH did not impair the steady-state stability of cyt c1 of the cyt bc1 complex or the stability of the membrane-bound electron carrier cyt cy (Fig. 1B). In agreement with the lack of individual subunits of cbb3-Cox, we also failed to detect cbb3-Cox-dependent oxygen uptake activity in membranes of the CcoH deletion strain (Fig. 1C). All phenotypes of the CcoH mutant were fully restored by expressing CcoH in trans (Fig. 1), indicating that deleting CcoH does not induce polar effects on the downstream ccoI and ccoS genes, which are also essential for the assembly of a functional cbb3-Cox (25). In comparison to that in wild-type membranes, we observed a slightly increased oxygen uptake activity in membranes from cells of the CW6 mutant expressing plasmid-borne ccoH, suggesting that under the conditions tested, CcoH might be a limiting factor for steady-state levels of cbb3-Cox activity.
FIG. 1.
CcoH is essential for cbb3-Cox stability and activity in R. capsulatus. (A) Intracytoplasmic membranes (ICMs; 50 μg of protein) of the indicated R. capsulatus strains were solubilized with dodecylmaltoside and separated by BN-PAGE. CW6/CcoH corresponds to a genomic ccoH deletion mutant carrying a copy of ccoH on a plasmid. After being transferred onto a polyvinylidene difluoride (PVDF) membrane, complexes were decorated with antibodies against the CcoN and CcoP subunits (α-CcoN and α-CcoP). (B) The ICMs shown in panel A were separated on Tris-tricine gels and either transferred onto PVDF membranes to detect the steady-state amount of CcoN (upper panel) or analyzed by heme staining, which revealed the membrane-bound c-type cytochromes of R. capsulatus (lower panel). Indicated are the subunits CcoP and CcoO of cbb3-Cox, cyt c1 of the bc1 complex, and cyt cy. (C) cbb3-Cox activities in different ICMs were measured as oxygen uptake activities in at least three independent experiments.
CcoH is a type I integral membrane protein.
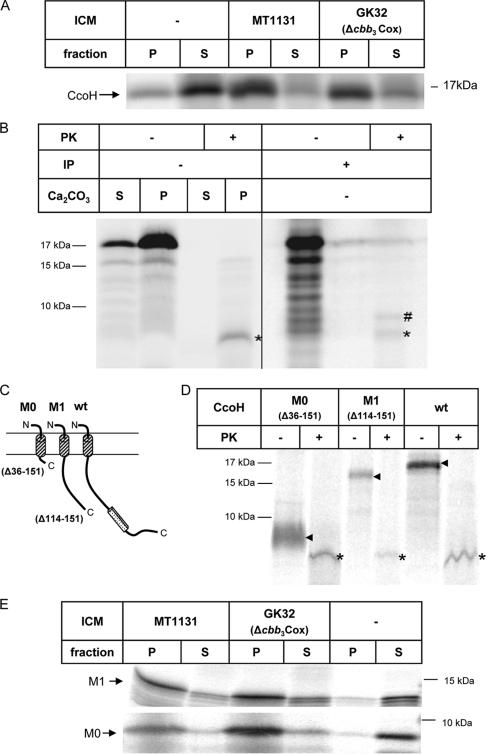
ccoH is predicted to encode a small protein of 151 amino acids, which presumably spans the membrane at least once with an N-terminally located transmembrane domain. However, a second hydrophobic stretch at the C terminus could constitute an additional transmembrane domain, as predicted by many secondary-structure prediction programs. For determining the topology of CcoH, we employed an R. capsulatus in vitro transcription-translation system together with inverted intracytoplasmic membrane vesicles (26, 34). CcoH was synthesized in vitro in the presence or absence of ICMs and subsequently subjected to alkaline carbonate extraction, which allows differentiation between integral membrane proteins and proteins that are soluble or only peripherally attached to the membrane (10). In vitro synthesis of CcoH resulted in a radioactively labeled protein of 17 kDa (Fig. 2A), which is in line with the predicted molecular mass of CcoH. In the absence of ICMs, CcoH was found predominantly in the supernatant (S) after carbonate extraction and ultracentrifugation. The small amount of CcoH found in the pellet fraction (P) even in the absence of ICMs is most likely the result of aggregation, which is frequently observed if small hydrophobic proteins are synthesized in vitro (7, 35). In the presence of wild-type ICMs, however, the majority of CcoH was carbonate resistant, i.e., recovered from the pellet fraction after centrifugation. This result demonstrates that CcoH is an integral membrane protein. Carbonate resistance of CcoH was also observed in membranes of the cbb3-Cox knockout strain GK32, suggesting that even in the absence of cbb3-Cox, in vitro-synthesized CcoH can be inserted stably into ICMs.
FIG. 2.
CcoH is an integral membrane protein with an Nout-Cin topology. (A) CcoH was in vitro synthesized in an R. capsulatus cell-free transcription-translation system in the absence or presence of ICMs. Subsequently, the reaction mixtures were treated with alkaline carbonate, and carbonate-resistant (pellet, P) and carbonate-sensitive (supernatant, S) materials were separated by ultracentrifugation. Membrane-associated or soluble proteins were found in the supernatants after CO3 treatment and centrifugation. (B) CcoH was in vitro synthesized in the presence of ICMs and carbonate extracted. The carbonate-resistant material was then resuspended in ICM buffer and treated with proteinase K. Samples were separated on a Tris-tricine gel. Immunoprecipitations (IP) were performed with anti-His antibodies, which recognized the N-terminal His tag of CcoH. Asterisks (*) indicate the proteinase K-protected fragments of CcoH, while a pound sign (#) indicates an unspecific band that was immunoprecipitated by anti-His antibodies. (C) Schematic representation of C-terminally truncated CcoH derivatives. wt, full-sized CcoH; CcoH-M0, a CcoH derivative that consists mainly of the transmembrane domain; CcoH-M1, a derivative that lacks the second hydrophobic segment. (D) Proteinase K digestion of the in vitro-synthesized CcoH derivatives in the presence of wild-type ICMs. Arrows indicate the in vitro-synthesized derivatives without proteinase K treatment, and asterisks indicate the proteinase K-protected fragments. (E) Carbonate extraction of in vitro-synthesized constructs performed in the presence of wild-type ICMs (MT1131) and ICMs lacking cbb3-Cox (GK32) and without ICMs (−).
To analyze the topology of CcoH in the membrane, we employed proteinase K protection assays of in vitro-synthesized CcoH. CcoH was in vitro synthesized in the absence of ICMs and subjected first to carbonate extraction. After ultracentrifugation, the supernatant and the pellet fractions were treated with proteinase K, which completely digested CcoH (Fig. 2B, left panel). In the presence of ICMs, proteinase K treatment of the carbonate-resistant material resulted in a proteinase K-protected fragment of about 5 kDa, which was not observed after proteinase K treatment of the supernatant, i.e., of the carbonate-sensitive material. The occurrence of a 5-kDa protease-protected fragment supports a topology of CcoH in which the N-terminal hydrophobic domain (amino acids 10 to 33) serves as a transmembrane domain, while amino acids 34 to 151 are accessible to proteinase K and thus reside on the cytoplasmic side of the inverted-membrane vesicles. In order to verify this topology, we made use of the N-terminal His tag present in the in vitro-synthesized CcoH. Anti-His6 antibodies immunoprecipitated both the full-size CcoH and the protease-protected 5-kDa band, demonstrating that the N-terminal His tag was protected against proteinase K digestion (Fig. 2B, right panel).
Because these data cannot completely rule out that the second hydrophobic domain is also involved in membrane binding of CcoH, we analyzed the proteinase K protection of two in vitro-synthesized CcoH truncations (Fig. 2C). In vitro synthesis of CcoH-M1, which lacks the second hydrophobic domain (amino acids 120 to 150), resulted in a slightly smaller in vitro-synthesized product (Fig. 2D), but after proteinase K treatment we observed the 5-kDa protected fragment, also seen for full-size CcoH. Proteinase K treatment of a CcoH derivative, which consisted almost exclusively of the first transmembrane domain (CcoH-M0, amino acids 1 to 35), also resulted in the occurrence of the 5-kDa protease-protected fragment (Fig. 2D). That the deletion of the second hydrophobic domain did not interfere with membrane integration of CcoH or its topology in the membrane was further confirmed by carbonate extraction, which demonstrated that the two C-terminally truncated CcoH derivatives were, like full-size CcoH, carbonate resistant (Fig. 2E).
In summary, these data demonstrate that the first hydrophobic domain of CcoH serves as a transmembrane domain, while the second hydrophobic domain is not required for stable membrane integration of CcoH in vitro. Thus, CcoH is a typical type I membrane protein with a single transmembrane domain and an Nout-Cin topology.
The second hydrophobic stretch of CcoH is required for its stability in vivo.
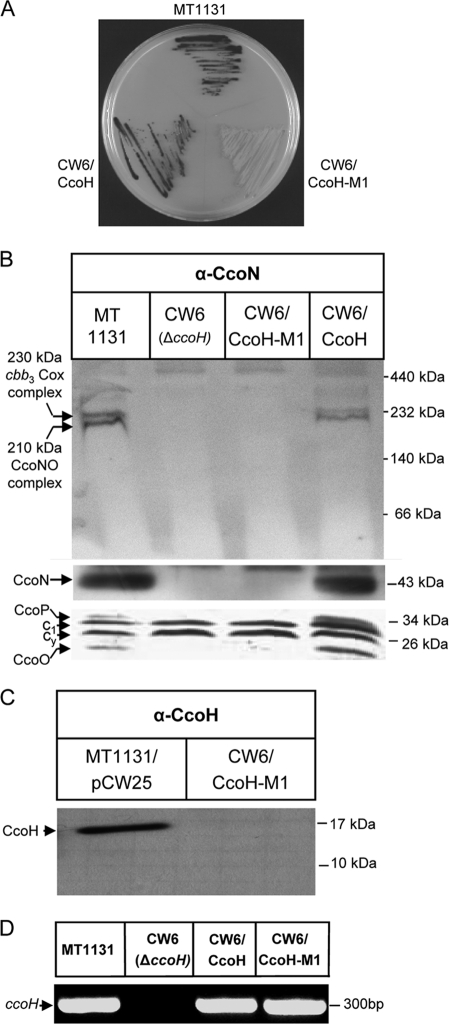
Although the second hydrophobic stretch of CcoH does not seem to be required for membrane integration, its high level of sequence conservation among different species suggests an important role for the function of CcoH. Its role was analyzed by testing whether the CcoH-M1 construct, which lacks the second hydrophobic stretch, could complement the CcoH deletion strain CW6. NADI staining allows the detection of active cbb3-Cox in whole colonies. DMPD is oxidized by cbb3-Cox and, together with α-naphthol, forms the dye indophenol blue (24). Colonies of the R. capsulatus wild-type strain MT1131 turned blue within 30 s after treatment with the NADI reagent, and a similar response was observed for the CW6 strain complemented with full-size CcoH (CW6/CcoH) (Fig. 3A). In contrast, CW6 expressing CcoH-M1 (CW6/CcoH-M1) did not show any response even after 30 min, indicating that the second hydrophobic domain of CcoH is essential for function. This was confirmed by biochemical analyses using BN-PAGE, immunodetection, and heme staining, which revealed that neither the 230-kDa cbb3-Cox complex nor the individual subunits of cbb3-Cox were detectable in membranes of CW6 expressing CcoH-M1 (Fig. 3B).
FIG. 3.
The second hydrophobic segment of CcoH is crucial for its function and stability. (A) The truncated CcoH-M1 derivative or full-size CcoH were expressed from a plasmid in the ccoH deletion strain CW6, and cbb3-Cox activities were determined by NADI staining of R. capsulatus colonies. The dark color indicates active cbb3-Cox. MT1131 corresponds to wild-type R. capsulatus. (B) Steady-state stabilities of cbb3-Cox complexes and individual subunits in membranes from different R. capsulatus strains. The upper panel shows the results of BN-PAGE with subsequent immunodetection using antibodies against the CcoN subunit. The middle panel shows the presence of the CcoN subunit after separation of membranes by SDS-Tris-tricine-PAGE and subsequent immunodetection. The lower panel depicts a heme-stained gel. (C) Detection of the truncated CcoH-M1 derivative expressed in strain CW6 by using anti-CcoH antibodies. (D) Results of RT-PCR using total RNA from different R. capsulatus strains showing ccoH expression.
Because CW6 and CW6/CcoH-M1 displayed exactly the same phenotype, we analyzed whether the truncated CcoH derivative was stable in R. capsulatus membranes. We were unable to detect the truncated CcoH version (Fig. 3C) by immunodetection using anti-CcoH antibodies, but RT-PCR using a specific primer pair for the central part of the ccoH gene revealed the presence of an identical PCR product using RNA isolated from wild-type, CW6/CcoH, or CW6/CcoH-M1 cells as a template (Fig. 3D). This indicates that ccoH-M1 is transcribed into mRNA but fails to yield a stable protein in the membrane in vivo. As the in vitro data (Fig. 2D and E) seemed to indicate that CcoH-M1 can be stably integrated into ICM, the apparent difference between the in vitro and in vivo data (Fig. 3) is probably due to the fact that the in vitro experiments were performed in the presence of a protease inhibitor cocktail and for a short period of time (i.e., 30 min).
CcoH stability in the membrane depends on cbb3-Cox.
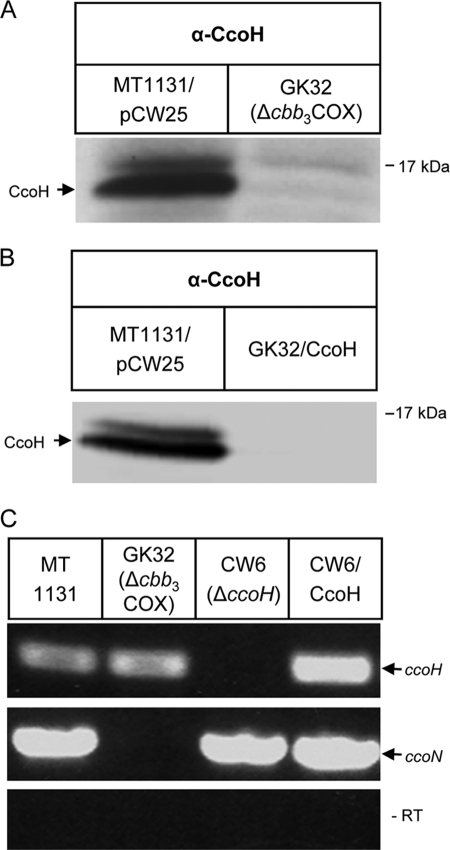
The instability of the truncated CcoH derivative in vivo could indicate that the lack of the second hydrophobic domain precludes the protein-protein interactions that are required for CcoH stability. Because CcoH is essential for cbb3-Cox assembly, we tested whether the stability of CcoH in the membrane was influenced by the absence of cbb3-Cox. By Western blotting using anti-CcoH antibodies, CcoH was detectable in wild-type R. capsulatus membranes but not in membranes of the cbb3-Cox deletion strain GK32 (Fig. 4A). CcoH was also not detectable in GK32 expressing multiple ccoH copies from a plasmid (Fig. 4B). RT-PCR revealed that the lack of the cbb3-Cox genes in GK32 did not influence transcription of the ccoH gene (Fig. 4B). As a control, we used a primer pair specific for ccoN, which encodes the catalytic subunit of cbb3-Cox. The ccoN mRNA was detectable by RT-PCR in wild-type cells, CW6, and CW6 expressing plasmid-borne ccoH (Fig. 4C). These data demonstrate a mutual dependence of cbb3-Cox and CcoH. In the absence of CcoH, cbb3-Cox is not stably assembled, and in the absence of cbb3-Cox, CcoH is not stable in membranes. The RT-PCR data indicate that this is not the result of a transcriptional coupling, because the mRNA levels are not significantly influenced in the respective mutants. Gene expression studies using a lacZ-based translational reporter fusion to ccoH showed comparable β-galactosidase activities in wild-type and GK32 cells (data not shown). Therefore, the lack of stability is posttranscriptional and is probably the result of a posttranslational degradation event.
FIG. 4.
CcoH stability is dependent on cbb3-Cox. (A) The presence of CcoH was analyzed by Western blotting in either wild-type strain MT1131 expressing a plasmid containing the complete ccoNOQP-ccoGHIS gene cluster or in the cbb3-Cox deletion strain GK32. (B) The same as panel A but with GK32 expressing a plasmid-borne copy of ccoH. (C) Results of RT-PCR performed by using total RNA derived from wild-type strain MT1131, cbb3-Cox knockout strain GK32, CcoH knockout strain CW6, and CW6 complemented with the ccoH gene. The reactions were performed with primer pairs binding to ccoH (upper lane) or ccoN (middle lane). A control PCR for detecting genomic DNA contaminations was performed with the same RNA preparations but without the reverse transcriptase step (lower lane, −RT).
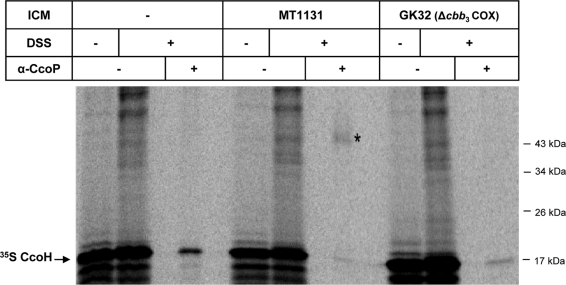
The stabilizing effect of cbb3-Cox on CcoH could most easily be explained by their direct physical contact. We therefore employed an in vitro cross-linking approach using the membrane-permeable chemical cross-linker DSS (disuccinimidyl suberate). CcoH was in vitro synthesized in the absence of ICMs or in the presence of either wild-type or GK32 ICMs. After the addition of DSS, the reaction mixture was immunoprecipitated using anti-CcoP antibodies. In the presence of wild-type ICMs, anti-CcoP antibodies immunoprecipitated a radioactively labeled band at about 50 kDa of the expected size of a CcoH-CcoP cross-linking product (Fig. 5). No cross-linking product was observed in the absence of membranes nor in the presence of cbb3-Cox deletion membranes. We also tried to detect a possible interaction between CcoN and CcoH by DSS cross-linking and immunoprecipitation, but the cross-linking product was too weak to make a definite statement about a possible CcoN-CcoH interaction. Nevertheless, the strong cross-link between CcoH and CcoP demonstrates that CcoH is in close proximity to the CcoP subunit of cbb3-Cox.
FIG. 5.
CcoH interacts with the CcoP subunit of cbb3-Cox. CcoH was in vitro synthesized in the absence of ICMs or in the presence of either wild-type strain MT1131 ICMs or ICMs derived from the cbb3-Cox knockout strain GK32. After in vitro synthesis and integration into the membrane, one part was treated with dimethyl sulfoxide (DMSO), while the other part was incubated with DSS dissolved in DMSO. The DSS-treated sample was further split, and one part was directly TCA precipitated, while the other part was subjected to immunoprecipitation using antibodies against the CcoP subunit of cbb3-Cox. The asterisk (*) indicates the CcoH-CcoP cross-linking product.
CcoH is stably associated with the cbb3-Cox complex.
The interaction between CcoP and CcoH would be in agreement with a function of CcoH during the assembly of the cbb3-Cox complex, which supports its proposed role as a dedicated cbb3-Cox assembly factor (25, 26). In addition, the observation that CcoH is stable only in the presence of cbb3-Cox suggests that CcoH might associate more permanently with this enzyme.
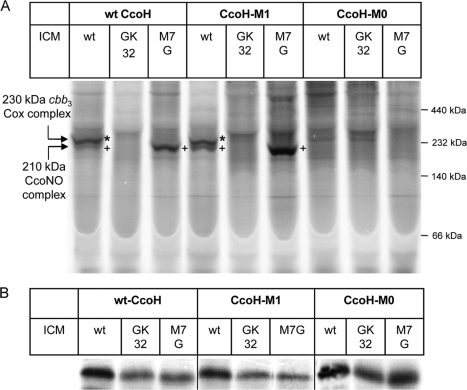
In order to address this question, we combined BN-PAGE with in vitro labeling. CcoH was in vitro synthesized and integrated into wild-type R. capsulatus membranes, which were subsequently separated by BN-PAGE. If CcoH bound to the cbb3-Cox complexes, it should radioactively label them and the complexes should be detectable by autoradiography. In the presence of wild-type ICMs, CcoH labeled the 230-kDa complex, which corresponds to the active, fully assembled cbb3-Cox complex, and weakly labeled the 210-kDa complex (Fig. 6A), which represents an intermediate containing at least the CcoN and CcoO subunits (26). As observed previously (26), the smallest complex, which represents a CcoP-CcoQ complex (34), was not detectable by BN-PAGE with in vitro labeling. Whether this is related to a low level of labeling efficiency or to the low level of abundance of the small complex is currently unknown. The specificity of in vitro labeling was controlled by using ICMs of the cbb3-Cox deletion strain GK32, which were not specifically labeled by CcoH. In contrast, in ICMs of the ccoP deletion strain M7G, which assembles the 210-kDa intermediate but not the 230-kDa complex, only labeling of the 210-kDa complex was observed. Since the 210-kDa intermediate does not contain the CcoP subunit, these data also indicate that CcoP is not the only contact partner of CcoH.
FIG. 6.
CcoH binds to a preassembled cbb3-Cox. (A) In vitro-synthesized wild-type CcoH (wt) or the truncated CcoH versions CcoH-M0 and CcoH-M1 were in vitro synthesized in the presence of the indicated membranes. MT1131 contains wild-type amounts of cbb3-Cox, while in GK32, cbb3-Cox is deleted. M7G corresponds to an R. capsulatus ccoP mutant that assembles the 210-kDa CcoNO subcomplex. After in vitro synthesis, samples were separated by BN-PAGE, and radioactively labeled bands were detected by phosphorimaging. Asterisks (*) indicate the fully assembled 230-kDa cbb3-Cox complex, while plus signs (+) indicate the 210-kDa subcomplex lacking the CcoP subunit. (B) Synthesis control of the in vitro reactions described for panel A. Note that the three panels correspond to the same gel, which was split into three parts. Therefore, the size differences among the three CcoH derivatives are not visible.
In vitro labeling also allowed us to determine whether the C-terminally truncated CcoH derivatives maintained their ability to bind to the complexes. CcoH-M1 was in vitro synthesized and integrated into wild-type, GK32, and M7G (ΔccoP) membranes. For full-sized CcoH, we observed labeling of both the 230-kDa and the 210-kDa complexes in wild-type membranes and labeling of only the 210-kDa complex in ΔccoP membranes. The labeling in M7G ICMs appeared to be stronger than the labeling in wild-type ICMs, which is probably related to the larger amount of the 210-kDa complex in M7G compared to that in wild-type strains. Nevertheless, these data indicate that CcoH-M1 is still able to bind to the cbb3-Cox complexes. Finally, we also tested the CcoH-M0 derivative, which lacks the complete cytoplasmic domain. However, here we observed no binding to either the 230-kDa nor the 210-kDa complex. The differences in labeling were not the result of large differences in the amounts of in vitro-synthesized proteins, as indicated in Fig. 6B. In summary, these data demonstrate that CcoH binds to both the fully assembled cbb3-Cox and to the 210-kDa cbb3-Cox intermediate. Because CcoP is missing in the 210-kDa complex, CcoH apparently binds not only to CcoP but also to another subunit of cbb3-Cox.
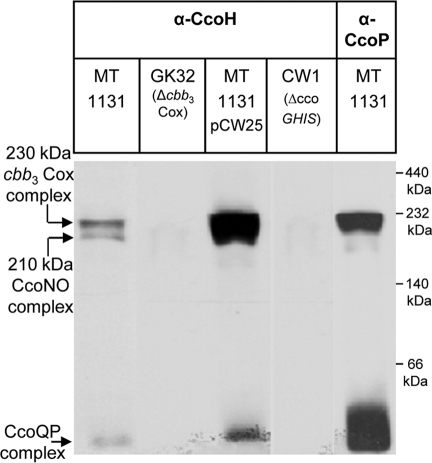
We could not determine by using the in vitro-labeling technique whether binding of in vitro-synthesized CcoH occurs via exchange with the endogenous CcoH or through an unoccupied binding site (26, 34).Therefore, we determined whether CcoH was detectable in the cbb3-Cox complexes by immunodetection. Wild-type membranes were separated by BN-PAGE, and after Western transfer, were decorated with anti-CcoH antibodies. The antibodies recognized two bands at 230-kDa and 210-kDa and a third band running below the 66-kDa marker band with BN-PAGE (Fig. 7). Strikingly, these were exactly the bands that were also recognized by antibodies against CcoN (Fig. 1 and 2) and CcoP (Fig. 6). All three complexes were undetectable in membranes of the cbb3-Cox deletion strain GK32 (Fig. 7) and became significantly stronger in membranes from wild-type cells expressing the complete ccoNOQP-ccoGHIS gene cluster (pCW25). The three bands were also undetectable in membranes of the ccoGHIS deletion mutant. Like the in vitro-labeling data (Fig. 6A), these data show that CcoH contacts not only the CcoP subunit but also other subunits of the cbb3-Cox complex, because the 210-kDa complex is recognized by anti-CcoH antibodies despite the fact that CcoP is not present in this subcomplex. In addition, these data demonstrate that CcoH contacts the cbb3-Cox complexes not only transiently during assembly but that it stays attached to the active 230-kDa cbb3-Cox complex.
FIG. 7.
CcoH is a component of the cbb3-Cox complexes. ICMs from the indicated strains were solubilized with DDM and separated by BN-PAGE as described for Fig. 1. Complexes were subsequently detected by antibodies against CcoH or CcoP. MT1131/pCW25 carries the ccoNOQP-ccoGHIS gene cluster on a plasmid. CW1 corresponds to the ccoGHIS deletion strain.
DISCUSSION
The assembly of a cofactor containing multisubunit protein complexes is a highly ordered process that is orchestrated by many auxiliary proteins. These proteins are referred to as assembly factors because they are usually not present in the mature complex, but in their absence, enzyme complexes fail to assemble. Although a single assembly factor has been shown to control the proper assembly of the heterotrimeric nitrate reductases (27), typically protein complexes assemble along an assembly line involving many of these dedicated assembly factors (18). Respiratory complexes like cytochrome oxidases have been used as model enzymes for analyzing these processes because the need for cofactor incorporation adds an additional level of complexity to their assembly.
In a previous study we proposed two possible roles for the putative assembly factor CcoH in cbb3-Cox assembly. CcoH could be required for stabilizing the CcoQ-CcoP and CcoN-CcoO subcomplexes. In addition, CcoH could facilitate the association of both subcomplexes in the functional 230-kDa cbb3-Cox complex (26). Our data now demonstrate that CcoH is indeed present in both complexes and that CcoH can be cross-linked to CcoP by the amine-specific cross-linker DSS. CcoH contains four lysine residues flanking the transmembrane domain, and lysine residues are also clustered on the cis side of the transmembrane segment of CcoP. Thus, the CcoH-CcoP contact could occur primarily via their respective transmembrane segments. The core complex of cbb3-Cox, consisting of CcoN, CcoO, and CcoP, has recently been crystallized from P. stutzeri (3), and the X-ray structure shows that the transmembrane region of CcoP is freely accessible and appears to have no contact with the other subunits of the cbb3-Cox complex (3). In addition, the transmembrane domains of both CcoP and CcoH show high levels of sequence conservation, which would favor a specific CcoH-CcoP interaction in addition to their role as membrane anchors.
Our data indicate that CcoP cannot be the only contact site for CcoH because CcoP is not detectable in the 210-kDa complex, which still contains CcoH. Whether this additional contact site is provided by CcoO or CcoN is currently unknown. Like CcoP, CcoO consists of a single transmembrane domain connected to the large periplasmic heme binding domain. Therefore, possible contacts between CcoH and CcoO would probably occur primarily via their respective transmembrane domains. However, the transmembrane domain of CcoO seems to be largely occupied by its interaction with the catalytic subunit, as initially suggested by molecular-modeling studies (16, 42) and now confirmed by the X-ray structure (3). Contact of CcoH with the cytosolic N terminus of CcoO is also unlikely, because the N terminus is rather short and not highly exposed in the crystal structure (3). Thus, an interaction between CcoH and CcoN in the 210-kDa complex appears to be more likely. Originally, two-dimensional predictions had pointed to the presence of 14 transmembrane segments, with the N and C termini facing the cytoplasm (52); however, there was also experimental evidence that the first two segments were not membrane integrated but rather located on top of the membrane surface (56). In P. stutzeri, CcoN consists of 12 transmembrane domains (3), but CcoN in P. stutzeri is also shorter than that in B. japonicum or R. capsulatus. In R. capsulatus, CcoN contains an N-terminal extension of about 50 amino acids, which could provide a contact site for the essential second hydrophobic segment of CcoH.
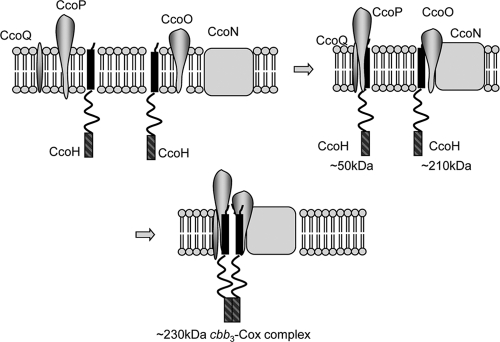
The second hydrophobic segment displays the highest level of sequence conservation within the CcoH family of proteins, highlighting its functional relevance. If this segment is deleted, cbb3-Cox is not assembled, and as a consequence, CcoH is not detectable. On the other hand, in vitro-synthesized CcoH lacking the second hydrophobic domain is able to bind to a preassembled cbb3-Cox complex and also to the CcoNO subcomplex. This finding suggests that the lack of the second hydrophobic domain does not completely block the binding of CcoH to the CcoNO subcomplex but rather prevents the formation of the stable 230-kDa cbb3-Cox holocomplex and is also in agreement with our assumption that it is primarily the transmembrane segment of CcoH that is responsible for making contact with the cbb3-Cox subunits. An attractive model for the role of CcoH during cbb3-Cox assembly is that one copy of CcoH binds via its transmembrane domain to CcoP in the 50-kDa CcoPQ complex (Fig. 8). A second CcoH also binds via its transmembrane domain to the 210-kDa CcoNO complex. These interactions stabilize both subcomplexes, and CcoH would then facilitate in the next step the assembly of both subcomplexes into the final 230-kDa cbb3-Cox complex, possibly via a dimerization of the C-terminal hydrophobic domains.
FIG. 8.
Model for the role of CcoH during assembly of cbb3 oxidase in R. capsulatus. After integration of the individual subunits of cbb3-Cox into the membrane, two subcomplexes are formed, the CcoQP subcomplex and the CcoNO subcomplex. Both subcomplexes also contain CcoH, which is essential for their formation or their stability. In the next step, both subcomplexes assemble to form the active 230-kDa cbb3-Cox complex. The second hydrophobic domain (hatched bars) of CcoH could facilitate this assembly and serve as a dimerization domain to allow the CcoHQP and the CcoHNO complexes to fuse. Please note that the indicated molecular masses have been deduced from analyses by BN-PAGE and therefore represent only approximations.
Our data reveal the crucial role of CcoH in cbb3-Cox assembly and also demonstrate that CcoH displays a rather unusual behavior for an assembly factor, because it can be detected as a stable component of the active cbb3-Cox complex in R. capsulatus membranes. Furthermore, the in vivo steady-state stabilities of cbb3-Cox and CcoH are dependent on each other, which is also unusual for an assembly factor. The lack of CcoH in the absence of cbb3-Cox is not due to impaired ccoH transcription but is most likely due to enhanced proteolysis of CcoH in the absence of cbb3-Cox in vivo. This assumption is in line with our observation that CcoH synthesized in the in vitro system is stable even in membranes lacking cbb3-Cox. The short incubation time and the presence of protease inhibitors are likely to prevent proteolysis in this system. In contrast to the rhizobia, in which ccoGHIS is suggested to form an operon with a single transcript (22), in Rhodobacter species the ccoGHIS genes contain internal promoters and can be expressed independently (25, 39), which indicates that ccoH expression is not necessarily linked to the expression of ccoG, ccoI, and ccoS. The stable association of CcoH with cbb3-Cox is surprising, because so far CcoH has never been found in purified-protein preparations (11, 12, 50, 57). However, in these preparations, cbb3-Cox was always purified as a three-subunit complex lacking the CcoQ subunit. CcoQ was also not detected in the recent X-ray structure of cbb3-Cox (3), which indicates that CcoQ is easily lost during purification. Thus, either CcoH is also lost during purification, or it is substoichiometric and not detected by SDS-PAGE Coomassie staining. It is interesting to note, however, that an additional transmembrane domain is detectable in the crystal structure, which obviously cannot be assigned to CcoQ by either side-chain recognition or by mass-spectrometric analysis of dissolved crystals (3).
Because the stable association of CcoH with cbb3-Cox was detected mainly by BN-PAGE in our study, it is important to emphasize that the presence of complexes in BN-polyacrylamide gels primarily reflects their stability. Therefore, we cannot entirely exclude that the 230-kDa cbb3-Cox complex represents a stable assembly intermediate, despite the fact that it is active (Fig. 8) (26, 34). It is also possible that CcoH is present in only a fraction of cbb3-Cox complexes in R. capsulatus and that this fraction is more stable than the cbb3-Cox complexes lacking CcoH. That CcoH is a stable component of cbb3-Cox is in line with the observation that CcoH is conserved in most bacterial species that contain a cbb3-Cox complex (8). However, in some species, e.g., Sulfurimonas denitrificans (43) and Helicobacter pylori (53), CcoH is missing. In many of these organisms, ccoH appears to be replaced by two genes (cog5456 and cog3198) (8), which show sequence homology to ccoH and which probably functionally replace ccoH. However, we could not find any homologous gene in the Helicobacter pylori genome. Interestingly, although the molecular masses of the Helicobacter pylori cbb3-Cox subunits are greater than those of the R. capsulatus subunits (31, 32), the Helicobacter pylori cbb3-Cox subunits run slightly below the R. capsulatus cbb3-Cox subunits in BN-PAGE (data not shown), which is expected if CcoH is stably associated with cbb3-Cox in R. capsulatus.
In summary, in our study we determined the important contribution of the predicted assembly factor CcoH to cbb3-Cox assembly. Our results reveal that CcoH displays a rather unusual behavior for an assembly factor in that it not only interacts with subunits of cbb3-Cox during intermediate stages of their assembly but that it stays associated with the active, fully assembled enzyme complex as well.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG-GRK1478 to H.-G.K. and P.H. and DFG-FOR 929 to H.-G.K. and B.W.), the German-French-University (DFH) Ph.D. College on Membranes and Membrane Proteins to H.-G.K. and P.H., and the NIH (GM 38237) and DOE (91ER 20052) to F.D. I.S. was supported by a fellowship from the Erasmus program of the European Union.
Footnotes
Published ahead of print on 15 October 2010.
REFERENCES
- 1.Bühler, D., R. Rossmann, S. Landolt, S. Balsiger, H. M. Fischer, and H. Hennecke. 2010. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J. Biol. Chem. 285:15704-15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bundschuh, F. A., A. Hannappel, O. Anderka, and B. Ludwig. 2009. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284:25735-25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buschmann, S., E. Warkentin, H. Xie, J. D. Langer, U. Ermler, and H. Michel. 2010. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science 329:327-330. [DOI] [PubMed] [Google Scholar]
- 4.Carr, H. S., and D. R. Winge. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36:309-316. [DOI] [PubMed] [Google Scholar]
- 5.Chang, H. Y., J. Hemp, Y. Chen, J. A. Fee, and R. B. Gennis. 2009. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc. Natl. Acad. Sci. U. S. A. 106:16169-16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daldal, F., S. Cheng, J. Applebaum, E. Davidson, and R. C. Prince. 1986. Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. U. S. A. 83:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deitermann, S., G. S. Sprie, and H. G. Koch. 2005. A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J. Biol. Chem. 280:39077-39085. [DOI] [PubMed] [Google Scholar]
- 8.Ducluzeau, A. L., S. Ouchane, and W. Nitschke. 2008. The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol. Biol. Evol. 25:1158-1166. [DOI] [PubMed] [Google Scholar]
- 9.Frangipani, E., and D. Haas. 2009. Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 298:234-240. [DOI] [PubMed] [Google Scholar]
- 10.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Horsman, J. A., B. Barquera, J. Rumbley, J. Ma, and R. B. Gennis. 1994. The superfamily of heme-copper respiratory oxidases. J. Bacteriol. 176:5587-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, K. A., M. Grooms, H. Myllykallio, C. Moomaw, C. Slaughter, and F. Daldal. 1994. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry 33:3120-3127. [DOI] [PubMed] [Google Scholar]
- 13.Greiner, P., A. Hannappel, C. Werner, and B. Ludwig. 2008. Biogenesis of cytochrome c oxidase—in vitro approaches to study cofactor insertion into a bacterial subunit I. Biochim. Biophys. Acta 1777:904-911. [DOI] [PubMed] [Google Scholar]
- 14.Hassani, B. K., C. Astier, W. Nitschke, and S. Ouchane. 2010. CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes. J. Biol. Chem. 285:19330-19337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helde, R., B. Wiesler, E. Wachter, A. Neubuser, H. K. Hoffschulte, T. Hengelage, K. L. Schimz, R. A. Stuart, and M. Muller. 1997. Comparative characterization of SecA from the alpha-subclass purple bacterium Rhodobacter capsulatus and Escherichia coli reveals differences in membrane and precursor specificity. J. Bacteriol. 179:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemp, J., C. Christian, B. Barquera, R. B. Gennis, and T. J. Martinez. 2005. Helix switching of a key active-site residue in the cytochrome cbb3 oxidases. Biochemistry 44:10766-10775. [DOI] [PubMed] [Google Scholar]
- 17.Hemp, J., H. Han, J. H. Roh, S. Kaplan, T. J. Martinez, and R. B. Gennis. 2007. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases. Biochemistry 46:9963-9972. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, J. M., and S. Funes. 2005. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene 354:43-52. [DOI] [PubMed] [Google Scholar]
- 19.Hiser, L., M. Di Valentin, A. G. Hamer, and J. P. Hosler. 2000. Cox11p is required for stable formation of the CuB and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 275:619-623. [DOI] [PubMed] [Google Scholar]
- 20.Horng, Y. C., S. C. Leary, P. A. Cobine, F. B. Young, G. N. George, E. A. Shoubridge, and D. R. Winge. 2005. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 280:34113-34122. [DOI] [PubMed] [Google Scholar]
- 21.Iwata, S., C. Ostermeier, B. Ludwig, and H. Michel. 1995. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376:660-669. [DOI] [PubMed] [Google Scholar]
- 22.Kahn, D., M. David, O. Domergue, M. L. Daveran, J. Ghai, P. R. Hirsch, and J. Batut. 1989. Rhizobium meliloti fixGHI sequence predicts involvement of a specific cation pump in symbiotic nitrogen fixation. J. Bacteriol. 171:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalimonchuk, O., and G. Rödel. 2005. Biogenesis of cytochrome c oxidase. Mitochondrion 5:363-388. [DOI] [PubMed] [Google Scholar]
- 24.Koch, H. G., O. Hwang, and F. Daldal. 1998. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 180:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch, H. G., C. Winterstein, A. S. Saribas, J. O. Alben, and F. Daldal. 2000. Roles of the ccoGHIS gene products in the biogenesis of the cbb3-type cytochrome c oxidase. J. Mol. Biol. 297:49-65. [DOI] [PubMed] [Google Scholar]
- 26.Kulajta, C., J. O. Thumfart, S. Haid, F. Daldal, and H. G. Koch. 2006. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J. Mol. Biol. 355:989-1004. [DOI] [PubMed] [Google Scholar]
- 27.Lanciano, P., A. Vergnes, S. Grimaldi, B. Guigliarelli, and A. Magalon. 2007. Biogenesis of a respiratory complex is orchestrated by a single accessory protein. J. Biol. Chem. 282:17468-17474. [DOI] [PubMed] [Google Scholar]
- 28.Marrs, B., and H. Gest. 1973. Genetic mutations affecting the respiratory electron-transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J. Bacteriol. 114:1045-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrs, B., and H. Gest. 1973. Regulation of bacteriochlorophyll synthesis by oxygen in respiratory mutants of Rhodopseudomonas capsulata. J. Bacteriol. 114:1052-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myllykallio, H., and U. Liebl. 2000. Dual role for cytochrome cbb3 oxidase in clinically relevant proteobacteria? Trends Microbiol. 8:542-543. [DOI] [PubMed] [Google Scholar]
- 31.Nagata, K., S. Tsukita, T. Tamura, and N. Sone. 1996. A cb-type cytochrome-c oxidase terminates the respiratory chain in Helicobacter pylori. Microbiology 142:1757-1763. [DOI] [PubMed] [Google Scholar]
- 32.Park, A. M., K. Nagata, E. F. Sato, T. Tamura, K. Shimono, and M. Inoue. 2003. Mechanism of strong resistance of Helicobacter pylori respiration to nitric oxide. Arch. Biochem. Biophys. 411:129-135. [DOI] [PubMed] [Google Scholar]
- 33.Pereira, M. M., F. L. Sousa, A. F. Verissimo, and M. Teixeira. 2008. Looking for the minimum common denominator in haem-copper oxygen reductases: towards a unified catalytic mechanism. Biochim. Biophys. Acta 1777:929-934. [DOI] [PubMed] [Google Scholar]
- 34.Peters, A., C. Kulajta, G. Pawlik, F. Daldal, and H. G. Koch. 2008. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. J. Bacteriol. 190:5576-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preisig, O., R. Zufferey, and H. Hennecke. 1996. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch. Microbiol. 165:297-305. [DOI] [PubMed] [Google Scholar]
- 36.Qin, L., C. Hiser, A. Mulichak, R. M. Garavito, and S. Ferguson-Miller. 2006. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 103:16117-16122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauhamäki, V., D. A. Bloch, M. I. Verkhovsky, and M. Wikström. 2009. Active site of cytochrome cbb3. J. Biol. Chem. 284:11301-11308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter, O. M., and B. Ludwig. 2003. Cytochrome c oxidase—structure, function, and physiology of a redox-driven molecular machine. Rev. Physiol. Biochem. Pharmacol. 147:47-74. [DOI] [PubMed] [Google Scholar]
- 39.Roh, J. H., and S. Kaplan. 2002. Interdependent expression of the ccoNOQP-rdxBHIS loci in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 184:5330-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Scolnik, P. A., M. A. Walker, and B. L. Marrs. 1980. Biosynthesis of carotenoids derived from neurosporene in Rhodopseudomonas capsulata. J. Biol. Chem. 255:2427-2432. [PubMed] [Google Scholar]
- 42.Sharma, V., A. Puustinen, M. Wikstrom, and L. Laakkonen. 2006. Sequence analysis of the cbb3 oxidases and an atomic model for the Rhodobacter sphaeroides enzyme. Biochemistry 45:5754-5765. [DOI] [PubMed] [Google Scholar]
- 43.Sievert, S. M., K. M. Scott, M. G. Klotz, P. S. Chain, L. J. Hauser, J. Hemp, M. Hugler, M. Land, A. Lapidus, F. W. Larimer, S. Lucas, S. A. Malfatti, F. Meyer, I. T. Paulsen, Q. Ren, and J. Simon. 2008. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74:1145-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sistrom, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J. Gen. Microbiol. 22:778-785. [DOI] [PubMed] [Google Scholar]
- 45.Smith, D., J. Gray, L. Mitchell, W. E. Antholine, and J. P. Hosler. 2005. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 280:17652-17656. [DOI] [PubMed] [Google Scholar]
- 46.Stiburek, L., and J. Zeman. 2010. Assembly factors and ATP-dependent proteases in cytochrome c oxidase biogenesis. Biochim. Biophys. Acta 1797:1149-1158. [DOI] [PubMed] [Google Scholar]
- 47.Svensson-Ek, M., J. Abramson, G. Larsson, S. Tornroth, P. Brzezinski, and S. Iwata. 2002. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 321:329-339. [DOI] [PubMed] [Google Scholar]
- 48.Reference deleted.
- 49.Swem, D. L., L. R. Swem, A. Setterdahl, and C. E. Bauer. 2005. Involvement of SenC in assembly of cytochrome c oxidase in Rhodobacter capsulatus. J. Bacteriol. 187:8081-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamegai, H., and Y. Fukumori. 1994. Purification and some molecular and enzymatic features of a novel ccb-type cytochrome c oxidase from a microaerobic denitrifier, Magnetospirillum magnetotacticum. FEBS Lett. 347:22-26. [DOI] [PubMed] [Google Scholar]
- 51.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 52.Toledo-Cuevas, M., B. Barquera, R. B. Gennis, M. Wikstrom, and J. A. Garcia-Horsman. 1998. The cbb3-type cytochrome c oxidase from Rhodobacter sphaeroides, a proton-pumping heme-copper oxidase. Biochim. Biophys. Acta 1365:421-434. [DOI] [PubMed] [Google Scholar]
- 53.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 54.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272:1136-1144. [DOI] [PubMed] [Google Scholar]
- 55.Wieseler, B., and M. Muller. 1993. Translocation of pre-cytochrome c2 into intracytoplasmic membrane vesicles of Rhodobacter capsulatus requires a peripheral membrane protein. Mol. Microbiol. 7:167-176. [DOI] [PubMed] [Google Scholar]
- 56.Zufferey, R., O. Preisig, H. Hennecke, and L. Thony-Meyer. 1996. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 271:9114-9119. [DOI] [PubMed] [Google Scholar]