Abstract
Salmonellosis caused by Salmonella enterica serovar Newport is a major global public health concern, particularly because S. Newport isolates that are resistant to multiple drugs (MDR), including third-generation cephalosporins (MDR-AmpC phenotype), have been commonly isolated from food animals. We analyzed 384 S. Newport isolates from various sources by a multilocus sequence typing (MLST) scheme to study the evolution and population structure of the serovar. These were compared to the population structure of S. enterica serovars Enteritidis, Kentucky, Paratyphi B, and Typhimurium. Our S. Newport collection fell into three lineages, Newport-I, Newport-II, and Newport-III, each of which contained multiple sequence types (STs). Newport-I has only a few STs, unlike Newport-II or Newport-III, and has possibly emerged recently. Newport-I is more prevalent among humans in Europe than in North America, whereas Newport-II is preferentially associated with animals. Two STs of Newport-II encompassed all MDR-AmpC isolates, suggesting recent global spread after the acquisition of the blaCMY-2 gene. In contrast, most Newport-III isolates were from humans in North America and were pansusceptible to antibiotics. Newport was intermediate in population structure to the other serovars, which varied from a single monophyletic lineage in S. Enteritidis or S. Typhimurium to four discrete lineages within S. Paratyphi B. Both mutation and homologous recombination are responsible for diversification within each of these lineages, but the relative frequencies differed with the lineage. We conclude that serovars of S. enterica provide a variety of different population structures.
Salmonellosis is a major global cause of diarrheal and extraintestinal disease in humans and animals (66). Salmonella enterica subspecies enterica (referred to herein as S. enterica) has been subdivided serologically into >1,500 serovars (35), but we focus on S. enterica serovar Newport (S. Newport) here because over the last decade it has been a very common cause of human salmonellosis in both the United States and Europe (13, 16, 27). Furthermore, multidrug-resistant S. Newport isolates that are also resistant to extended-spectrum cephalosporins (MDR-AmpC) have now been reported from several countries (3, 24, 36) and are a serious problem among both food animals and humans (17, 25, 36, 48, 67). MDR-AmpC isolates are resistant to β-lactams, including third-generation cephalosporins, aminoglycosides, tetracyclines, sulfonamides and chloramphenicol (12, 36). Resistance to β-lactams is caused by plasmids carrying the ampC gene blaCMY-2, which encodes the CMY-2 β-lactamase (11, 65).
Most of our current understanding of the population structure of S. enterica relies on a series of seminal publications from R. K. Selander's group in the 1990s. These publications showed that some serovars consisted of monophyletic groups—so-called clonal groupings—but many other serovars confounded isolates from multiple lineages and were therefore polyphyletic (5, 51, 55, 56). More recent studies have indicated that genetic diversity within S. enterica reflects considerable homologous recombination in addition to mutation (7, 28, 47). As a result, the sequence diversity of S. enterica resembles a starburst radial expansion and lacks phylogenetic information (28). Individual monophyletic lineages, some of which equate to a serovar, are arranged at the tips of the starburst, and their evolutionary histories are only beginning to be elucidated (40, 45).
S. Newport is polyphyletic, according to multilocus enzyme electrophoresis (MLEE) (5), and the same conclusion can be drawn from multilocus sequence typing (MLST) studies (39, 59, 62). Two distinct lineages in S. Newport were reported to be differentially associated with humans and domesticated animals (1, 5). In contrast, microarray analysis has indicated that S. Newport is monophyletic (49). However, these conclusions are based on relatively small numbers of isolates, predominantly from North America, and information on isolates from Europe, where S. Newport is also common, is lacking.
Here, we analyzed 384 S. Newport isolates from Europe, North America, and elsewhere in order to study the evolution and population structure of the serovar by MLST (62). These data were compared with MLST data from four other serovars, namely, S. enterica serovar Enteritidis (including the avian-adapted variants S. enterica serovar Gallinarum [61] and S. Gallinarum var. Pullorum), S. enterica serovar Kentucky, S. enterica serovar Paratyphi B (including the d-tartrate-positive variant S. Paratyphi B var. Java), and S. enterica serovar Typhimurium.
MATERIALS AND METHODS
Bacterial isolates.
S. Newport strains were isolated in France (52 isolates), Germany (70), the United States (224), and other countries (35) (Table 1). Three isolates that lacked geographic information were also analyzed. The strains were isolated between 1918 and 2005 from humans, cattle, swine, reptiles, food, and other sources. We also analyzed reference S. Newport strains from the SARB Salmonella reference collection (SARB36, SARB37, and SARB38) (6) that were obtained from Fidelma Boyd, University College Cork, Cork, Ireland. MLST information had previously been published for 84 of the 384 isolates (39, 62), and MLST information was obtained from the Salmonella MLST website (http://mlst.ucc.ie/dbs/Senterica) for 9 others in mid-2008. MLST data for serovars Enteritidis, Kentucky, Paratyphi B, and Typhimurium were also downloaded from the Salmonella MLST website at the same time (see Table S1 in the supplemental material).
TABLE 1.
Sources of S. Newport isolates
| Yr of isolation (no. of isolates) | Host | No. |
||
|---|---|---|---|---|
| Europe | North America | Othera | ||
| 1940-1959 (33)b | Human | 6 | 4 | 10 |
| Rat | 1 | |||
| Reptile | 7 | |||
| Swine | 1 | 1 | ||
| Unknown | 3 | |||
| 1960-1979 (117) | Chicken | 1 | ||
| Frog legs | 3 | |||
| Equine | 1 | 1 | ||
| Human | 30 | 77 | 2 | |
| Lion | 1 | |||
| Swine | 1 | |||
| 1980-2005 (226) | Bovine | 20 | ||
| Chicken | 2 | 10 | ||
| Food/feed | 4 | 9 | ||
| Human | 60 | 75 | ||
| Reptile | 20 | 1 | ||
| Swine | 13 | |||
| Turkey | 9 | |||
| Unknown | 3 | |||
| Unknown (8) | Human | 2 | 2 | |
| Swine | 1 | |||
| Unknown | 3 | |||
| Total | 132 | 225 | 27 | |
Isolates from Africa, Asia, South America, or an unknown country.
One strain was isolated in 1918.
Antimicrobial susceptibility typing.
Isolates from the United States were screened using the Clinical and Laboratory Standards Institute (CLSI) protocol (14, 15) by the broth microdilution method as described previously (39). Other isolates were tested for antibiotic resistance following the same protocol, except that amikacin, cephalothin, cefoxitin, and ceftriaxone were tested by the CLSI disc diffusion method.
MLST.
Genomic DNA was extracted using the Jetflex Genomic DNA purification kit (Genomed) from liquid cultures grown overnight at 37°C in Luria-Bertani (LB) broth. Fragments of seven housekeeping genes, (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) were amplified and sequenced as described previously (44). The sequences were assembled and trimmed using Bionumerics 4.5 (Applied Maths). Alleles and sequence type (ST) numbers were assigned by submitting the sequences and strain information to the Salmonella MLST website.
Phylogenetic and evolutionary analyses.
The nucleotide sequence alignments for all gene fragments from unique STs were used to infer clonal relationships with ClonalFrame (19) from the 50% consensus of 10 runs, each with 100,000 iterations following a burn-in phase of 100,000 iterations. A concatenated sequence was also generated from each unique ST. The concatenated sequence alignment was analyzed by the Neighbor-net algorithm (9) that is implemented in SplitsTree 4.0 (42), using the best-fit substitution model chosen by Modeltest 3.7 (GTR+I+G) (50). Recombination within the concatenated sequences was tested using Reticulate (43) and the pairwise homoplasy index (Φw) test of recombination (8).
A minimal-spanning tree (MSTREE) was generated from the allelic profiles of the isolates with Bionumerics 4.5 in order identify ST complexes. These were then subjected to the test described by Feil et al. (33) in order to assess the relative roles of recombination and mutation in the evolution of each group. ClonalFrame was also used to assess recombination and mutation frequencies within each group and the recombination flux between groups, as previously described (18). To do this, we extracted from the ClonalFrame output the genetic fragments for which the probability of recombination was above 95%. Each such putatively imported fragment was then compared to the corresponding region from all other STs. If no match with up to two differences was found, the origin of the import was designated “external.” If multiple singleton STs or STs from independent eBurstGroups met these criteria, the origin was designated “ambiguous.” Otherwise, the origin was attributed to the matching eBurstGroup or ST.
Characteristics of housekeeping genes.
The mean frequencies of nonsynonymous substitutions per nonsynonymous site (dN) and synonymous substitutions per synonymous site (dS) were computed for each gene fragment from all serovars using DnaSP version 4.0 (53). The average pairwise nucleotide diversity per site (π) was calculated using MEGA version 4.0 (60) with Jukes-Cantor correction for aligned sequences from each gene, as well as the concatenated sequence alignments in each group.
RESULTS
Population structures of S. Newport and other serovars.
Fourteen STs based on the combination of alleles at seven housekeeping gene fragments had previously been described within S. Newport (39, 62). We identified 35 additional STs, for a total of 49, by examining a total of 384 isolates from a greater diversity of sources (see Table S2 in the supplemental material). We then used ClonalFrame on sequence alignments of gene fragments for one representative from each of these 49 STs plus 61 STs from other serovars in order to infer their phylogenetic relatedness. ClonalFrame identifies regions that are likely to have arisen by homologous recombination and accounts for them when reconstructing the clonal genealogy. The lengths of the branches in the evolutionary tree are measured in coalescent time units, which are equal to the (unknown) effective population size multiplied by the average duration of a generation. Clusters of STs that diverged at <0.17 coalescent units and contained at least 3 STs were considered to represent natural groupings and were assigned group designations. We also assigned group designations to other STs containing at least 10 isolates. STs that did not meet these criteria are referred to by their ST designations and are considered to represent singletons.
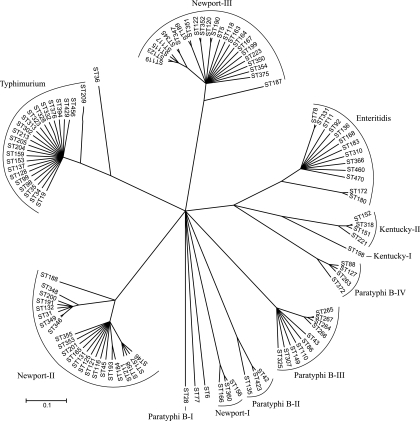
According to the ClonalFrame analysis, S. Newport contains three groups, which we designated Newport-I, Newport-II, and Newport-III (Fig. 1). Newport-I contained only three STs and 8% of the isolates. The Newport-II and Newport-III lineages were more diverse and more frequent, containing 58% of the isolates in 23 STs and 34% in 23 STs, respectively (Table 2; see Table S2 in the supplemental material).
FIG. 1.
A 50% consensus tree based on 10 independent runs of ClonalFrame 1.1. The scale is measured in coalescent time units.
TABLE 2.
Groups identified using ClonalFrame for each S. enterica serovar
| Serotype | No. of isolates | ClonalFrame group | No. of STs | ST(s) |
|---|---|---|---|---|
| Enteritidisa | 140 | Enteritidis | 13 | 11, 470, 183, 366, 78, 310, 168, 136, 460, 92, 331, 180, 172 |
| Enteritidis | 2 | Singletons | 2 | 6, 77 |
| Kentucky | 13 | Kentucky-I | 1 | 198 |
| Kentucky | 159 | Kentucky-II | 4 | 151,152, 221, 318 |
| Newport | 32 | Newport-I | 3 | 156, 166, 360 |
| Newport | 222 | Newport-II | 23 | 31, 132, 348, 188, 191, 200, 346, 349, 193, 45, 116, 121, 125, 131, 165, 353, 355, 46, 157, 211, 158, 201, 184 |
| Newport | 130 | Newport-III | 23 | 118, 189, 122, 199, 164, 163, 345, 351, 120, 190, 223, 167, 5, 187, 347, 352, 354, 115, 119, 117, 375, 123, 350 |
| Paratyphi Bb | 16 | Paratyphi B-I | 1 | 28 |
| Paratyphi B | 5 | Paratyphi B-II | 3 | 42, 423, 135 |
| Paratyphi B | 29 | Paratyphi B-III | 10 | 86, 43, 267, 266, 265, 264, 149, 307, 110, 325 |
| Paratyphi B | 16 | Paratyphi B-IV | 4 | 88, 127, 263, 372 |
| Typhimurium | 362 | Typhimurium | 22 | 19, 128, 376, 209, 205, 204, 159, 137, 429, 313, 35, 99, 456, 153, 213, 302, 98, 323, 332, 328, 34, 394 |
| Typhimurium | 2 | Singleton | 1 | 36 |
| Total | 1128 | 110 |
Including S. Gallinarum and S. Gallinarum var. Pullorum.
Serovar Paratyphi B also included S. Paratyphi B var. Java.
The other serovars contained highly variable numbers of groups (Table 2). Sixty-six S. Paratyphi B isolates yielded 18 STs in four distinct groups. In contrast, 172 S. Kentucky isolates contained only five STs within two groups, and 140/142 S. Enteritidis and 362/364 S. Typhimurium isolates clustered into a single serovar-specific group containing 13 and 22 STs, respectively.
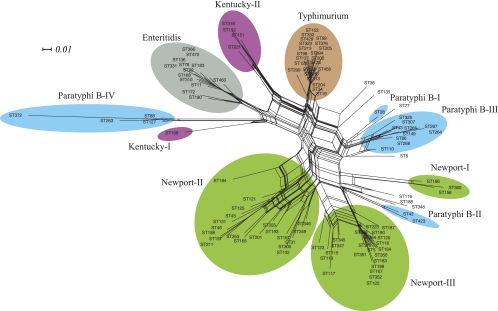
Similar group assignments were obtained when the concatenated sequences were examined using Neighbor-net (Fig. 2), with minor exceptions. Neighbor-net marks conflicting signals in nucleotide sequences that arise by recombination or recurrent mutations (homoplasies) as parallel paths in a phylogenetic network (9). Most STs from S. Enteritidis and S. Typhimurium radiated from a single point, according to Neighbor-net, suggesting that diversity in these groups was largely due to mutations. However, parallelograms were the rule for the other groups, indicating substantial recombination within each of them. Possibly, the fact that three STs from Newport-II, one from Paratyphi B-II, and one from S. Enteritidis were treated as singletons by Neighbor-net also reflects homologous recombination.
FIG. 2.
Neighbor-net on concatenated sequence alignment. The scale represents phylogenetic distances between the STs using a GTR+I+G substitution model with parameters estimated using Modeltest 3.7.
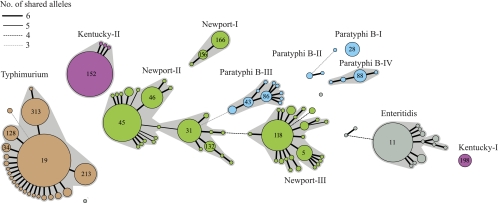
Bacterial groups have been defined in multiple bacterial species (26, 29, 31, 41) by the eBurst population genetics algorithm, which calculates distances based on the number of shared alleles. We used an allele-based approach to identify the group structure among our isolates, namely, an MSTREE that is based on principles similar to those for eBurst (Fig. 3). Groups were defined as clusters of at least three STs with internal links of 5/7 identical alleles between pairs of STs. As mentioned above, STs with at least 10 isolates were also assigned group designations. The overall compositions of the groups were generally consistent with the sequence-based groupings according to ClonalFrame (Fig. 1). However, at the allelic level, the ClonalFrame group Newport-II seemed to consist of two subgroups, which were connected via an intermediate ST that shared 5/7 alleles (double locus variant [DLV]) with STs in both of the subgroups (Fig. 3). Furthermore, ST172 plus ST180 are separated from the S. Enteritidis group within the MSTREE because they differ by three or more alleles, as are ST135 from Paratyphi B-II and ST350 from Newport-III.
FIG. 3.
An MSTREE from the allelic profiles of isolates. Numbers in circles are ST designations.
An MSTREE shows one of multiple possible networks. We therefore examined the network structure by visualizing cross-links between pairs of STs that shared three to six identical alleles (see Fig. S1 in the supplemental material). Such cross-links were observed within each group and may indicate alternative paths of evolution. We also observed cross-links between Newport-II and Newport-III, as well as between ST110 of Paratyphi B-III and three STs (ST31, ST191, and ST346) of Newport-II. These observations indicate that homologous recombination has occurred on multiple occasions between groups within S. enterica and supports the similar conclusions from the Neighbor-net analyses.
Thus, except for minor discrepancies, both sequence and allelic similarities indicate that three groups exist within S. Newport, four within S. Paratyphi B, two within S. Kentucky, and one each within S. Typhimurium and S. Enteritidis. Additional exceptional isolates fall outside these groups. Nucleotide variation within these isolates is largely synonymous (see Table S3 in the supplemental material), indicating the predominance of purifying selection. Furthermore, the degree of sequence variation is moderate in concatenated sequences from each of these groups (π < 0.006) (see Table S4 in the supplemental material).
Comparison of Newport groupings with different methodologies.
According to MLEE, almost all of 105 Newport isolates from the Americas fell into one of two clusters, designated newport I and II (5). We tested three of these isolates from a reference strain collection (6) with our MLST scheme and were therefore able to show that the MLEE newport I and newport II groups correspond to Newport-III and Newport-II from this study, respectively (Table 3). We also examined the two currently available genomes of Newport strains, isolates SL254 (accession no. CP001113) and SL317 (accession no. NC_011080). MLST genes identify SL254 as a member of Newport-II (ST45) and SL317 as a member of Newport-III (ST5). The fimA, mdh, and manB alleles from a simpler MLST scheme (1) were also extracted from the genomes, showing that SL254 corresponds to ST11 in Newport A and SL317 corresponds to ST78 in Newport B within that scheme (Table 3).
TABLE 3.
Correlation of Newport group designations between different studies
| Reference strain | Group designation |
Reference | |
|---|---|---|---|
| Other studies | This study | ||
| SARB36 (Np8) | MLEE newport I | ST5 (Newport-III) | 5 |
| SARB37 (Np11) | MLEE newport II | ST31 (Newport-II) | 5 |
| SARB38 (Np15) | MLEE newport II | ST46 (Newport-II) | 5 |
| SL254 (ST11) | MLST Newport A | ST45 (Newport-II) | 1 |
| SL317 (ST78) | MLST Newport B | ST5 (Newport III) | 1 |
Source of diversity within each group.
We attempted to determine whether the concatenated sequence alignments contained traces of recombination by measuring compatibility scores on one concatenated sequence per ST with Reticulate (43) and the probability of a null hypothesis of recombination with the Φw test (8). These tests were not possible for most of the groups because they do not contain enough informative sites and could only be evaluated for Newport-II and Newport-III, Enteritidis, and Paratyphi B-III (see Table S5 in the supplemental material).
Intermediate compatibility scores and low Φw probabilities were observed for Newport-II and Newport-III, which suggests a history of recombination in agreement with the Neighbor-net analyses. High compatibility scores, indicating a lack of recombination, were observed for Enteritidis and Paratyphi B-III, as were high Φw probabilities (see Table S5 in the supplemental material). Similarly, recombination was rare within Enteritidis according to the Neighbor-net analysis, but unlike these results, the Neighbor-net analysis indicated intermediate levels of recombination for Paratyphi B-III.
The relative frequencies of recombination and mutation were calculated within each group using both ClonalFrame (19) and the method of Feil et al. (33). For the latter approach, we examined each sequential step of the MSTREE, beginning with the predicted founder of each group. At each step, allelic changes involving at least 3 nucleotides were attributed to recombination and allelic changes of 1 or 2 nucleotides to mutation. (This cutoff was based on the bimodal patterns of mismatch frequency distributions for each gene, in some of which pairs of sequences were found with two nucleotide differences but never with three [data not shown].) However, alleles that differed by only 1 or 2 nucleotides were also scored as recombinants if they were present in STs outside the local lineage (30, 32, 33). All of the data from the MLST website in July 2008 were used for scoring these events. The founder of each group was considered to be the ST with the highest number of single-locus variants or, where two STs had equal numbers of single-locus variants, the ST with the highest number of isolates. These data were then used to calculate the ratios of recombination to mutation events (R/M) per allele and per site, as previously described (30, 32, 33).
ClonalFrame has a tendency to underestimate recombination relative to mutation (19), whereas the method described above would score repeat mutations (homoplasies) as recombination events and is therefore likely to have the opposite bias. However, the results from the two approaches showed similar trends, except in two lineages ( Table 4). Recombination has been more frequent than mutation within Kentucky-II, Newport-II, Paratyphi B-II, Paratyphi B-III, and Paratyphi B-IV, whereas mutation was more frequent within Enteritidis and Typhimurium (Table 4). Newport-I and Newport-III were the two exceptions where the R/M per allele was <1 with ClonalFrame and >1 by the method of Feil et al. (33). When expressed as R/M per site, recombination introduced more diversity than mutation in all groups according to the method of Feil et al. and in all groups except Newport-I and Typhimurium according to ClonalFrame (Table 4). These observations indicate that both ClonalFrame and the method of Feil et al. are more sensitive indicators of recombination within organisms with limited diversity than are the algorithms used by compatibility scores, or the Φw test, which did not detect extensive recombination in all of the groups.
TABLE 4.
Estimates of relative frequencies of recombination and mutation events for each group
| Groupa | ClonalFrame |
Feil et al. (33) |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of mutation events | No. of recombination events | R/M per allele | R/M per site | No. of mutation events | No. of recombination events | R/M per allele | R/M per site | |
| Enteritidis | 11 | 5 | 0.45 | 1.36 | 10 | 4 | 0.40 | 1.60 |
| Kentucky-II | 1 | 3 | 3.00 | 19.00 | 1 | 2 | 2.00 | 14.00 |
| Newport-I | 2 | 0 | 0 | 0 | 0 | 2 | >2.00 | >2.00 |
| Newport-II | 12 | 17 | 1.42 | 7.08 | 7 | 20 | 2.86 | 8.14 |
| Newport-III | 18 | 13 | 0.72 | 2.56 | 9 | 16 | 1.78 | 4.11 |
| Paratyphi B-II | 4 | 4 | 1.00 | 4.25 | 1 | 4 | 4.00 | 5.00 |
| Paratyphi B-III | 3 | 8 | 2.67 | 8.00 | 2 | 7 | 3.50 | 13.00 |
| Paratyphi B-IV | 2 | 3 | 1.50 | 17.00 | 1 | 2 | 2.00 | 34.00 |
| Typhimurium | 16 | 4 | 0.25 | 0.81 | 16 | 9 | 0.56 | 1.69 |
Groups Kentucky-I and Paratyphi B-I consist of only a single ST each and therefore were excluded.
ClonalFrame was also used to infer recombination flux between the groups (Table 5), as previously described (18). For each branch of a reconstructed genealogy, ClonalFrame identifies fragments that are likely to have been imported, but it does not search for the origin of these putative recombination events. It is possible, however, to attribute an origin to each import, as described in Materials and Methods. Many imports had ambiguous origins (more than one of the lineages studied here could be the origin) or external origins (none of the lineages under study could be the origin). Nevertheless, the origin of most imports in Newport-II was unambiguously attributed to Newport-III (11 out of 17 events) and vice versa (7 out of 13 events), which may indicate that extensive recombination is occurring or has recently occurred between the two groups. These potential exchanges are also suggested by extensive cross-linking in the MSTREE (see Fig. S1 in the supplemental material).
TABLE 5.
Recombination flux between different groups
| Group | No. of imports originating from: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| En | Kn-II | Np-I | Np-II | Np-III | Pb-II | Pb-III | Pb-IV | Tm | Ambiguous/external | |
| Enteritidis (En) | 1 | 1 | 3 | |||||||
| Kentucky-II (Kn-II) | 2 | 1 | ||||||||
| Newport-I (Np-I) | ||||||||||
| Newport-II (Np-II) | 2 | 1 | 11 | 1 | 2 | |||||
| Newport-III (Np-III) | 7 | 6 | ||||||||
| Paratyphi B-II (Pb-II) | 1 | 3 | ||||||||
| Paratyphi B-III (Pb-III) | 1 | 7 | ||||||||
| Paratyphi B-IV (Pb-IV) | 1 | 2 | ||||||||
| Typhimurium (Tm) | 1 | 3 | ||||||||
Associations of the S. Newport groups with hosts.
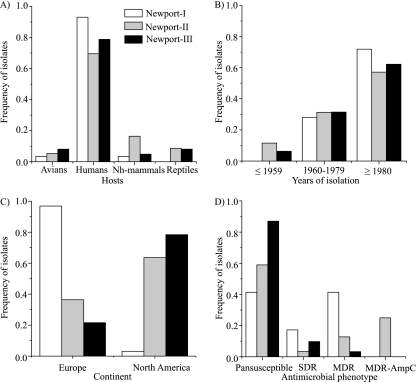
Prior, less extensive analyses based on MLEE and MLST have suggested a differential host association of Newport groups with humans and swine (5) or cattle (1). Our results support a significant differential association with hosts for the three S. Newport groups (Fig. 4A; see Table S6a in the supplemental material). Seventy-five percent of our isolates were from humans, resulting in a null hypothesis that approximately 75% of the isolates of each group within S. Newport should have been isolated from humans. The frequencies that were isolated from humans were slightly lower for Newport-II (144/207; 70%) and slightly higher for Newport-III (97/123; 79%) (Fig. 4A). In contrast, 27/29 Newport-I isolates (93%) were from humans and only 2 isolates were from avian or nonhuman mammals. A second striking feature of the results was that 16% of the Newport-II isolates were from nonhuman mammals versus much lower frequencies for Newport-I (3%) and Newport-III (5%) (Fig. 4A). As a result, 34/41 (83%) isolates from nonhuman mammals were in Newport-II. Newport-II also contained 64% of the reptile isolates. The remaining (36%) reptile isolates were grouped within Newport-III. We conclude that all three groups can readily infect humans but that Newport-I seems to preferentially infect humans and S. Newport isolates from nonhuman mammals or reptiles usually belong to Newport-II. Our data contradict previous conclusions from MLEE and MLST based on smaller samples from the Americas that suggested a strong preferential association with humans for Newport-III (1, 5). Instead, the relative frequency of all Newport-III isolates from humans was only slightly elevated, and it was isolated from all other sources at appreciable frequencies, as well.
FIG. 4.
Frequencies of isolates among Newport lineages from different hosts (excluding 25 isolates from food, feed, or other, unknown sources) (A), dates (excluding 8 isolates with no information) (B), continents (excluding 27 isolates from other geographic locations or lacking information) (C), and antimicrobial resistance phenotypes (excluding 19 isolates that were not tested and one isolate that was not assigned to any of the four phenotypic categories) (D).
Lack of a temporal association with a Newport group.
With the exception of one S. Newport human isolate from France collected in 1918, all others were isolated between 1940 and 2005. Over this period, there was no significant association between the S. Newport groups and dates of isolation (Fig. 4B; see Table S6b in the supplemental material). We note, however, that Newport-I, which is genetically quite homogeneous, may be of recent origin because all 32 strains were isolated since the 1960s (Fig. 4B).
Geographic association with S. Newport groups.
There was a very clear and significant association between the S. Newport group and the continent of isolation (see Table S6c in the supplemental material). Thirty-one of 32 Newport-I isolates were from Europe, whereas most Newport-II and Newport-III isolates were from North America (Fig. 4C). However, 5/7 European isolates in ST156 of Newport-I were isolated from tourists returning from Egypt and were probably acquired in Egypt.
Antimicrobial susceptibility patterns and association with the S. Newport groups.
We determined the susceptibilities of 365 S. Newport isolates to 16 antimicrobials. Antibiotic resistance fell into 46 different combinations of susceptibility or resistance to individual antimicrobials (see Table S7 in the supplemental material), which were simplified into three phenotype categories: pansusceptible, resistance to any single drug (SDR), and resistance to two or more drugs belonging to at least two different classes (MDR). Two hundred forty-three isolates (67%) were pansusceptible, whereas 122 were SDR or MDR, most commonly to sulfamethoxazole (27%), streptomycin (26%), tetracycline (25%), and/or ampicillin (24%) (Table 6; see Table S7 in the supplemental material).
TABLE 6.
Antimicrobial resistance among S. Newport isolates from different host types
| Antimicrobial agent | Resistance breakpointa | No. of resistant isolates (no. intermediate resistant) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Human (n = 258) | Bovine (n = 20) | Swine (n = 17) | Chicken (n = 13) | Turkey (n = 9) | Reptile (n = 27) | Otherb (n = 21) | Total (n = 365) | ||
| Ampicillin | ≥32 | 56 (1) | 16 (0) | 7 (0) | 4 (0) | 1 (0) | 0 (0) | 4 (0) | 88 (1) |
| Amoxicillin-clavulanic acid | ≥32/16 | 27 (2) | 16 (0) | 7 (0) | 3 (0) | 0 (0) | 0 (0) | 4 (0) | 57 (2) |
| Ceftriaxone | ≥8 | 27 (0) | 16 (0) | 6 (0) | 3 (0) | 0 (0) | 0 (0) | 4 (0) | 56 (0) |
| Cephalothin | ≥32 | 37 (1) | 16 (0) | 7 (0) | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 68 (1) |
| Chloramphenicol | ≥32 | 39 (1) | 17 (0) | 6 (0) | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 70 (1) |
| Trimethoprim-sulfamethoxazole | ≥4/76 | 17 (0) | 3 (0) | 2 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (0) | 25 (0) |
| Cefoxitin | ≥32 | 27 (0) | 16 (0) | 7 (0) | 3 (0) | 0 (0) | 0 (0) | 4 (0) | 57 (0) |
| Gentamicin | ≥16 | 6 (0) | 4 (0) | 2(0) | 1 (0) | 2 (0) | 0 (0) | 0 (0) | 15 (0) |
| Kanamycin | ≥64 | 13 (0) | 8 (0) | 3 (0) | 2 (0) | 1 (0) | 0 (0) | 0 (0) | 27 (0) |
| Nalidixic acid | ≥32 | 6 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 7 (0) |
| Sulfamethoxazole | ≥512 | 63 (0) | 18 (0) | 8 (0) | 4 (0) | 2 (0) | 1 (0) | 4 (0) | 100 (0) |
| Streptomycin | ≥64 | 59 (0) | 18 (0) | 8 (0) | 4 (0) | 1 (0) | 2 (0) | 4 (0) | 96 (0) |
| Tetracycline | ≥16 | 56 (1) | 19 (0) | 8 (0) | 5 (0) | 0 (0) | 0 (0) | 5 (0) | 93 (1) |
| Ceftiofur | ≥8 | 27 (0) | 16 (0) | 7 (0) | 3 (0) | 0 (0) | 0 (0) | 4 (0) | 57 (0) |
MICs in μg/ml. The MICs (μg/ml) for intermediate resistance were as follows: ampicillin, 16; amoxicillin-clavulanic acid, 16/8; ceftriaxone, 2; cephalothin, 16; chloramphenicol, 16; cefoxitin, 16; gentamicin, 8; kanamycin, 32; streptomycin, 16; tetracycline, 8; and ceftiofur, 4. All of the isolates were sensitive to amikacin and ciprofloxacin (resistance breakpoints, ≥32 and ≥4 μg/ml, respectively).
Isolates from animal feed, food, fertilizer, frog legs, horse, lion, meat, and rat.
The three groups showed striking and significant differences in antimicrobial resistance (Fig. 4D; see Table S6d in the supplemental material). Eighty-seven percent (106/122) of Newport-III isolates were pansusceptible versus only 41% (12/29) of Newport-I and 59% (125/213) of Newport-II. All 56 MDR-AmpC isolates belonged to Newport-II, within which they represented ∼26% of all isolates. Only 11% of Newport-II isolates were MDR, and 4% were SDR.
DISCUSSION
S. Newport is a polyphyletic serovar.
We have tested a large number of S. Newport strains (384 isolates), isolated since 1940 and from a variety of geographic sources, including both Europe and North America. MLST data from these strains demonstrate that S. Newport consists of three distinct lineages, which we designated Newport-I, Newport-II, and Newport-III. The same lineages were distinguished whether the MLST data were analyzed at the sequence level or at the allelic level (Fig. 1 to 3 and Table 2). The only minor differences between these approaches were that Neighbor-net excluded three STs from Newport-II (Fig. 2) and that allelic analyses subdivided Newport-II into two linked subgroups (Fig. 3). Our results confirm prior analyses (1, 5, 39, 59, 62) that concluded that S. Newport is polyphyletic. However, those prior analyses defined only Newport-II and Newport-III, largely because they only tested S. Newport isolates from the Americas, where Newport-I is rare.
Newport-I has allelic profiles distinct from those of Newport-II or Newport-III, except that it shares hisD allele 12 and thrA allele 12 with some STs in Newport-II and purE allele 5 and thrA allele 12 with some STs in Newport-III. (These two lineages also share alleles, as indicated by the cross-linking patterns in Fig. S1 in the supplemental material). Newport-I is currently comprised of only three STs, which might indicate that it evolved recently by lateral acquisition of genes encoding antigenic determinants that are used for serotyping. Indeed, Newport-I was not found among the few strains that were isolated before the 1960s. Alternatively, Newport-I might have recently undergone an extreme evolutionary bottleneck or selective sweep that removed most nucleotide diversity from the lineage (57). It is also possible that Newport-I maintains a different host reservoir that has not been sampled by us, leading to an underrepresentation of nucleotide diversity in our study.
Association of S. Newport groups with hosts, geographic locations, and antimicrobial resistance.
In agreement with prior reports (1, 5), most S. Newport isolates from nonhuman mammals are Newport-II (Fig. 4A). However, Newport-II is commonly isolated from human infections, as well as from birds and reptiles, and does not seem to be strongly host adapted. Similarly, Newport-III was also isolated from various hosts at frequencies that reflect the numbers of isolates that were tested. In contrast, almost all Newport-I isolates were from humans, and Newport-I was isolated only at very low frequencies from birds or nonhuman mammals, suggesting human-to-human transmission. Alternatively, such a pattern might arise if the primary reservoirs for Newport-I had not been sampled, e.g., water or plants. We note that Newport-I is currently rarer than the other two lineages, and most isolates from humans are either Newport-II or Newport-III.
Both Newport-II and Newport-III are common in Europe, as well as in North America, but at somewhat different frequencies: 36% of Newport-II isolates were from Europe and 63% were from North America, whereas 78% of Newport-III isolates were from North America and only 22% were isolated in Europe (see Table S6c in the supplemental material). In contrast, Newport-I is almost entirely specific to Europe (Fig. 4C), where 97% of these strains were isolated. However, five Newport-I isolates (ST156) were likely acquired by European travelers in Egypt.
Possibly one of the most striking aspects of this investigation was the patterns of resistance to antimicrobials of the individual lineages; 87% of Newport-III isolates were pansusceptible, and antibiotic resistance does not seem to have become common within this lineage. Most (59%) MDR isolates were Newport-II, as were all MDR-AmpC isolates, which were exclusively associated with ST45 and ST116.
MDR-AmpC S. Newport first appeared in 1998 in the United States (23) and has subsequently spread extensively among cattle and humans (17, 67). Antibiotic resistance determinants in MDR-AmpC isolates are encoded on large (>150-kb) IncA/C2 plasmids, such as pSN254, which has been fully sequenced (63). The use of therapeutic or prophylactic antibiotics might have selected for an expansion of MDR-AmpC S. Newport on farms and promoted its spread among cattle. In particular, the use of ceftiofur, a third-generation cephalosporin that is licensed within the United States for use in cattle, could have selected for S. Newport carrying the blaCMY-2 plasmid. Further selection for a lineage containing this plasmid may reflect the common prophylactic use of tetracycline, because tetracycline resistance is also encoded on the same plasmid. It has been suggested that all MDR-AmpC strains have a recent global origin (2, 25, 39), and our assignment of all MDR-AmpC S. Newport isolates to two closely related STs within Newport-II is consistent with the recent clonal expansion from a single S. Newport strain. pSN254 is not conjugative, unlike other IncA/C plasmids (34), which might explain why the MDR-AmpC phenotype has apparently not spread to other STs of S. Newport. In contrast to Newport-II, MDR in ST156 within Newport-I is associated with variant K6, H, or L of Salmonella genomic island 1 (SGI1) (21, 22), which indicates that genetic rearrangements within SGI1 can occur even within a single sequence type. Similar antibiotic resistance patterns were also found in ST166 within Newport-I, raising the possibility that they also possess variants of SGI1. The molecular basis for the other antibiotic resistance patterns within S. Newport is unknown but could possibly be unraveled by using the neutral population genetic patterns revealed by MLST to investigate patterns of vertical and horizontal descent of genes encoding these resistance patterns.
Differences in R/M ratios between lineages.
S. enterica has traditionally been considered by population geneticists to be one of the most clonal bacteria (58). This tradition is reinforced by the use of serotyping for epidemiological purposes, which is often interpreted as if all isolates of a given serovar are closely related. However, it has been known since the MLEE studies of the mid-1990s that some serovars are polyphyletic, i.e., that they have multiple evolutionary origins and are unlikely to be very similar, except for the serotyping antigens (5, 51, 55, 56). Similarly, it has been known for a number of years that genetic recombination occurs within S. enterica (7, 28, 47), which negates the assumption that S. enterica is highly clonal. The results presented here illustrate both of these points and also highlight differences in diversity and polyphyletic versus monophyletic origins between individual serovars of S. enterica.
S. Typhimurium and S. Enteritidis seem to be largely monophyletic, and almost all isolates of each serovar cluster in a serovar-specific lineage (Fig. 1 to 3 and Table 2). In contrast, S. Kentucky consists of two unrelated lineages with little internal diversity, while S. Paratyphi B consists of at least four unrelated lineages. S. Newport with its three lineages thus falls between S. Kentucky and S. Paratyphi B in terms of population structure and possesses as much genetic diversity as any of the other lineages, or more (Fig. 1 to 3 and Table 2; see Table S4 in the supplemental material). These results are largely consistent with previous studies with MLEE (5, 54, 55) and MLST (59, 62), but they provide much greater detail.
As suggested above for Newport-I, a new serovar-specific lineage can arise from the acquisition of serotyping antigens by horizontal exchange. However, additional, currently unknown lineages will probably be discovered with time simply because of the sampling bias in the current data set. The STs and lineages described here represent the status of the data set in late 2008. Since then, the data set has grown to include additional STs and lineages as more isolates and isolates from other geographical regions have been investigated. For example, additional STs were discovered within the S. Typhimurium lineage among isolates from Mexico and Africa (45, 64). Similarly, S. Kentucky now (September 2010) includes isolates from three lineages plus two singleton STs. Also, due to an increasing lack of one-to-one correspondence between a serovar and an ST or lineage, we have abandoned the lineage designations presented here and have assigned numerical designations, so-called eBurstGroups that do not include the serovar name (M. Achtman, unpublished data.). These are used in Table S8 in the supplemental material, which provides a detailed list of the potential sources of recombination events. A correspondence table of lineage designations to eBurstGroups is presented in Table S9 in the supplemental material.
How does diversity accumulate within a lineage? Our data suggest that the accumulation of diversity reflects a mixture of mutation plus homologous recombination, whose relative frequencies differ with the lineage. Both mutation and recombination events were identified in the genealogies of all the lineages (Table 4), but the relative R/M frequencies per allele ranged from 0.25 to 0.56 (ClonalFrame estimate- Feil method) for Enteritidis and Typhimurium to 1.0 to 4.0 for Paratyphi B-II. Similarly, the numbers of nucleotides introduced by recombination versus mutation (R/M per site) ranged from 0.81 to 1.69 (Enteritidis and Typhimurium) to >10.0 (Kentucky-II and Paratyphi B-IV). Newport-II was intermediate between these extremes, with an R/M per allele value of 1.42 to 2.86 and an R/M per site value of 7.08 to 8.14. It was unclear whether point mutations were more frequent than recombination in Newport-I and Newport-III because the two methods yielded conflicting results. Only a subset of these lineages could be tested for the frequency of recombination with other population genetic algorithms due to the paucity of informative sites, but both additional tests that were used suggested that Enteritidis is more clonal than Newport-II and Newport-III (see Table S5 in the supplemental material). These results agree with previous studies (7, 28, 47) showing that recombination has been important in generating new alleles within S. enterica. They are also consistent with other recent analyses that have identified a range of ratios of mutation to recombination among different bacterial species (37, 38), as well as a range of different mutation rates between species (46).
How can one account for such different ratios of recombination to mutation in distinct lineages? In principle, one might expect recombination to be more common in S. enterica lineages that cause gastrointestinal diseases in a variety of hosts because of increased opportunities due to mixed infections. However, a lifestyle where little opportunity for horizontal exchange should exist is apparently not limiting for vectors of such exchange, because individual haplotypes of the host-restricted and invasive serovar Typhi have acquired a variety of plasmids and/or lysogenic prophages (40). Alternatively, different lineages might differ in their propensities for importing DNA via transduction or conjugation due to lineage-specific differences in immunity to bacteriophages, such as are due to CRISPRs (4), or patterns of susceptibility to bacteriophages, such as are encoded by certain plasmids (52). A further mechanism that could affect the frequency of homologous recombination is the existence of lineage-specific differences for defense mechanisms against genetic exchange, such as restriction-modification systems (R-M systems). Three distinct R-M systems have been identified in S. Typhimurium, and different specificities of such R-M systems exist in different serovars of S. enterica (10). Thus, a variety of mechanisms that could account for differential abilities for homologous recombination are known. However, at least some of these mechanisms seem to be specific to individual haplotypes within the lineages and are unlikely to result in the dramatic differences between entire lineages that are described here. As with antibiotic resistance, additional experiments are needed to determine whether lineage-wide patterns exist for the ability to acquire DNA by conjugation or transduction.
Lineages within S. Newport.
Having addressed the sources of diversity within a lineage, we now return to the sources of Newport-II and Newport-III, which are less distinct from each other than either is from Newport-I. Newport-II and Newport-III share multiple alleles, as revealed by cross-link analysis (see Fig. S1 in the supplemental material), suggesting that they may have arisen from a single lineage that has now differentiated. Support for this possibility is provided by the ongoing process of subdivision of Newport-II into two lineages (Fig. 3). Alternatively, these cross-links represent preferential recombination between the lineages, possibly because they share a niche. Similar to what has been proposed for Campylobacter jejuni versus Campylobacter coli (20), Newport-II and Newport-III may even merge via recombination in the future. ClonalFrame identified Newport-III as the source of imports for 11/17 recombination events in Newport-II (Table 5) and Newport-II as the source for 7/13 imports in Newport-III. However, most imported sequences are also found in multiple other S. enterica serovars that could have been the source of the imports (see Table S8 in the supplemental material).
In conclusion, three distinct lineages, Newport-I, Newport-II, and Newport-III, exist within S. Newport. These lineages are differentially associated with human isolates from Europe, animal isolates, and human isolates from North America, respectively. The MDR phenotype, especially MDR-AmpC, is associated with Newport-II, whereas most isolates in Newport-III are pansusceptible. The population structure of S. enterica varies with the serovar, and many serovars contain two or more divergent lineages. These differences partly reflect different relative ratios of recombination to mutation. S. enterica represents a pool of diverse strains that have been assigned to >1,500 serovars based on the antigenic profiles of the surface antigens (35). However, a serovar does not necessarily indicate a group of genetically identical isolates, and genes encoding surface antigens have probably been exchanged extensively between serovars, giving rise to multiple lineages. Therefore, MLST will need to be applied to a globally representative collection of S. enterica isolates in order to provide a detailed understanding of S. enterica population structure.
Supplementary Material
Acknowledgments
We thank J. Wain, Laboratory of Gastrointestinal Pathogens, Health Protection Agency, London, United Kingdom; Derek Brown, Scottish Salmonella, Shigella and Clostridium difficile Reference Laboratory, Stobhill Hospital, Glasgow, United Kingdom; and other scientists who have contributed to the Salmonella MLST database (http://mlst.ucc.ie/dbs/Senterica). We also thank A. Schroeter, M. Jaber, and D. Avsaroglu from the Federal Institute for Risk Assessment (BfR), Berlin, Germany, for their help and support in phenotypic characterization of S. Newport isolates.
This work was partly supported by grants 05/FE1/B882 (Science Foundation of Ireland), R01 GM084318-01A1 (National Institutes of Health), and BfR-45-004 (Federal Institute for Risk Assessment) and by the Med-Ved-Net (European Union-funded Network of Excellence for the Prevention and Control of Zoonoses).
Footnotes
Published ahead of print on 8 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alcaine, S. D., Y. Soyer, L. D. Warnick, W. L. Su, S. Sukhnananda, J. Richards, E. D. Fortes, P. McDonough, T. P. Root, N. B. Dumas, Y. Grohn, and M. Wiedmann. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaine, S. D., S. S. Sukhnanand, L. D. Warnick, W. L. Su, P. McGann, P. McDonough, and M. Wiedmann. 2005. Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob. Agents Chemother. 49:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlet, G., T. J. Barrett, P. Butaye, A. Cloeckaert, M. R. Mulvey, and D. G. White. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945-1954. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 5.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, A. Cravioto, and R. K. Selander. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. U. S. A. 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 7.Brown, E. W., M. K. Mammel, J. E. LeClerc, and T. A. Cebula. 2003. Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens. Proc. Natl. Acad. Sci. U. S. A. 100:15676-15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant, D., and V. Moulton. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255-265. [DOI] [PubMed] [Google Scholar]
- 10.Bullas, L. R., C. Colson, and B. Neufeld. 1980. Deoxyribonucleic acid restriction and modification systems in Salmonella: chromosomally located systems of different serotypes. J. Bacteriol. 141:275-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2002. Outbreak of Multidrug-Resistant Salmonella Newport in United States, January-April 2002. MMWR Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2008. Salmonella surveillance: annual summary, 2006. U.S. Department of Health and Human Services, Atlanta, GA.
- 14.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 3rd ed. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 15.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 16.Cobbold, R. N., D. H. Rice, M. A. Davis, T. E. Besser, and D. D. Hancock. 2006. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 228:585-591. [DOI] [PubMed] [Google Scholar]
- 17.Devasia, R. A., J. K. Varma, J. Whichard, S. Gettner, A. B. Cronquist, S. Hurd, S. Segler, K. Smith, D. Hoefer, B. Shiferaw, F. J. Angulo, and T. F. Jones. 2005. Antimicrobial use and outcomes in patients with multidrug-resistant and pansusceptible Salmonella Newport infections, 2002-2003. Microb. Drug Resist. 11:371-377. [DOI] [PubMed] [Google Scholar]
- 18.Didelot, X., M. Barker, D. Falush, and F. G. Priest. 2009. Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 32:81-90. [DOI] [PubMed] [Google Scholar]
- 19.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingle, K. E., F. M. Colles, D. Falush, and M. C. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doublet, B., K. Praud, F.-X. Weill, and A. Cloeckaert. 2009. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J. Antimicrob. Chemother. 63:282-289. [DOI] [PubMed] [Google Scholar]
- 22.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 24.Egorova, S., L. Kaftyreva, P. A. Grimont, and F. X. Weill. 2007. Prevalence and characterization of extended-spectrum cephalosporin-resistant nontyphoidal Salmonella isolates in adults in Saint Petersburg, Russia (2002-2005). Microb. Drug Resist. 13:102-107. [DOI] [PubMed] [Google Scholar]
- 25.Egorova, S., M. Timinouni, M. Demartin, S. A. Granier, J. M. Whichard, V. Sangal, L. Fabre, A. Delaune, M. Pardos, Y. Millemann, E. Espie, M. Achtman, P. A. D. Grimont, and F. X. Weill. 2008. Ceftriaxone-resistant Salmonella enterica serotype Newport, France. Emerg. Infect. Dis. 14:954-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.European Centre for Disease Prevention and Control. 2009. Enter-net quarterly Salmonella reports. European Centre for Disease Prevention and Control (ECDC). http://ecdc.europa.eu/en/publications/Publications/Forms/AllItems.aspx.
- 28.Falush, D., M. Torpdahl, X. Didelot, D. F. Conrad, D. J. Wilson, and M. Achtman. 2006. Mismatch induced speciation in Salmonella: model and data. Philos. Trans. R. Soc. Lond B Biol. Sci. 361:2045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic comparisons. Proc. Natl. Acad. Sci. U. S. A. 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contribution of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 33.Feil, E. J., J. Maynard Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fricke, W. F., T. J. Welsh, P. F. McDermott, M. K. Mammel, J. E. LeClerc, D. G. White, T. A. Cebula, and J. Ravel. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guibourdenche, M., P. Roggentin, M. Mikoleit, P. I. Fields, J. Bockemuhl, P. A. D. Grimont, and F. X. Weill. 2010. Supplement 2003-2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161:26-29. [DOI] [PubMed] [Google Scholar]
- 36.Gupta, A., J. Fontana, C. Crowe, B. Bolstroff, A. Stout, S. van Duyne, M. P. Hoekstra, J. M. Whichard, T. J. Barrett, and F. J. Angulo. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 37.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. The impact of homologous recombination on the generation of diversity in bacteria. J. Theor. Biol. 239:210-219. [DOI] [PubMed] [Google Scholar]
- 38.Hanage, W. P., B. G. Spratt, K. M. E. Turner, and C. Fraser. 2006. Modelling bacterial speciation. Philos. Trans. R. Soc. Lond B Biol. Sci. 361:2039-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harbottle, H., D. G. White, P. F. McDermott, R. D. Walker, and S. Zhao. 2006. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 44:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holt, K. E., J. Parkhill, C. J. Mazzoni, P. Roumagnac, F.-X. Weill, I. Goodhead, R. Rance, S. Baker, D. Maskell, J. Wain, C. Dolecek, M. Achtman, and G. Dougan. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honsa, E., T. Fricke, A. J. Stephens, D. Ko, F. Kong, G. L. Gilbert, F. Huygens, and P. M. Giffard. 2008. Assignment of Streptococcus agalactiae isolates to clonal complexes using a small set of single nucleotide polymorphisms. BMC Microbiol. 8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 43.Jakobsen, I. B., and S. Easteal. 1996. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biosci. 12:291-295. [DOI] [PubMed] [Google Scholar]
- 44.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 45.Kingsley, R. A., C. L. Msefula, N. R. Thomson, S. Kariuki, K. E. Holt, M. A. Gordon, D. Harris, L. Clarke, S. Whitehead, V. Sangal, K. Marsh, M. Achtman, M. E. Molyneux, M. Cormican, J. Parkhill, C. A. Maclennan, R. S. Heyderman, and G. Dougan. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19:2279-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morelli, G., X. Didelot, B. Kusecek, S. Schwarz, D. Falush, C. Bahlawane, S. Suerbaum, and M. Achtman. 2010. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 6:e1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Octavia, S., and R. Lan. 2006. Frequent recombination and low level of clonality within Salmonella enterica subspecies I. Microbiology 152:1099-1108. [DOI] [PubMed] [Google Scholar]
- 48.Poppe, C., L. Martin, A. Muckle, M. Archambault, S. McEwen, and E. Weir. 2006. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can. J. Vet. Res. 70:105-114. [PMC free article] [PubMed] [Google Scholar]
- 49.Porwollik, S., E. F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 186:5883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 51.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. Farmer III. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ridley, A. M., P. Punia, L. R. Ward, B. Rowe, and E. J. Threlfall. 1996. Plasmid characterization and pulsed-field electrophoretic analysis demonstrate that ampicillin-resistant strains of Salmonella enteritidis phage type 6a are derived from Salm. enteritidis phage type 4. J. Appl. Bacteriol. 81:613-618. [DOI] [PubMed] [Google Scholar]
- 53.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 54.Selander, R. K., P. Beltran, N. H. Smith, R. M. Barker, P. B. Crichton, D. C. Old, J. M. Musser, and T. S. Whittam. 1990. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect. Immun. 58:1891-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. T. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selander, R. K., J. Li, E. F. Boyd, F.-S. Wang, and K. Nelson. 1994. DNA sequence analysis of the genetic structure of populations of Salmonella enterica and Escherichia coli, p. 17-49. In F. G. Priest, A. Ramos-Cormenzana, and R. Tindall (ed.), Bacterial systematics and diversity. Plenum, New York, NY.
- 57.Smith, N. H., S. V. Gordon, R. Rua-Domenech, R. S. Clifton-Hadley, and R. G. Hewinson. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4:670-681. [DOI] [PubMed] [Google Scholar]
- 58.Spratt, B. G. 2004. Exploring the concept of clonality in bacteria. Methods Mol. Biol. 266:323-352. [DOI] [PubMed] [Google Scholar]
- 59.Sukhnanand, S., S. Alcaine, L. D. Warnick, W.-L. Su, J. Hof, M. P. J. Craver, P. McDonough, K. J. Boor, and M. Wiedmann. 2005. DNA sequence-based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. J. Clin. Microbiol. 43:3688-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599 [DOI] [PubMed] [Google Scholar]
- 61.Thomson, N. R., D. J. Clayton, D. Windhorst, G. Vernikos, S. Davidson, C. Churcher, M. A. Quail, M. Stevens, M. A. Jones, M. Watson, A. Barron, A. Layton, D. Pickard, R. A. Kingsley, A. Bignell, L. H. B. Clark, D. Ormond, Z. Abdellah, K. Brooks, I. Cherevach, T. Chillingworth, J. Woodward, H. Norberczak, A. Lord, C. Arrowsmith, K. Jagels, S. Moule, K. Mungall, M. Sanders, S. Whitehead, J. A. Chabalgoity, D. Maskell, T. Humphrey, M. Roberts, P. A. Barrow, G. Dougan, and J. Parkhill. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insight into evolutionary and host adaptation pathways. Genome Res. 18:1624-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torpdahl, M., M. N. Skov, D. Sandvang, and D. L. Baggesen. 2005. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J. Microbiol. Methods 63:173-184. [DOI] [PubMed] [Google Scholar]
- 63.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. LeClerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiesner, M., M. B. Zaidi, E. Calva, M. Fernandez-Mora, J. J. Clava, and C. Silva. 2009. Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotype in Mexican Salmonella enterica serovar Typhimurium strains. BMC Microbiol. 9:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.World Health Organization. 2005. Drug-resistant Salmonella. WHO, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs139/en/index.html.
- 67.Zhao, S. D., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, B. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase bla(CMY) genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.