Abstract
RcsB is the response regulator of the complex Rcs two-component system, which senses perturbations in the outer membrane and peptidoglycan layer. BglJ is a transcriptional regulator whose constitutive expression causes activation of the H-NS- and StpA-repressed bgl (aryl-β,d-glucoside) operon in Escherichia coli. RcsB and BglJ both belong to the LuxR-type family of transcriptional regulators with a characteristic C-terminal DNA-binding domain. Here, we show that BglJ and RcsB interact and form heterodimers that presumably bind upstream of the bgl promoter, as suggested by mutation of a sequence motif related to the consensus sequence for RcsA-RcsB heterodimers. Heterodimerization of BglJ-RcsB and relief of H-NS-mediated repression of bgl by BglJ-RcsB are apparently independent of RcsB phosphorylation. In addition, we show that LeuO, a pleiotropic LysR-type transcriptional regulator, likewise binds to the bgl upstream regulatory region and relieves repression of bgl independently of BglJ-RcsB. Thus, LeuO can affect bgl directly, as shown here, and indirectly by activating the H-NS-repressed yjjQ-bglJ operon, as shown previously. Taken together, heterodimer formation of RcsB and BglJ expands the role of the Rcs two-component system and the network of regulators affecting the bgl promoter.
The nucleoid-associated protein H-NS is a pleiotropic regulator that generally functions as a repressor (silencer) of transcription. The biological role of H-NS has been best studied in Escherichia coli and Salmonella enterica and includes control of stress responses, pathogenicity, and horizontal transfer of DNA (reviewed in references 15, 18, 44, and 54). In addition, H-NS has been proposed to be important in nucleoid organization (45). Numerous studies have addressed the mechanism of DNA binding and transcriptional regulation by H-NS. The protein supposedly binds as a dimer to specific nucleation sites usually located within an AT-rich sequence context. Then, H-NS forms extended complexes by polymerization along the DNA (now defined as “stiffening”) and by building DNA-H-NS-DNA bridges (“bridging”) (8, 35). Formation of such H-NS-DNA complexes next to promoters represses transcription by occluding RNA polymerase or, as shown in some cases, by trapping RNA polymerase at the promoter (44). In addition, H-NS-like proteins, such as StpA, can contribute to gene regulation and silencing (16, 42).
Repression (silencing) by H-NS can be relieved by various mechanisms (44, 54). Most commonly, repression by H-NS is relieved by the binding of specific transcriptional regulators, which compete with H-NS for binding or which restructure the H-NS-nucleoprotein complex. Other mechanisms include locus-specific changes of the DNA structure (bending) (17), enhancement of the transcription rate (43), and possibly direct modulation of H-NS activity by changes in the physiological conditions, such as osmolarity, temperature, and pH (35).
The bgl (aryl-β,d-glucoside) operon of E. coli is a classical example of a locus that is tightly repressed by H-NS. Efficient repression of bgl by H-NS involves synergistic binding of H-NS to regulatory elements located upstream of the promoter and downstream within the transcription unit (43). Historically, spontaneous mutations mapping in cis to the bgl promoter that relieve repression have attracted attention, and it has been speculated that such mutations are a means to control bgl expression at the level of the population under selective environmental conditions (36, 47). Later, it was found that repression of the bgl operon by H-NS can be relieved by the LysR-type transcription factor LeuO (see below) and by BglJ (25, 58). BglJ ia a transcription factor with a C-terminal helix-turn-helix motif of the LuxR type and is encoded in an operon together with YjjQ, another LuxR-type transcription factor proposed to be important for virulence of avian-pathogenic E. coli (APEC) (34, 55). Another prominent member of the family of LuxR-type transcription factors is RcsB, the response regulator of the complex Rcs (regulation of capsule synthesis) two-component system, which senses outer membrane stress and perturbations in the peptidoglycan layer (20, 31). RcsB is a pleiotropic transcription factor involved in the control of motility, cell division, outer membrane protein expression, capsule synthesis, acid stress response, and the small regulatory RNA RprA (5, 29, 39). RcsB, as a homodimer, activates transcription of several genes by binding upstream of the −35 promoter region, including ftsA, osmC, osmB, bdm, and rprA (2, 10, 23, 56). In addition, RcsB forms heterodimers with RcsA, which is likewise a LuxR-type transcription factor. RcsA-RcsB heterodimers activate the capsule synthesis operons cps and yjbEFGH, positively autoregulate rcsA (21, 59), and repress fhlCD, encoding the master regulators of bacterial flagellum biogenesis (22). Furthermore, interaction of RcsB with the acid stress regulator GadE was recently described (5), and in Salmonella enterica serovar Typhi, interaction of RcsB with TviA was found to control Vi antigen synthesis (62).
LeuO is a pathogenicity determinant in S. enterica and is important for biofilm formation in Vibrio cholerae (32, 41, 57). It is a regulator of many genes, including those for outer membrane proteins, drug efflux, the small regulatory RNA DsrA, and the RNA-based immunity system CRISPR (28, 33, 53, 60). LeuO also activates expression of the yjjQ-bglJ operon (55). However, while the relevance of LeuO and YjjQ for pathogenicity indicates that their genes are expressed under certain in vivo conditions in the host environment, both the leuO gene and the yjjQ-bglJ operon are repressed by H-NS under laboratory growth conditions (6, 55).
In this work, we addressed the mechanism by which BglJ counteracts repression of the bgl promoter by H-NS. A screen for mutants in which derepression of bgl by BglJ is abrogated yielded an rcsB mutant, and we demonstrate here that the two-component response regulator RcsB is essential for BglJ to act as an H-NS antagonist at the bgl locus. Further analyses demonstrated that BglJ and RcsB form heterodimers and suggested that these heterodimers directly bind to the bgl upstream regulatory element (URE). In addition, we show that LeuO also binds to the bgl URE and directly activates the bgl promoter.
MATERIALS AND METHODS
Strains, plasmids, and media.
All strains and plasmids used in this study are listed in Table 1. Cloning of plasmids, construction of strains by transduction, and gene replacement followed standard protocols (1, 9, 61), as briefly described in the supplemental material.
TABLE 1.
E. coli K-12 strains
| Strain | Relevant genotypea | Construction/source |
|---|---|---|
| BW30270 | MG1655 rph+ | Coli Genetic Stock Center no. 7925 |
| KL788 | λ− Thr-1 Δ(gpt-lac)5 tsx-35 sulA3 e14− Rac-0 rfbD1 mgl-51 recA441(Ts) relA1 rpsL31(strR) kdgK51 mtl-1 spoT1 thi-1 lexA71::Tn5 creC510 (stored as S1152) | Coli Genetic Stock Center no. 6218 |
| M182 stpA::tet | Δlac74 galU galK strA stpA::tet (stored as S159) | 64 |
| SU101 | λ lysogen with PsulAlexA-op+/+ lacZ fusion in JL1434 | 12 |
| SU202 | λ lysogen withPsulAlexA-op408/+ lacZ fusion in JL1434 | 12 |
| S1734 | yjjQ/bglJ-Y6::miniTn10-cat (= bglJC) in S764 | 38 |
| S524 | CSH50 ΔlacZ-Y217 (gpt-pro)+ | 14 |
| S2176 | S524 yjjQ/bglJ-Y6::miniTn10-cat (= bglJC) | S524 × T4GT7 (S1734) |
| S2817 | S524 attB::[Specr wt-Pbgl(+25) lacZ] (Bgl− and Lac−) | S524 × pKEKB30 |
| S2822 | S524 attB::[Specr wt-Pbgl(+25) lacZ] bglJC (Bgl+ and Lac+) | S2817 × T4GT7 (S1734) |
| S2828 | S2822 rcsB-2828::miniTn10-tet (Bgl− and Lac−) | S2822 × λNK1323 |
| S3918 | S524 ΔrcsB::Specr | S524 × T4GT7 (S3278) |
| S3919 | S524 bglJC ΔrcsB::Specr | S2176 × T4GT7 (S3278) |
| S541 | CSH50 ΔlacZ-Y217 Δbgl-AC11 | 14 |
| S3010 | S541 Δhns::kanKD4 | 43 |
| S3278 | S541 ΔrcsB::Specr | × PCR S774/S775(pKESD8) |
| S3377 | S541 ΔrcsB::Specr Δ(yjjP-yjjQ-bglJ)::catKD3 | S3278 × PCR S783/S676 (pKD3) |
| S1185 | S541 sulA3 | 13 |
| S3360 | S541 sulA3 lexA71::Tn5 | S1184 × T4GT7(KL788) Kanr |
| S3373 | S3360 ΔrcsBFRT | × PCR S819/S820 pKD3 × pCP20 |
| S3384 | S3360 ΔrcsBFRT Δ(yjjP-yjjQ-bglJ)FRT | S3373 × PCR S783/S676 pKD3; × pCP20 |
| S3434 | S3384 attB::(SpecrlacIq T1 PsulAlexA-op+/+ lacZ) | S3384/pLDR8 × pKES163 |
| S3442 | S3384 attB::(SpecrlacIq T1 PsulAlexA-op408/+ lacZ) | S3384/pLDR8 × pKES164 |
| S3974 | BW30270 ilvG+ (valine resistant) | BW30270/pKD46 × annealed oligonucleotides T96/T97 |
| S4197 | BW30270 ilvG+ ΔlacZ | S3974 × pFDY217 |
| T15 | S4197 rcsB::kanKD4 | × PCR S819/S820(pKD4) |
| T70 | S4197 Δ(yjjP-yjjQ-bglJ)::cat | × PCR S676/S783(pKD3) |
| T71 | S4197 ΔleuOFRT | × PCR T209/T210(pKD3); × pCP20 |
| T314 | S4197 ΔleuOFRT Δ(yjjP-yjjQ-bglJ)FRT | T71 × T4GT7 (T70); × pCP20 |
| T568 | T314 attB::(Specr Pbgl t1RATbglG lacZ) | T314/pLDR8 × pKENV61 |
| T576 | T314 attB::(Specr Pbgl-mut2 t1RATbglG lacZ) | T314/pLDR8 × pKES220 |
| T578 | T314 attB::(Specr Pbgl-mut3 t1RATbglG lacZ) | T314/pLDR8 × pKES221 |
| T580 | T314 attB::(Specr Pbgl-mut1 t1RATbglG lacZ) | T314/pLDR8 × pKES222 |
| T727 | T314 attB::(Specr Pbgl t1RATbglG lacZ) ΔrcsBFRT | T568 × T4GT7 (T15); × pCP20 |
| T729 | T314 attB::(Specr Pbgl t1RATbglG lacZ) ΔhnsFRT | T568 × T4GT7 (S3010); × pCP20 |
| T731 | T314 attB::(Specr Pbgl-mut2 t1RATbglG lacZ) ΔhnsFRT | T576 × T4GT7 (S3010); × pCP20 |
| T733 | T314 attB::(Specr Pbgl-mut3 t1RATbglG lacZ) ΔhnsFRT | T578 × T4GT7 (S3010); × pCP20 |
| T735 | T314 attB::(Specr Pbgl-mut1 t1RATbglG lacZ) ΔhnsFRT | T580 × T4GT7 (S3010); × pCP20 |
| T757 | T314 attB::(Specr Pbgl t1RATbglG lacZ) ΔhnsFRTstpA::tet | T729 × T4GT7 (M182 stpA::tet) |
JL1434 is lexA71::Tn5 (Def)sulA211 Δlac169/F′ lacIq lacZΔM15::Tn9. S764 is CSH50 bgl+-C234 ΔlacOP::[SpecR Pbgl-C234(+54) bglGorf] lon-107::miniTn10-tet yjjQ/bglJ-Y6::miniTn10-cat. rcsB-S2828 carries a miniTn10-cm insertion in rcsB with a target site duplication of 9 bp at positions 217 to 225 relative to the rcsB translation start site. sulA3 carries a single-nucleotide A-to-G exchange 35 bp upstream of ATG. The construction of strains by procedures (×) including transduction with phage T4GT7, integration of reporter constructs into attB, replacement of genes with resistance cassettes, and deletion of the resistance cassettes using plasmid pCP20 was performed as described previously (9, 55, 61). ΔlacZ was introduced into strain S3974 by gene replacement using the rep(Ts) plasmid pFDY217, as described previously (3).
Transposon mutagenesis.
Transposon mutagenesis using the phage λNK1323 miniTn10::tet transposon system was performed as described previously (40). Briefly, strain S2822 carrying the bgl promoter dual reporter constructs was infected with λNK1323 lysate (40), and transposon mutants were selected at 41°C on MacConkey lactose tetracycline plates. Lac-negative colonies were restreaked on MacConkey lactose tetracycline plates, as well as on BTB salicin tetracycline indicator plates (14), and Lac- and Bgl-negative mutants were further analyzed by a semirandom two-step PCR (ST-PCR protocol), as described previously (7, 38). In one of the mutants, the miniTn10::tet transposon mapped within the rcsB open reading frame (at position bp 225 relative to the translation start, with a 9-bp target site duplication, TACATCAAG). This allele was designated rcsB::miniTn10-tet and stored as strain S2828 (Table 1).
β-Galactosidase assay.
Cultures were grown overnight in LB medium with antibiotics. Then, 8-ml cultures were inoculated to an optical density at 600 nm (OD600) of 0.05 to 0.1 and grown to an OD600 of 0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was added, where indicated, to a final concentration of 1 mM to the overnight and the exponential cultures for induction. The bacteria were harvested, and β-galactosidase activities were determined independently at least three times, as described previously (40). Standard deviations were less than 10%, unless otherwise indicated.
Coimmunoprecipitation.
Coimmunoprecipitation to analyze the interaction of RcsB with BglJ was performed using transformants of strain S3377 [ΔrcsB Δ(yjjP-yjjQ-bglJ)] with plasmids expressing tagged variants of the RcsB and BglJ proteins. For expression of RcsB with a C-terminal hemagglutinin (HA) tag, strain S3377 was transformed with plasmid pKEAP38. For expression of BglJ with a C-terminal FLAG tag, plasmid pKERV10 was used. The untransformed strain S3377, single transformants (carrying either pKEAP38 or pKERV10), and the double transformants (carrying pKEAP38 and pKERV10) were grown overnight in LB without (empty strain) or with suitable antibiotics. Then, 100 ml of the same medium was inoculated from the overnight culture to an OD600 of 0.05 and grown to an OD600 of 0.3, when IPTG (1 mM final concentration) was added for induction of protein expression. Cells were harvested after 2 h of induction, pelleted by centrifugation, washed once with lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 0.5 mM EDTA, 10% glycerol), and again pelleted by centrifugation. The cells were resuspended in 1 ml lysis buffer (with 1 mM phenylmethylsulfonyl fluoride [PMSF] freshly added) and lysed by sonication. The cell lysates were cleared by centrifugation. A fraction of the lysate equivalent to 200 μg of soluble protein was diluted to a volume of 950 μl with lysis buffer and incubated with 5 μl rabbit anti-HA IgG (Sigma-Aldrich; H6908; 1:200 dilution for immunoprecipitation) for 3 h at 4°C in a tube rotator. Then, 5 mg of protein A-Sepharose beads (GE Healthcare) was added. The samples were incubated for 2 h at 4°C in a tube rotator to allow binding, and the beads were pelleted by centrifugation and washed 3 times with 1 ml lysis buffer. After the final wash, 50 μl Laemmli buffer (49) was added, and the proteins were separated on 12% SDS-PAGE gels and then blotted onto a PVDF membrane (GE Healthcare). For Western analysis, the membranes were blocked with 3% nonfat dry milk powder in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), and incubated with rat anti-HA (Roche, 1:500) and mouse anti-Flag (Sigma-Aldrich, 1:5,000) antibodies. As secondary antibodies, IRDye 800CW-conjugated goat anti-mouse antibody (Li-Cor BioSciences; 1:10,000) and IRDye 680-conjugated goat anti-rat antibody (Molecular Probes; 1:5,000) were used. The blots were scanned with an Odyssey imaging system (Li-Cor Biosciences).
DNase I footprinting.
For DNase I footprinting, the bgl promoter and upstream region (positions −202 to +30 relative to the transcription start site) were amplified by PCR. For 5′-end labeling of the top strand with T4-polynucleotide kinase and [γ-32P]ATP, primers T79 (5′ OH) and T110 (5′ phosphate) were used, while for labeling of the bottom strand, primers T109 (5′ phosphate) and T80 (5′ OH) were used. The binding reaction of LeuO (in the indicated amounts) to the labeled fragments (approximately 200,000 cpm) was performed at 30°C for 20 min in a volume of 20 μl in binding buffer (100 mM KCl, 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 10% glycerol, 500 mM imidazole), 50 ng/μl bovine serum albumin (BSA), and 5 ng/μl herring sperm DNA. Then, 2 μl DNase I (Roche Molecular Biochemicals; 5 ng/μl in binding buffer) was added, and the reaction was stopped exactly 1 min later by the addition of 20 μl of phenol. The samples were extracted with chloroform-isoamyl alcohol, and the DNA was ethanol precipitated. The dried samples were resuspended in 6 μl of sequencing gel loading buffer (79% formamide, 0.1% [wt/vol] bromphenol blue, 0.1% [wt/vol] xylene cyanol, and 5 mM EDTA) and separated on 6% denaturing sequencing gels (6% Long Ranger [Lonza], 7 M urea, 0.8× Tris-borate-EDTA [TBE]) next to a sequencing ladder. The sequencing ladder was generated using the T7 sequencing kit (USB Corporation) and [α-32P]dCTP, with the same primers used for generation of the bgl PCR fragment.
RESULTS
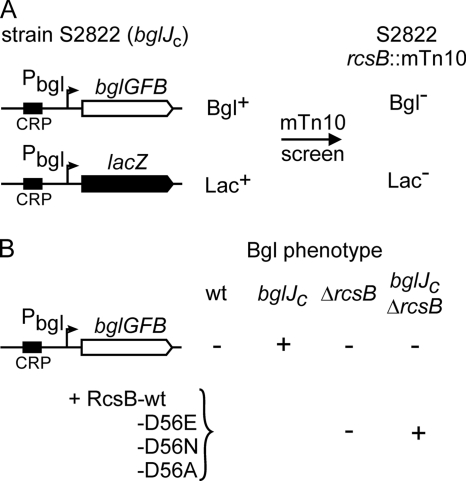
Derepression of bgl by BglJ requires RcsB.
Transcription factor BglJ relieves repression of the bgl operon by H-NS (25). To identify factors that are involved in derepression of bgl by BglJ, we performed a transposon mutagenesis screen using strain S2822, which carries the bgl operon and a bgl promoter-lacZ fusion as dual reporters (Fig. 1A). In addition, the strain constitutively expresses bglJ (referred to as bglJc below) because of the insertion of a miniTn10 transposon upstream of bglJ (allele yjjQ/bglJ-Y6::miniTn10) (38). Thus, this dual-reporter strain for monitoring activity of the bgl promoter is Bgl and Lac positive. Transposon mutants were screened for a Bgl- and Lac-negative phenotype, and one of the transposon mutations mapped in rcsB (Fig. 1A). To verify that a mutation of rcsB interferes with derepression of the bgl promoter by BglJ, an rcsB deletion was introduced into strain S2176, which carries the wild-type bgl operon and expresses bglJ constitutively (bglJc). Analysis of the Bgl phenotype on indicator plates demonstrated that the deletion of rcsB abrogates repression of bgl by BglJ (Fig. 1B). Further, complementation of the ΔrcsB mutant with plasmid pKETS6 carrying rcsB under the control of the IPTG-inducible tac promoter restored the Bgl-positive phenotype in the bglJc strain, but not in the wild type. This suggests that both RcsB and BglJ are required to relieve the H-NS-mediated repression of bgl. Interestingly, complementation of the ΔrcsB strain was also possible with RcsB mutants carrying exchanges in the conserved aspartate (residue 56) of the N-terminal receiver domain. Mutation RcsB-D56E mimics the active phosphorylated state, and mutations D56N and D56A mimic the inactive state of RcsB (27, 51). These results suggest that BglJ and RcsB act together independently of RcsB phosphorylation.
FIG. 1.
Activation of the bgl operon by BglJ requires RcsB. (A) Schematic of a transposon mutagenesis screen for mutants in which activation (relief of H-NS-mediated repression) of bgl by BglJ is abrogated. Strain S2822 carries the bgl operon and a bgl promoter-lacZ fusion as dual reporters for bgl expression. In addition, the strain carries allele yjjQ/bglJ-Y6::miniTn10-cat (bglJc) for constitutive expression of bglJ. A miniTn10-tet (mTn10) transposon mutagenesis screen yielded Bgl- and Lac-negative mutants, one of which carried a transposon insertion in rcsB (assigned to strain S2828 [Table 1]). (B) RcsB is required for derepression of bgl by BglJ. The Bgl phenotypes of the E. coli K-12 wild type (wt) (strain S524) and its isogenic derivatives, which constitutively express bglJ (bglJC; strain S2176) or carry a deletion of rcsB (ΔrcsB; strain S3918), as well as the double mutant bglJC ΔrcsB (strain S3919), was determined on BTB salicin indicator plates. Shown is complementation of the ΔrcsB mutants with plasmids encoding wild-type RcsB (pKETS6) or the RcsB mutants D56E (pKETS7), D56N (pKETS8), and D56A (pKES235).
Interaction of the LuxR-type transcription factors RcsB, BglJ, and YjjQ.
The LuxR-type response regulator RcsB is known to interact with RcsA and GadE (see the introduction). As derepression of the bgl operon by BglJ requires RcsB, we analyzed whether BglJ also interacts with RcsB and whether BglJ forms homodimers. In addition, we analyzed whether BglJ interacts with YjjQ (as BglJ and YjjQ are encoded in one operon [55]).
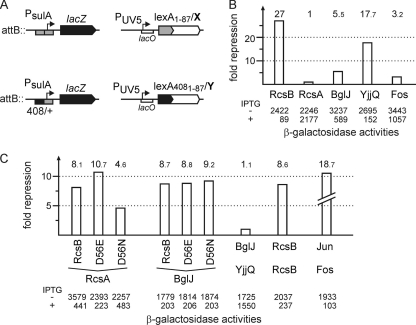
Interaction was tested using the bacterial LexA-based two-hybrid system (12) and by coimmunoprecipitation (see below). The LexA-based two-hybrid system is based on repression of the sulA promoter by LexA (Fig. 2A). The reporter for homodimer formation consists of the native sulA promoter fused to lacZ. In a lexA mutant, this promoter is constitutively active but can be repressed by fusion of the N-terminal LexA-DNA-binding domain (LexA1-87) to a protein that forms homodimers (Fig. 2A) (12). The sulA promoter-lacZ reporter for heterodimer formation carries a hybrid lexA operator (op408/+) with a mutation in one half-site (Fig. 2A). This operator can only be bound by heterodimers in which one partner includes a LexA1-87 wild-type DNA-binding domain and the other protein partner contains a LexA4081-87 mutant DNA-binding domain (Fig. 2A) (12). For the current analysis of homodimer and heterodimer formation, the LexA-based two-hybrid-system was transferred to a ΔrcsB and Δ(yjjP-yjjQ-bglJ) strain background (see the supplemental material). In addition, plasmids that express fusions of the wild-type LexA1-87 DNA-binding domain to BglJ, RcsB, YjjQ, and RcsA were constructed. RcsB and BglJ were also fused to the mutant LexA4081-87 DNA-binding domain (Fig. 2 and Table 1).
FIG. 2.
Interaction of BglJ with RcsB and YjjQ. (A) In the LexA-based two-hybrid system, the sulA promoter-lacZ fusion with the wild-type LexA operator was used to analyze homodimerization, and the sulA promoter-lacZ reporter fusion with a hybrid lexA408/+ operator served as a reporter for heterodimerization. For analysis of homodimerization, a fusion of the respective protein (X) to the wild-type LexA DNA-binding domain (lexA1-87/X) was expressed from a plasmid under the control of the IPTG-inducible lacUV5 promoter (PUV5). For heterodimerization analysis, fusions of protein X to the wild-type LexA DNA-binding domain (lexA1-87/X) and of protein Y to the LexA408 mutant DNA-binding domain (lexA4081-87/Y) were coexpressed from compatible plasmids. (B) Analysis of homodimer formation in RcsB, RcsA, BglJ, and YjjQ was performed with transformants of strain S3434 with plasmids pKEMK17 (lexA1-87-rcsB), pKES192 (lexA1-87-rcsA), pKEAP30 (lexA1-87-bglJ), pKEAP27 (lexA1-87-yjjQ), and pMS604 (lexA1-87-fos) as controls. Cultures were grown in LB tetracycline medium to an OD600 of 0.5. IPTG was added to 1 mM final concentration where indicated (+). The β-galactosidase activity was determined to monitor repression of the sulA promoter by the LexA1-87-X fusion protein. The fold repression (indicated by the bars), as a measure for dimerization, was calculated as the ratio of the β-galactosidase activities measured without and with induction of the LexA fusion proteins. (C) Analysis of heterodimer formation was performed with strain S3442, which was cotransformed with plasmids coding for LexA1-87-X and LexA4081-87-Y fusions, respectively. The cultures were grown in LB with antibiotics, and IPTG was added where indicated. The fold repression of the sulA promoter-lacZ fusion with the hybrid lexA operator (lexA-op408/+) is a measure of heterodimerization (indicated by the bars). The following plasmids were used: LexA1-87-RcsB (pKEMK17), LexA1-87-RcsA (pKES192), LexA1-87-BglJ (pKEAP30), LexA1-87-YjjQ (pKEAP27), and LexA1-87-Fos (pMS604) (12), as well as LexA4081-87-RcsB (pKEAP28), LexA4081-87-RcsBD56E (pKES150), LexA4081-87-RcsBD56N (pKES151), LexA4081-87-BglJ (pKEAP29), and LexA4081-87-Jun (pDP804) (12) as controls.
In the homodimerization assay, induction of the LexA1-87-RcsB fusion caused strong repression of the sulA promoter-lacZ reporter (in strain S3434), as expected (Fig. 2B). In contrast, the LexA1-87-RcsA fusion caused no repression (Fig. 2B), as anticipated from earlier studies, which suggested that RcsA forms heterodimers with RcsB but no homodimers (39). In comparison, the LexA1-87-BglJ fusion protein caused very moderate repression, indicating that BglJ forms weak homodimers. However, the LexA1-87-YjjQ fusion caused strong repression, suggesting efficient homodimer formation by YjjQ. As a control, a LexA1-87-Fos fusion known for its low capacity for homodimer formation was included. Induction of this fusion resulted in only weak repression (Fig. 2B).
In the heterodimerization assay, coinduction of LexA4081-87-RcsB with LexA1-87-RcsA resulted in strong repression (Fig. 2C). This result is in agreement with earlier data suggesting that RcsA and RcsB form heterodimers (39). Coexpression of BglJ (fused to LexA1-87) and RcsB (fused to LexA4081-87) likewise resulted in strong repression, suggesting that BglJ and RcsB form heterodimers (Fig. 2C). Interestingly, YjjQ (fused to LexA1-87) and BglJ (fused to LexA4081-87) caused no repression (Fig. 2C), suggesting that the two LuxR-type transcription factors BglJ and YjjQ do not interact, although they are encoded in one operon. Since neither deletion of yjjQ nor plasmid-directed expression of YjjQ plays a role in regulation of the bgl operon (data not shown), YjjQ was not included in further analyses. As additional positive controls for heterodimerization, interaction analyses of LexA1-87-Fos and LexA4081-87-Jun, as well as of RcsB (using LexA1-87-RcsB and LexA4081-87-RcsB), were included, which, as expected, caused repression (Fig. 2C).
Furthermore, we analyzed whether heterodimer formation of RcsB with BglJ and RcsA, respectively, depends on phosphorylation of RcsB. Heterodimer formation of RcsA with RcsB-D56E was enhanced compared to that of wild-type RcsB, while it was reduced with the RcsB-D56N mutant (Fig. 2C). In contrast, heterodimer formation of BglJ and RcsB was not affected by the mutation of the presumptive RcsB phosphorylation site (Fig. 2C). These data indicate that interaction of RcsB with RcsA is modulated by phosphorylation of RcsB and thus by induction of the Rcs signaling cascade. In contrast, the interaction of RcsB with BglJ is presumably not affected by RcsB phosphorylation, in agreement with the complementation analysis shown above (Fig. 1B).
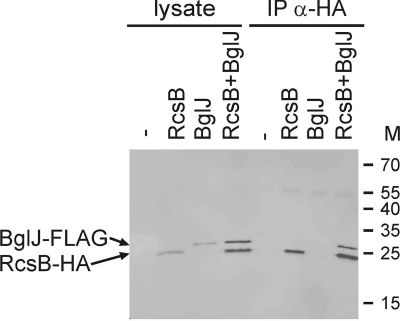
In a second set of experiments, heterodimer formation of BglJ with RcsB was analyzed by coimmunoprecipitation. To this end, compatible vectors for coexpression of BglJ-Flag and RcsB-HA in strain S3377 [ΔrcsB Δ(yjjP-yjjQ-bglJ)] were used. These plasmid-encoded BglJ-Flag and RcsB-HA proteins are functional, as tested by complementation of the respective mutants using bgl as a reporter (data not shown). Coimmunoprecipitation of cell lysates was performed with an HA tag-specific antibody (rabbit anti-HA IgG). For visualization of the proteins by Western blotting, fluorescent secondary antibodies were used, allowing simultaneous detection of the FLAG- and HA-tagged proteins in one gel (Fig. 3). Analysis of the cell lysates demonstrated that the proteins were well expressed (Fig. 3, lysates). After coimmunoprecipitation with an HA-specific antibody, RcsB-HA was detectable irrespective of whether it was expressed in the absence or presence of BglJ-FLAG (Fig. 3). However, BglJ-FLAG was precipitated only when it was coexpressed with RcsB-HA (Fig. 3). This demonstrated that the coimmunoprecipitation was specific and suggests that BglJ-FLAG interacts with RcsB-HA (Fig. 3).
FIG. 3.
Coimmunoprecipitation of BglJ-FLAG with RcsB-HA. (A) Coimmunoprecipitation of BglJ-FLAG with RcsB-HA was performed for lysates prepared from strain S3377 [ΔrcsB Δ(yjjP-yjjQ-bglJ)] (−) and for transformants of strain S3377 with plasmids pKEAP38 (RcsB-HA) (RcsB), pKERV10 (BglJ-FLAG) (BglJ), or both plasmids. Immunoprecipitation (IP) was performed with rabbit anti-HA IgG. The lysates and the coimmunoprecipitates were separated on SDS-PAGE and analyzed by Western blotting. For simultaneous detection of BglJ-FLAG and RcsB-HA, the Western blot was developed with rat anti-HA (α-HA) and mouse anti-FLAG as primary antibodies and fluorescence labeled anti-mouse and anti-rat secondary antibodies.
Mapping of a BglJ-RcsB box in the bgl regulatory region.
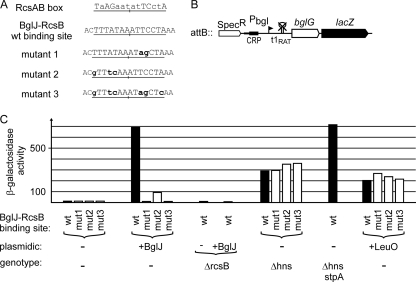
The data indicate that BglJ-RcsB heterodimers relieve repression of bgl by H-NS by binding next to the bgl promoter. For RcsA-RcsB heterodimers, a consensus sequence (termed the RcsAB box) was proposed (39, 59). This RcsAB box is nonpalindromic (see Fig. S1 in the supplemental material) and is presumably recognized by binding of RcsB to one half-site and by binding of RcsA to the other half-site (22). Interestingly, within the bgl regulatory region, a perfect match to one half-site of the RcsAB box is located at positions −88 to −95 (relative to the transcription start site) (Fig. 4A). Assuming that the right half-site of the RcsAB box is bound by RcsB (see Fig. S1 in the supplemental material), this match indicates that the RcsB subunit of the BglJ-RcsB heterodimer may bind to this motif and that BglJ contacts adjacent base pairs.
FIG. 4.
Effect of mutation of the BglJ-RcsB binding site on derepression of bgl by BglJ-RcsB. The expression levels directed by bgl promoter-lacZ fusions (shown schematically in panel B) with wild-type and mutant BglJ-RcsB binding sites (A) were determined for exponential cultures grown in LB (with appropriate antibiotics and 1 mM IPTG) (C). (A) The BglJ-RcsB binding site is underlined, and mutations are indicated in lowercase boldface letters. (B) The bgl-lacZ fusions were integrated at the phage λ attB site (the strains are listed in Table 1). The bgl promoter (Pbgl) is indicated by a flagged arrowhead, the cyclic AMP (cAMP) receptor protein-cAMP complex (CRP-cAMP) binding site is shown as a black box, and the mutation of terminator t1 (t1RAT) is indicated by a crossed hairpin loop. (C) Expression levels were determined in strain T314 [Δ(yjjP-yjjQ-bglJ) ΔleuO] (−) transformed with the empty vector pKESK22, with plasmid pKETS1 for expression of BglJ in trans (+BglJ), or with plasmid pKEDR13 for expression of LeuO (+LeuO). The β-galactosidase activities with LeuO provided in trans were determined 6 times independently, as the standard deviation was up to 40%. In addition, the expression levels of the bgl-lacZ fusions were analyzed in transformants of Δhns, ΔrcsB, and Δhns stpA mutant derivatives, as indicated.
To test the relevance of this presumptive BglJ-RcsB binding site, site-specific mutations were introduced in the most conserved bases matching the right half-site of the RcsAB box (Fig. 4A, mutant 1). In addition, the left half-site of the presumptive BglJ-RcsB box was mutated (Fig. 4A, mutant 2), and mutations in both half-sites were combined (Fig. 4A, mutant 3). The effect of these mutations on derepression of bgl by BglJ-RcsB was tested using a bgl-lacZ reporter construct that carries all elements required for repression by H-NS (Fig. 4B). However, expression of this reporter is independent of regulatory elements for sugar-specific regulation, as it carries a mutation of terminator t1 (43, 46). Note that sugar-specific regulation of the bgl operon is promoter independent and is mediated by the specific transcriptional antiterminator BglG, which allows transcription read-through at terminator t1. BglG activity is regulated by phosphorylation that is dependent on the availability of the specific substrate and other sugars (26).
For expression analyses, the bgl-lacZ reporter constructs with the putative wild-type and mutated BglJ-RcsB binding sites, respectively, were integrated at the λ-attB site of strain T314 [ΔlacZ, Δ(yjjP-yjjQ-bglJ), and ΔleuO, as LeuO also derepresses the bgl operon (see below)]. To analyze derepression of the bgl-lacZ reporter, BglJ was provided in trans using plasmid pKETS1 carrying bglJ under the control of the inducible tac promoter. Note that there is some ambiguity about the translation start codon of bglJ. Plasmid pKETS1 includes the most 5′ AUG, which maps within yjjQ. This plasmid directs the expression of active BglJ protein (see below), while plasmids pKETS9 and pKETS10, which include the second or third start codon, provide no functional BglJ (data not shown), suggesting that translation of the bglJ gene begins at the very first start codon of the open reading frame.
The bgl-lacZ reporter construct with the presumptive wild-type BglJ-RcsB box directed low levels of β-galactosidase activity, as expected (12 units) (Fig. 4C). When BglJ was provided in trans, expression increased 57-fold to 690 units (Fig. 4C). In a ΔrcsB mutant, expression was low (9 units), and expression remained low (8 units) when BglJ was provided in trans, demonstrating again that derepression of bgl by BglJ requires RcsB (Fig. 4C). Next, the expression levels directed by BglJ-RcsB binding-site mutants 1 to 3 (Fig. 4A) were tested in the absence or presence of BglJ. In the cases of mutants 1 and 3, which both carry exchanges corresponding to the conserved bases of the right half-site, induction of plasmid-encoded BglJ had no effect (10 to 13 units in all cases) (Fig. 4C). This demonstrates that mutations in the presumptive BglJ-RcsB binding site abrogate derepression of bgl by BglJ-RcsB. Interestingly, binding site mutant 2 also affected derepression of the bgl promoter-lacZ fusion by BglJ-RcsB, as the expression level increased only 8-fold, from 12 to 92 units, when BglJ was expressed (Fig. 4C). Mutant 2 carries mutations in the left half of the putative BglJ-RcsB box, which is presumably contacted by the BglJ subunit of the BglJ-RcsB heterodimer (Fig. 4A). Taken together, these data demonstrate that the putative BglJ-RcsB motif is important for derepression of bgl by BglJ-RcsB heterodimers.
As a further control, expression of the bgl-lacZ reporter constructs with the wild-type BglJ-RcsB box and its mutants was tested in isogenic Δhns strains. Expression levels were high (290 to 360 units) (Fig. 4C), as expected, as H-NS represses the bgl promoter. Further, in the Δhns mutant, activity was similarly high, irrespective of whether the BglJ-RcsB box was mutated, demonstrating that the site-specific mutations do not affect promoter activity or repression by H-NS (Fig. 4C). Interestingly, the expression level directed by the bgl-lacZ fusion was lower in the Δhns mutant (290 units) than when plasmid-encoded BglJ was provided in the wild type (690 units). This indicated that the bgl promoter is not fully active in the hns mutant. In agreement with previous studies, which had demonstrated that StpA partially represses bgl in hns mutants (24, 42, 63), the expression level directed by the bgl-lacZ reporter was 715 units in the Δhns stpA double mutant and thus similar to that upon derepression of bgl by BglJ-RcsB. However, growth of the Δhns stpA double mutant was significantly slower than that of the hns mutant. Similarly, expression of plasmidic BglJ resulted in significantly slower growth in the hns mutant and caused a severe growth reduction in the hns stpA double mutant (data not shown). Therefore, we could not test whether BglJ-RcsB further enhances bgl promoter activity in the absence of H-NS and StpA. However, taken together, the data suggest that the BglJ-RcsB heterodimer binds within the upstream regulatory region and antagonizes repression of bgl by H-NS, and also by StpA.
Derepression of bgl by BglJ-RcsB and that by LeuO are independent of each other.
In addition to BglJ, the LysR-type transcription factor LeuO abrogates H-NS-mediated repression of bgl (58). LeuO also activates the yjjQ-bglJ operon (55). To test whether derepression of bgl by LeuO is independent of BglJ and whether the mutations in the BglJ-RcsB binding site interfere with derepression of bgl by LeuO, expression levels were also tested with LeuO provided in trans. For this, plasmid pKEDR13 carrying leuO under the control of the inducible tac promoter was used. Induction of plasmid-encoded LeuO caused derepression of the bgl-lacZ fusion (directing 200 units of β-galactosidase activity) (Fig. 4C), demonstrating that LeuO activates bgl independently of BglJ-RcsB [the strain background is Δ(yjjP-yjjQ-bglJ) ΔleuO]. Similarly high expression levels were directed by the BglJ-RcsB binding-site mutants when LeuO was present (200 units for the wild type compared to 215 to 265 units for the RcsB site mutants) (Fig. 4C). This demonstrates that the mutations of the presumptive BglJ-RcsB box do not interfere with derepression of bgl by LeuO. However, LeuO did not cause full activation (approximately 200 units in the presence of LeuO compared to 690 units in the presence of BglJ), possibly because plasmid-directed expression of the pleiotropic LeuO affects the accuracy of the β-galactosidase assay (Fig. 4C). Nonetheless, the data suggest, that LeuO derepresses bgl by directly binding to the upstream regulatory region of the bgl promoter. Importantly, these data suggest that mutations in the presumptive BglJ-RcsB binding site do not abolish LeuO binding. To further validate this assumption, the LeuO binding site was mapped by DNase I footprinting using purified C-terminally His-tagged LeuO. The LeuO footprint showed protection of approximately 60 bp, extending from positions −101 to −160 relative to the bgl promoter transcription start site (see Fig. S2 in the supplemental material). Thus, the LeuO binding site maps just adjacent to the BglJ-RcsB site (Fig. 5). Extended footprints are typical of LeuO (11, 28, 60) and other LysR-type transcription factors (37). However, it remains unknown whether LeuO and BglJ-RcsB can bind simultaneously.
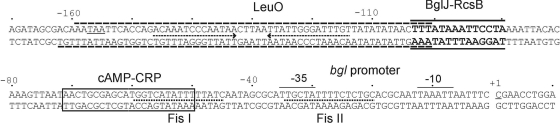
FIG. 5.
Sequence of the bgl promoter and upstream regulatory region. Indicated are the −35, −10, and transcription start sites of the promoter; the CRP binding site (boxed) (48); and the Fis binding sites (dotted lines) (4), as well as the LeuO (dashed lines) and BglJ-RcsB (solid lines) binding sites characterized here. H-NS binds to the upstream regulatory element and the promoter, but the H-NS nucleation sites have so far not been mapped. The stop codon of the phoU gene located upstream of bgl is underlined, and the inverted arrows indicate an inverted repeat that may represent the phoU transcriptional terminator.
DISCUSSION
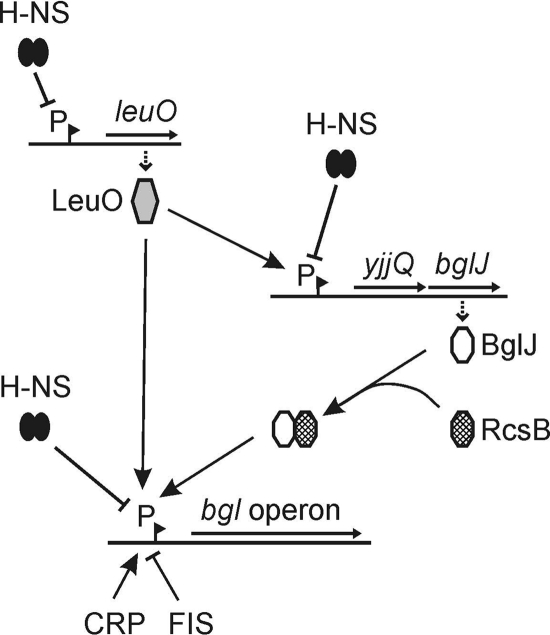
BglJ, a LuxR-type transcriptional regulator, and LeuO, a LysR-type transcriptional regulator, relieve repression of the bgl operon by H-NS. Here, we have shown that activation of bgl by BglJ depends on the two-component response regulator RcsB. BglJ and RcsB interact and presumably form heterodimers, as demonstrated by two-hybrid analysis and coimmunoprecipitation. These BglJ-RcsB heterodimers bind to the bgl URE, as suggested by site-specific mutation of a sequence motif, one half of which is related to the RcsA-RcsB consensus sequence. In addition, we have shown that LeuO likewise binds to the bgl URE and activates bgl directly. Taken together, the data suggest that binding of BglJ-RcsB and LeuO, respectively, interferes with formation of a repressing nucleoprotein complex by H-NS and thus results in activation of bgl. Furthermore, interaction of BglJ and RcsB extends the Rcs two-component signaling transduction system and the network of transcription regulators that affect the bgl operon (Fig. 6 shows a model).
FIG. 6.
Model illustrating regulation of the bgl promoter and regulatory interactions of the transcriptional regulators LeuO and BglJ-RcsB (for details, see Discussion). Pointed arrowheads indicate activation, and blunt arrowheads indicate repression. Promoters (P) are indicated by pointed flags. In addition to being controlled by H-NS, StpA, LeuO, and BglJ-RcsB, the bgl promoter is CRP dependent and is repressed by FIS (4, 48, 52).
The Rcs two-component system is widely conserved in enterobacteria and has an impact on biofilm formation and virulence in various species (29, 39). The response regulator RcsB functions both as a homodimer and as an RcsA-RcsB heterodimer. RcsB also interacts with GadE, which is likewise a LuxR-type transcription factor, and possibly with TviA in S. enterica (5, 62). Our finding that BglJ and RcsB also form a heterodimer that acts as a transcriptional activator of the bgl operon expands the role of the Rcs system and underscores the notion that RcsB activity, in addition to being modulated by phosphorylation, is controlled by interaction with other transcriptional regulators. Furthermore, the activity of the heterodimer BglJ-RcsB is presumably independent of RcsB phosphorylation, while the activity of the RcsA-RcsB heterodimer is phosphorylation dependent (39). This extra level of combinatorial control of the response regulator RcsB is likely to have an impact on the regulatory repertoire attributable to the Rcs two-component signal transduction system.
There are several parallels between RcsA and BglJ. Both RcsA-RcsB and BglJ-RcsB heterodimers function as activators, or rather, as H-NS antagonists. BglJ-RcsB counteracts H-NS-mediated repression of the bgl operon (see above), while RcsA-RcsB activates the cps-wza and yjb operons, encoding enzymes for capsule synthesis, and the H-NS-repressed rcsA gene (21, 29, 39). Activation by RcsA-RcsB and BglJ-RcsB, respectively, involves binding sites that map 100 bp or more upstream of the transcription start site (see Fig. S1 in the supplemental material). A further parallel is that both the rcsA gene and the bglJ gene (within the yjjQ-bglJ operon) are repressed by H-NS. Expression of rscA is autoregulated, while expression of the yjjQ-bglJ operon is activated by the LysR-type transcription factor LeuO. Similarly, the complex regulation of the acid stress response gene gadA, which is activated by GadE and RcsB, involves repression by H-NS (5). This indicates that RcsB, with its interacting partners, may play an important role as an H-NS antagonist.
Furthermore, we demonstrated that LeuO directly binds to the bgl upstream regulatory region and relieves repression independently of BglJ-RcsB. Considering the fact that LeuO also relieves H-NS-mediated repression of the yjjQ-bglJ operon (55), this suggests that LeuO can affect bgl expression in two ways, directly by activating the bgl promoter and indirectly by activating expression of the yjjQ-bglJ operon. However, the leuO gene is also repressed by H-NS and only moderately induced by branched amino acid starvation in a ppGpp-dependent manner (19). The latter may not lead to sufficiently high expression levels of LeuO under laboratory conditions, as these stress conditions seem not to affect LeuO target genes. Accordingly, up-to-date analyses of regulation by LeuO have been performed with chromosomal or plasmidic alleles under the control of constitutive or inducible promoters (11, 28, 53, 55, 60).
Taken together, LeuO and BglJ-RcsB form a small regulatory network that relieves H-NS-mediated repression of the bgl operon (Fig. 6). However, expression of leuO and bglJ is repressed by H-NS, at least under laboratory growth conditions. As LeuO is a virulence factor in S. enterica (32, 57) and as YjjQ, which is coencoded with BglJ, is presumably important for infection by avian-pathogenic E. coli (34), it is conceivable that certain conditions in the host environment induce their expression, which in turn should also relieve repression of the bgl operon and allow its induction by substrate (aryl-β,d-glucosides). The bgl operon is a very tightly controlled locus which may possibly serve a very specialized function related to extraintestinal pathogenicity (50), in agreement with the finding that the bgl operon is induced in a septicemic strain when it infects the mouse liver (30).
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Schn371/10-1) and by the Cologne Graduate School of Genetics and Functional Genomics.
We thank Kathleen Plamper for excellent technical assistance and Maria Fabisch, Sonja Klemme, Julia Kleinmanns, and Selman Öztürk for contributing to the construction of plasmids and β-galactosidase assays.
Footnotes
Published ahead of print on 15 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2005. Current protocols In molecular biology. John Wiley & Sons, Inc., Hoboken, NJ.
- 2.Boulanger, A., A. Francez-Charlot, A. Conter, M. P. Castanie-Cornet, K. Cam, and C. Gutierrez. 2005. Multistress regulation in Escherichia coli: expression of osmB involves two independent promoters responding either to σS or to the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187:3282-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caramel, A., and K. Schnetz. 1998. Lac and Lambda repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 284:875-883. [DOI] [PubMed] [Google Scholar]
- 4.Caramel, A., and K. Schnetz. 2000. Antagonistic control of the E. coli bgl promoter by FIS and CAP in vitro. Mol. Microbiol. 36:85-92. [DOI] [PubMed] [Google Scholar]
- 5.Castanie-Cornet, M. P., K. Cam, B. Bastiat, A. Cros, P. Bordes, and C. Gutierrez. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 38:3546-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. C., M. Y. Chou, C. H. Huang, A. Majumder, and H. Y. Wu. 2005. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 280:5101-5112. [DOI] [PubMed] [Google Scholar]
- 7.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 8.Dame, R. T., M. C. Noom, and G. J. L. Wuite. 2006. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 444:387-390. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De la Cruz, M. A., M. Fernandez-Mora, C. Guadarrama, M. A. Flores-Valdez, V. H. Bustamante, A. Vazquez, and E. Calva. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 66:727-743. [DOI] [PubMed] [Google Scholar]
- 12.Dmitrova, M., G. Younes-Cauet, P. Oertel-Buchheit, D. Porte, M. Schnarr, and M. Granger-Schnarr. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205-212. [DOI] [PubMed] [Google Scholar]
- 13.Dole, S., Y. Klingen, V. Nagarajavel, and K. Schnetz. 2004. The protease Lon and the RNA-binding protein Hfq reduce silencing of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 186:2708-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dole, S., S. Kühn, and K. Schnetz. 2002. Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates. Mol. Microbiol. 43:217-226. [DOI] [PubMed] [Google Scholar]
- 15.Dorman, C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5:157-161. [DOI] [PubMed] [Google Scholar]
- 16.Doyle, M., M. Fookes, A. Ivens, M. W. Mangan, J. Wain, and C. J. Dorman. 2007. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315:251-252. [DOI] [PubMed] [Google Scholar]
- 17.Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, F. C., and S. Rimsky. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, M., A. Majumder, K. J. Tsai, and H. Y. Wu. 2000. ppGpp-dependent leuO expression in bacteria under stress. Biochem. Biophys. Res. Commun. 276:64-70. [DOI] [PubMed] [Google Scholar]
- 20.Farris, C., S. Sanowar, M. W. Bader, R. Pfuetzner, and S. I. Miller. 2010. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192:4894-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrières, L., S. N. Aslam, R. M. Cooper, and D. J. Clarke. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153:1070-1080. [DOI] [PubMed] [Google Scholar]
- 22.Francez-Charlot, A., B. Laugel, G. A. Van, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 23.Francez-Charlot, A., M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2005. Osmotic regulation of the Escherichia coli bdm (biofilm-dependent modulation) gene by the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Free, A., M. E. Porter, P. Deighan, and C. J. Dorman. 2001. Requirement for the molecular adapter function of StpA at the Escheirchia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42:903-918. [DOI] [PubMed] [Google Scholar]
- 25.Giel, M., M. Desnoyer, and J. Lopilato. 1996. A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K-12. Genetics 143:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Görke, B. 2003. Regulation of the Escherichia coli antiterminator protein BglG by phosphorylation at multiple sites and evidence for transfer of phosphoryl groups between monomers. J. Biol. Chem. 278:46219-46229. [DOI] [PubMed] [Google Scholar]
- 27.Gupte, G., C. Woodward, and V. Stout. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernández-Lucas, I., A. L. Gallego-Hernandez, S. Encarnacion, M. Fernandez-Mora, A. G. Martinez-Batallar, H. Salgado, R. Oropeza, and E. Calva. 2008. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 190:1658-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, Y. H., L. Ferrieres, and D. J. Clarke. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157:206-212. [DOI] [PubMed] [Google Scholar]
- 30.Khan, M. A., and R. E. Isaacson. 1998. In vivo expression of the β-glucoside (bgl) operon of Escherichia coli occurs in mouse liver. J. Bacteriol. 180:4746-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laubacher, M. E., and S. E. Ades. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190:2065-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrenz, M. B., and V. L. Miller. 2007. Comparative analysis of the regulation of rovA from the pathogenic Yersiniae. J. Bacteriol. 189:5963-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, G., C. Ewers, C. Laturnus, I. Diehl, J. Dai, E.-M. Antão, K. Schnetz, and L. H. Wieler. 2008. Characterization of a yjjQ mutant of avian pathogenic E. coli (APEC). Microbiology 154:1082-1093. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., H. Chen, L. J. Kenney, and J. Yan. 2010. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 24:339-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madan, R., R. Kolter, and S. Mahadevan. 2005. Mutations that activate the silent bgl operon of Escherichia coli confer a growth advantage in stationary phase. J. Bacteriol. 187:7912-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddocks, S. E., and P. C. F. Oyston. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609-3623. [DOI] [PubMed] [Google Scholar]
- 38.Madhusudan, S., A. Paukner, Y. Klingen, and K. Schnetz. 2005. Independent regulation of H-NS mediated silencing of the bgl operon at two levels: upstream by BglJ and LeuO and downstream by DnaKJ. Microbiology 151:3349-3359. [DOI] [PubMed] [Google Scholar]
- 39.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY.
- 41.Moorthy, S., and P. I. Watnick. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller, C. M., G. Schneider, U. Dobrindt, L. Emody, J. Hacker, and B. E. Uhlin. 2010. Differential effects and interactions of endogenous and horizontally acquired H-NS-like proteins in pathogenic Escherichia coli. Mol. Microbiol. 75:280-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagarajavel, V., S. Madhusudan, S. Dole, A. R. Rahmouni, and K. Schnetz. 2007. Repression by binding of H-NS within the transcription unit. J. Biol. Chem. 282:23622-23630. [DOI] [PubMed] [Google Scholar]
- 44.Navarre, W. W., M. McClelland, S. J. Libby, and F. C. Fang. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456-1471. [DOI] [PubMed] [Google Scholar]
- 45.Noom, M. C., W. W. Navarre, T. Oshima, G. J. L. Wuite, and R. T. Dame. 2007. H-NS promotes looped domain formation in the bacterial chromosome. Curr. Biol. 17:R913-R914. [DOI] [PubMed] [Google Scholar]
- 46.Radde, N., J. Gebert, U. Faigle, R. Schrader, and K. Schnetz. 2008. Modeling feedback loops in the H-NS mediated regulation of the Escherichia coli bgl operon. J. Theor. Biol. 250:298-306. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds, A. E., J. Felton, and A. Wright. 1981. Insertion of DNA activates the cryptic bgl operon of E. coli K12. Nature 293:625-629. [DOI] [PubMed] [Google Scholar]
- 48.Reynolds, A. E., S. Mahadevan, S. F. J. LeGrice, and A. Wright. 1986. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J. Mol. Biol. 191:85-95. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Sankar, S. T., G. Neelakanta, V. Sangal, G. Plum, M. Achtman, and K. Schnetz. 2009. Fate of the H-NS repressed bgl operon in evolution of Escherichia coli. PLoS Genet. 5:journal.pgen.1000405. [DOI] [PMC free article] [PubMed]
- 51.Scharf, B. E. 2010. Summary of useful methods for two-component system research. Curr. Opin. Microbiol. 13:246-252. [DOI] [PubMed] [Google Scholar]
- 52.Schnetz, K., and J. C. Wang. 1996. Silencing of Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 24:2422-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimada, T., K. Yamamoto, and A. Ishihama. 2009. Involvement of leucine-response transcription factor LeuO in regulation of the genes for sulfa-drug efflux. J. Bacteriol. 191:4562-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoebel, D. M., A. Free, and C. J. Dorman. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533-2545. [DOI] [PubMed] [Google Scholar]
- 55.Stratmann, T., S. Madhusudan, and K. Schnetz. 2008. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 190:926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sturny, R., K. Cam, C. Gutierrez, and A. Conter. 2003. NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites. J. Bacteriol. 185:4298-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018-1024. [DOI] [PubMed] [Google Scholar]
- 58.Ueguchi, C., T. Ohta, C. Seto, T. Suzuki, and T. Mizuno. 1998. The leuO gene-product has a latent ability to relieve the bgl silencing in Escherichia coli. J. Bacteriol. 180:190-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wehland, M., and F. Bernhard. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesi in enteric bacteria. J. Biol. Chem. 275:7013-7020. [DOI] [PubMed] [Google Scholar]
- 60.Westra, E. R., U. Pul, N. Heidrich, M. M. Jore, M. Lundgren, T. Stratmann, R. Wurm, A. Raine, M. Mescher, H. L. Van, M. Mastop, E. G. Wagner, K. Schnetz, O. J. Van Der, R. Wagner, and S. J. Brouns. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77:1380-1393. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, G. G., K. Y. K. Young, G. J. Edlin, and W. Konigsberg. 1979. High-frequency generalised transduction by bacteriophage T4. Nature 280:80-82. [DOI] [PubMed] [Google Scholar]
- 62.Winter, S. E., M. G. Winter, P. Thiennimitr, V. A. Gerriets, S. P. Nuccio, H. Russmann, and A. J. Baumler. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol. Microbiol. 74:175-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf, T., W. Janzen, C. Blum, and K. Schnetz. 2006. Differential dependence of StpA on H-NS in auto-regulation of stpA and in regulation of bgl. J. Bacteriol. 188:6728-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, A., S. Rimsky, M. E. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acids dynamics. EMBO J. 15:1340-1349. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.