Abstract
Acetyl phosphate (AcP) is a small-molecule metabolite that can act as a phosphoryl group donor for response regulators of two-component systems (TCSs). The serious human respiratory pathogen Streptococcus pneumoniae (pneumococcus) synthesizes AcP by the conventional pathway involving phosphotransacetylase and acetate kinase, encoded by pta and ackA, respectively. In addition, pneumococcus synthesizes copious amounts of AcP and hydrogen peroxide (H2O2) by pyruvate oxidase, which is encoded by spxB. To assess possible roles of AcP in pneumococcal TCS regulation and metabolism, we constructed strains with combinations of spxB, pta, and ackA mutations and determined their effects on ATP, AcP, and H2O2 production. Unexpectedly, ΔackA mutants were unstable and readily accumulated primary suppressor mutations in spxB or its positive regulator, spxR, thereby reducing H2O2 and AcP levels, and secondary capsule mutations in cps2E or cps2C. ΔackA ΔspxB mutants contained half the cellular amount of ATP as a ΔspxB or spxB+ strain. Acetate addition and anaerobic growth experiments suggested decreased ATP, rather than increased AcP, as a reason that ΔackA mutants accumulated spxB or spxR suppressors, although experimental manipulation of the AcP amount was limited. This finding and other considerations suggest that coping with endogenously produced H2O2 may require energy. Starting with a ΔspxB mutant, we constructed Δpta, ΔackA, and Δpta ΔackA mutants. Epistasis and microarray experiment results were consistent with a role for the SpxB-Pta-AckA pathway in expression of the regulons controlled by the WalRKSpn, CiaRHSpn, and LiaSRSpn TCSs involved in sensing cell wall status. However, AcP likely does not play a physiological role in TCS sensing in S. pneumoniae.
Streptococcus pneumoniae (pneumococcus) is a major human respiratory pathogen that causes over 1.6 million deaths worldwide annually (19, 23, 51). S. pneumoniae is a Gram-positive, ovococcal-shaped bacterium that is usually found as a diplococcus or as short chains of cells (3, 58). S. pneumoniae colonizes, often asymptomatically, the nasopharynx of 10 to 20% of healthy adults and 40 to 60% of young children (51). Colonization is the major reservoir for transmission of S. pneumoniae among humans, its major host (9, 23, 51). Colonization can also progress to serious invasive diseases, including pneumonia, otitis media (earache), meningitis, and bacteremia in susceptible individuals (7, 29, 30, 51). During colonization and invasive disease, pneumococcus encounters diverse environments and changes in temperature and availability of oxygen, metal ions, and nutrients (2, 5, 17, 18, 21, 24, 39, 47). Pneumococcus must also contend with innate and humoral immune responses (23, 51). Many genes involved in bacterial adaptation, survival, and virulence encode enzymes that mediate central metabolism and maximize energy production (44). These links are only starting to be studied systematically in S. pneumoniae, and they provide promise in identifying targets for antibiotic development to combat emerging multidrug resistance (12, 29).
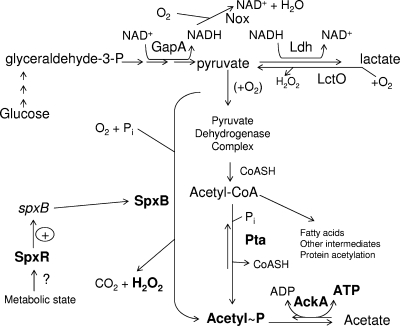
S. pneumoniae is an aerotolerant anaerobe with fermentative metabolism. It lacks a tricarboxylic acid (TCA) cycle and an electron transport chain, and its central metabolism consists primarily of glycolysis (Fig. 1) (20, 28, 48). Pneumococcus catabolizes glucose and other sugars by glycolysis to generate two molecules of ATP by substrate-level phosphorylation and produce pyruvate, which is converted to acetyl coenzyme A (CoA) and other metabolites, including acetyl phosphate (AcP) (Fig. 1) (14). AcP can be converted by acetate kinase (AckA) to ATP and acetate to generate an additional molecule of ATP (53, 54). Under aerobic conditions with glucose as the primary carbon source, pneumococcus produces mainly lactate and a small amount of acetate as fermentation products, whereas a mixture of acids and ethanol are produced from other sugars, such as galactose (56).
FIG. 1.
Model of central metabolism in S. pneumoniae growing aerobically in glucose-containing BHI broth. An abbreviated version of the Embden-Meyerhof-Parnas glycolytic pathway is shown; this pathway converts glucose to pyruvate and generates two ATP molecules. Pyruvate is converted to lactate by lactate dehydrogenase (Ldh), regenerating NAD+, and lactate can be converted back to pyruvate by lactate oxidase (LctO). NADH oxidase (Nox) also converts NADH back to NAD+. Pyruvate is converted to acetyl-CoA by the pyruvate dehydrogenase complex. Acetyl∼P (AcP) is formed from acetyl-CoA by Pta or from pyruvate by pyruvate oxidase (SpxB), which also produces H2O2. Transcription of the spxB gene is positively regulated by the SpxR CBS-HotDog domain regulator. Acetyl∼P is converted to acetate and another molecule of ATP by AckA. See the text for additional information and references.
Three metabolic enzymes lead to the synthesis of AcP in S. pneumoniae (Fig. 1). SpxB pyruvate oxidase uses molecular oxygen and inorganic phosphate to convert pyruvate directly into AcP and H2O2 (40, 42, 45, 47). SpxB is present in only a limited number of bacterial species (26, 42) and is the major source of the significant amount (≈1 mM) of endogenous H2O2 secreted in S. pneumoniae cultures (40, 42, 45, 47). Previously, we demonstrated that spxB transcription is positively regulated by the novel CBS-HotDog domain protein SpxR, which may link the SpxB amount to energy and metabolic state (42). Cellular AcP amounts are reduced by 80% in exponentially growing ΔspxB mutants, and it was inferred, but not shown directly, that cellular ATP amount may also be decreased (40). Despite this large drop in cellular AcP amount (40) and a decrease in H2O2 production of 90% (42), we detected only limited changes in global transcription patterns in a ΔspxB mutant compared to its spxB+ parent (42). This result suggested that AcP does not play a major role in pneumococcal signaling, although we could not rule out that the total cellular AcP amount remained large enough in ΔspxB mutants to mask changes in signaling or that compensatory metabolic pathways operate in ΔspxB mutants. Curiously, ΔspxB mutants treated with high (20 mM) concentrations of exogenously added H2O2 seem to have greatly reduced ATP pools and die rapidly (40, 42).
The two other pneumococcal enzymes that synthesize AcP comprise the phosphotransacetylase (Pta)-AckA pathway, which is widely distributed in other bacteria (Fig. 1) (11, 53, 54). There has been controversy about whether S. pneumoniae possesses an active pyruvate dehydrogenase under aerobic growth conditions (56). In work to be presented elsewhere, we demonstrate the presence of a functional pyruvate dehydrogenase (unpublished data). Acetyl-CoA produced by this complex is converted by Pta to AcP (Fig. 1) (reviewed in reference 53). AckA converts AcP produced by Pta or SpxB to acetate and ATP, or in the reverse direction, acetate to ADP and AcP (Fig. 1). In bacteria lacking electron transport and a TCA cycle, such as S. pneumoniae, AckA is thought to play an important role in providing additional ATP from pyruvate (53, 54). The Pta-AckA pathway has been studied extensively in Escherichia coli and Salmonella species (reviewed in references 53 and 54). In these and other bacteria, response regulators from several two-component regulatory systems (TCSs) are phosphorylated by AcP in vitro and in vivo, and it has been suggested that AcP phosphorylation links regulation of these TCSs to metabolic state (13, 15, 25, 54). In addition, acetyl-CoA from the Pta-AckA pathway serves as an acetyl group donor to numerous proteins, including RNA polymerase, thereby modulating their activities and functions (53, 54). Despite its likely importance to metabolism and signaling, the Pta-AckA pathway has not been characterized in S. pneumoniae. Moreover, precedents from other bacteria like E. coli and Salmonella species cannot be applied to pneumococcus, because those organisms possess a different central metabolism and lack the SpxB pyruvate oxidase that produces AcP.
This study provides the first characterization of the contribution of the Pta-AckA pathway to pneumococcal metabolism, specifically, ATP and AcP production, and its links to the reaction catalyzed by SpxB. By constructing strains containing combinations of mutations resulting in a defective SpxB-Pta-AckA pathway, we show that cellular ATP amounts are affected by abolishment of AcP production and lack of AckA function. Surprisingly, we found that ΔackA mutants are unstable and rapidly accumulate suppressor mutations that inactivate SpxB or SpxR. The basis for this suppression and the possible role of AcP in TCS signaling were tested by a combination of epistasis analysis, microarrays, and determinations of ATP and AcP amounts. The results were consistent with an energy-dependent process that protects S. pneumoniae from endogenously produced H2O2 and a role for the SpxB-Pta-AckA pathway in expression of regulons controlled by a specific subset of TCSs, although a physiological role for AcP in pneumococcal response regulator phosphorylation seems unlikely.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table S1 of the supplemental material. All cultures were grown statically with limited aeration in brain heart infusion broth (BHI; Becton-Dickinson [BD]) or on blood agar (BA) plates containing Trypticase soy agar II modified (BD) and 5% (vol/vol) defibrinated sheep blood (Remel) at 37°C in an atmosphere of 5% CO2. Growth was monitored directly by measuring the optical density at 620 nm (OD620) using a Spectronic 20 Genesys spectrophotometer. Overnight cultures of strains were started by inoculating frozen bacterial stocks into 17-mm-diameter polystyrene plastic tubes containing 5 ml of BHI broth and then performing 100-fold serial dilutions over five tubes. Overnight cultures were incubated between 15 and 18 h, and cultures with an OD620 of 0.1 to 0.4 were used to inoculate final BHI broth cultures lacking antibiotics to an OD620 of 0.005.
Construction and verification of mutant strains.
The mutant strains (listed in Table S1 of the supplemental material) were constructed by transformation of competent S. pneumoniae cells with linear PCR amplicons as described previously (37, 43). Briefly, antibiotic resistance markers were introduced into amplicons by overlapping PCR using the primers listed in Table S2 of the supplemental material. S. pneumoniae cells were induced to competence by the addition of synthetic competence stimulatory peptide. Transformants were selected on BA plates containing antibiotics at the concentrations listed in Table S1. Markerless deletions and point mutations were constructed using the two-step Janus method of allelic replacement in a rpsL1 (streptomycin-resistant) genetic background (42, 46). In the first step, the Kanr-rpsL+ Janus cassette was crossed into genes of interest from PCR amplicons by selection for kanamycin resistance and screening for streptomycin sensitivity. In the second step, the Janus cassette was replaced by transformation with a PCR amplicon containing the desired mutations by selection for streptomycin resistance and screening for kanamycin sensitivity. Final transformants were isolated as single colonies three times on BA plates containing antibiotics and propagated for storage in BHI broth containing antibiotics as needed. All constructs were confirmed by PCR amplification and sequencing as described previously (42).
Quellung reaction.
To determine the presence or absence of capsule, the Quellung reaction was performed on colonies from BA plates or on BHI broth cultures (28). Briefly, an overnight colony was transferred from a plate to a glass microscope slide and mixed with 1.5 μl of 1× phosphate-buffered saline (PBS; Ambion), or 1.5 μl of a broth culture at an OD620 of 0.05 was transferred onto a microscope slide. A 1.5-μl volume of type 2 capsule serum (Statens Serum Institute) was added and mixed with the sample. A glass coverslip was placed on the sample, and the slide was observed on a Nikon E-400 phase-contrast microscope with a 100× oil immersion objective. If capsule was present, the cell surface appeared to swell after addition of the serum, whereas if capsule was absent, there was no change in the appearance of the cells.
Hydrogen peroxide release assay.
The rate of H2O2 production was determined using the Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen) as previously described (42).
Biofilm characterization.
Biofilms on the sides and bottoms of tubes containing overnight bacterial cultures were visualized and quantitated with crystal violet. Overnight BHI broth cultures in 17-mm-diameter polystyrene plastic tubes were grown as described above. Culture supernatants of unattached cells that had reached an OD620 of 0.5 to 0.6 were removed from overnight tubes, and biofilms were gently washed twice with 500 μl of room temperature PBS. A 500-μl mixture of 0.15% (wt/vol) crystal violet, 8.2% (vol/vol) ethanol, 0.4% (vol/vol) methanol was added and incubated at room temperature for 20 min. The staining solution was removed, and biofilms were rinsed four times with 1 ml of PBS. Retained crystal violet stain was dissolved with 2 ml of 95% (vol/vol) ethanol for 20 min, and a sample was diluted 10-fold in 95% (vol/vol) ethanol. The A550 of the dissolved crystal violet was determined in 96-well plates by using a microplate reader (Molecular Dynamics) and normalized to the OD620 of the culture supernatants.
ATP and AcP determinations.
ATP and AcP amounts were assayed using the Cell Titer Glo reagent (Promega) as described previously (40, 41) with the following modifications. Twenty-milliliter cultures of desired strains were grown in 50-ml polystyrene tubes to early exponential phase (OD620, 0.2). In contrast to the protocol described in reference 40, cells were not heated during ATP extraction, and the same initial extraction procedure was used for ATP and AcP determinations. Cells were collected by centrifugation for 5 min at 10,000 × g at 4°C and washed twice with 250 μl of cold buffer A (10 mM sodium phosphate [pH 7.5], 10 mM MgCl2, and 1 mM EDTA). Aliquots (50 μl) of cell suspension were set aside on ice for protein assays. A 50-μl volume of cold 3.0 M HClO4 was added to the remaining 200 μl of cell suspension and incubated on ice for 30 min. After incubation, tubes were centrifuged for 2 min at 8,000 × g at 4°C. The supernatant was transferred to an empty tube, neutralized by the addition of 75 μl of saturated KHCO3 (36 g KHCO3 in 100 ml H2O), and centrifuged at 8,000 × g at 4°C.
For ATP determinations, duplicate 50-μl samples of supernatant were each mixed with 50 μl of Cell Titer Glo reagent (reconstituted according to the manufacturer's instructions) in wells of a white, opaque 96-well flat-bottom plate (Falcon). Plates were incubated for 10 min at room temperature, and luminescence was measured using a Spectra Max fluorescence plate reader running Soft Max Pro software (Molecular Devices). ATP concentrations were determined by comparison with standard curves of known concentrations of ATP (0.5 to 10.0 μM) freshly diluted in buffer A to which acid and base had been added as described above. All sample concentrations fell within the ranges of standard curves (data not shown).
For AcP assays, KHCO3-neutalized supernatants were transferred to empty tubes, and 15.0 mg of powdered activated charcoal (Sigma) was added to bind ADP and ATP in the extracts. Tubes were vortexed and incubated on ice for 15 min. Charcoal was removed by filtration through a 13-mm-diameter syringe filter (0.22-μm pore size), and extracts were collected into new tubes. Fifty-microliter samples were assayed as described above as a control for ATP removal by the charcoal treatment. Remaining AcP in samples was enzymatically converted to ATP. Samples of 100 μl were mixed with 1.0 μl of 100 mM MgCl2, 30 μl of 100 μM ADP, and 1.0 μl of 0.4-μg/μl purified E. coli acetate kinase (Sigma) (final concentrations of 1 mM MgCl2, 30 μM ADP, and 4 μg acetate kinase per ml). Reaction mixtures were incubated at 30°C for 90 min. After incubation, duplicate 50-μl aliquots were each mixed with 50 μl of Cell Titer Glo reagent, and ATP amounts were determined as described above, except the buffer for the standards was treated with activated charcoal and filtered like the test samples before known amounts of ATP were added.
All measurements were normalized to protein amounts in the extracts. Fifty-microliter samples that were set aside for protein assays were centrifuged at 16,000 × g at 4°C for 2 min. Cell pellets were suspended in 200 μl of 1% (wt/vol) SDS and 0.1% (vol/vol) Triton X-100 and mixed vigorously by vortexing. Protein amounts were determined in duplicate 5-μl samples using the DC protein assay kit (Bio-Rad) according to the manufacturer's instructions.
To validate these assays, a series of control experiments was performed. The first control ensured that AcP was fully recovered within the experimental error range in the extraction and that conversion of AcP to ATP was driven to completion. This control was done by adding a range of known amounts of AcP at the beginning of the extraction procedure and assaying the conversion of added AcP to ATP. The second control confirmed that the ATP produced in the AcP assay was obtained from the enzymatic reaction. Two separate reactions, one lacking purified AckA and one in which the extract was heated to 100°C for 5 min, were assayed and found to lack ATP production. In the third control experiment, we assayed ATP amounts in S. pneumoniae and E. coli cultures in side-by-side comparisons using two different extraction procedures that had been published previously for E. coli (6, 25). Similar results were obtained for S. pneumoniae when using either procedure, and we obtained results comparable to those published previously for E. coli (6, 25, 36).
Cellular content and concentrations of ATP and AcP (see Table 2 below) were estimated using the following parameters: (i) 0.42 ± 0.02 mg of protein (mean ± standard error; n = 8) was recovered from 20 ml of strain IU1781 at an OD620 (1-cm path length) of 0.2 by the procedure described above. A comparable amount of protein was recovered from unencapsulated strain IU1945. (ii) An OD620 (1-cm path length) of 0.2 corresponded to 5.5 × 107 CFU per ml for strain IU1781. A similar CFU per ml was obtained for strain IU1945. (iii) Strain IU1781 chains (≈11 cells per chain in BHI broth [3]) were disrupted to mostly diplococci by the vigorous mixing during serial dilution in PBS and brief heating in nutrient broth-soft agar used in the determinations of CFU per ml. Therefore, each CFU of strain IU1781 corresponded to a diplococcus of two cells, as with strain IU1945 (3). Disruption of strain IU1781 chains accounted for the observations that the CFU per ml per OD620 unit and protein yield (cell mass) per OD620 unit were similar for strains IU1781 and IU1945. (iv) The volume of strain IU1781 cells, ≈0.6 × 10−15 liter, was based on the cell dimensions described in reference 3. Consistent with these assumptions, the ATP content per mg of protein and ATP concentrations were similar for strains IU1781 and IU1945 (data not shown). Based on these considerations, the amount of protein recovered by this method was ≈200 μg per 1 × 109 pneumococcal cells.
RNA extraction and microarray analysis.
Final cultures were started at an OD620 of ≈0.001 in BHI broth and grown to early exponential phase (OD620, ≈0.1). Rapid lysis RNA extraction, purification, cDNA synthesis, labeling, hybridization to S. pneumoniae R6 microarrays (Ocimum Biosolutions), array washing, and data collection were performed as described previously (24, 28). Data were collected from one to four independent biological replicates, including dye swaps, and analyzed using software from the TM4 microarray software suite (www.tm4.org). Result files generated by GenePix Pro 6.0 software (Molecular Devices) were converted to the TIGR MultiExperiment Viewer file format using the ExpressConverter 2.1 software. Lowess (block) data normalization was performed using the TIGR MIDAS 2.21 software. Expression ratios and Bayesian P values were calculated as described previously (24, 28). The cutoff for significant changes in relative transcript amounts was set at ±1.8-fold, with a Bayesian P value of <0.001, when available.
Microarray data accession number.
Microarray data (intensity and expression ratio) were deposited in the GEO database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE23404.
RESULTS
Deletion of ackA leads to accumulation of suppressor mutations.
The S. pneumoniae serovar 2 D39 genome contains a single ackA ortholog (spd_1853) (28), whose gene product matches the amino acid sequence of E. coli acetate kinase (AckAEco) over its whole length (data not shown). ackASpn is preceded by a gene encoding an adenine-specific DNA methylase, characteristic of a restriction system methylase. A DNA methylase gene precedes ackA in several low-GC Gram-positive species (31). ackASpn is followed by a strong predicted transcription terminator in a 151-bp intercistronic region that precedes essential rnpA (encoding the protein subunit of RNase P). A linear amplicon containing a ΔackA::Kanr rpsL+ deletion/insertion mutation was synthesized that removed all but the first and last 60 bp of the ackA gene (see Materials and Methods; see also Tables S1 and S2 in the supplemental material). Transformation of this ΔackA amplicon into the D39 rpsL1 parent strain led to the appearance of three types of colonies: very small, smooth colonies; medium-size, rough colonies of about the same size as the parent; and larger mucoid colonies. This size variation suggested the accumulation of spontaneous suppressor mutations.
Consistent with this interpretation, restreaking the very small ΔackA colonies on BA plates led to the appearance of all three colony types. In contrast, the medium and large colonies were stable during restreaking and were characterized further (described below). Expression of a copy of the ackA+ gene from a fucose-inducible promoter (PfcsK) in the ectopic CEP site (IU4352 [see Table S1]) (50) complemented the instability of the ΔackA mutant on BA plates. In this background, transformants containing the ΔackA mutation formed normal-looking colonies when 0.2% (wt/vol) fucose was added to plates but formed the three sizes of colonies described above when fucose was omitted (data not shown). Finally, it should be noted that the rpsL1 allele was included in these strains to allow markerless mutant construction by allele exchange (see Materials and Methods; see also Tables S1 and S2 in the supplemental material) (42, 46). In many constructs, rpsL1 was complemented directly by the presence of the Kanr rpsL+ cassette. In other strains, the rpsL1 allele did not affect the phenotypes reported here (data not shown).
Biofilm formation by the medium-size colonies was caused by mutations in the capsule locus.
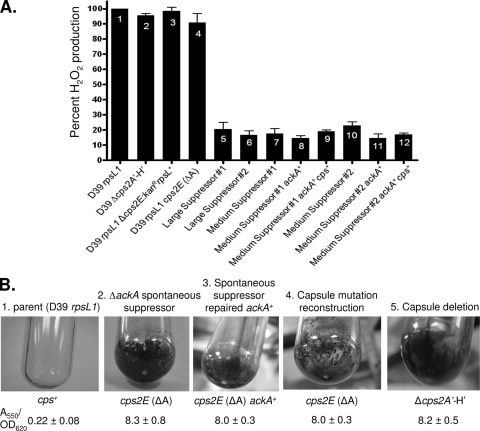
The ΔackA mutants that formed medium-size rough colonies had two distinct phenotypes. They produced only ≈10% H2O2 compared to the parent strain (Fig. 2A, bars 7 and 10), and they formed biofilms in BHI broth cultures in polystyrene tubes, as visualized by crystal violet staining (Fig. 2B, tube 2). Repair of the ΔackA deletion back to the wild-type ackA+ allele did not decrease biofilm formation (Fig. 2B, tube 3), indicating that the biofilm formation phenotype was caused by accumulation of another mutation in this class of ΔackA mutant. In S. pneumoniae, there is an inverse relationship between biofilm formation and capsule production in culture (reviewed in reference 33). Encapsulated strains, such as the parent D39 rpsL1 strain (Fig. 2B, tube 1), do not form robust biofilms in culture, whereas unencapsulated mutants form biofilms (Fig. 2B, tube 5) (1, 33, 35). The Quellung antibody test for serotype 2 capsule confirmed that the medium-size ΔackA mutants were indeed unencapsulated (data not shown). In contrast, the very small unstable ΔackA mutants and large mucoid suppressors produced capsule.
FIG. 2.
H2O2 production (A) and biofilm formation (B) by ΔackA suppressor mutants. (A) H2O2 production was determined relative to the parent strain (IU1781; bar 1) in limited aeration (static) cultures as described in Materials and Methods, and standard errors from multiple independent determinations are indicated. Bars 2 to 4, constructed unencapsulated mutants; bars 5 and 6, mucoid large colony suppressors; bars 7 and 10, medium-size rough colony suppressors; bars 8, 9, 11, and 12, medium-sized rough colony suppressors in which the ΔackA and cps2E (ΔA) mutations were repaired to the ackA+ and cps2E+ alleles. Relevant genotypes are shown and are also listed in Table S1 of the supplemental material. Bar 2, strain IU1945; bar 3, IU3286; bar 4, IU3309; bar 5, IU3254; bar 6, IU3255; bar 7, IU3250; bar 8, IU3252; bar 9, IU3311; bar 10, IU3251; bar 11, IU3253; bar 12, IU3313. (B) Biofilms formed by overnight cultures in polystyrene tubes were stained with crystal violet and quantitated based on the A550 as described in Materials and Methods. Panels from left: 1, parent encapsulated strain IU1781; 2, unencapsulated strain IU3250, a rough, medium-sized colony ΔackA suppressor containing the spxB (Trp339STOP) and cps2E (ΔA) mutations; 3, unencapsulated strain IU3252, medium-sized rough colony suppressor IU3250 repaired back to ackA+; 4, unencapsulated strain IU3309, an ackA+ spxB+ strain containing the constructed cps2E (ΔA) allele; 5, unencapsulated strain IU1945, an ackA+ spxB+ strain containing the large Δcps2A′-Δcps2H′ deletion. See Table S1 for full genotypes. A550 values after crystal violet staining were normalized to the density (OD620, ≈0.5 to 0.6) of the culture supernatants containing unattached cells. Averages are based on three or more independent determinations, and standard errors are indicated.
The capsule biosynthesis locus in S. pneumoniae D39 contains 17 genes (reviewed in reference 57). Previous studies showed that mutations that limit or prevent capsule biosynthesis only occur in the first five genes, cps2ABCDE, which are involved in initiating sugar repeat unit formation and regulating capsule chain length and amount (55, 57). Mutations in the other cps2 genes eliminate side chain assembly, transport, and polymerization and are lethal to cells, unless suppressed by mutations that inactivate cps2E (55), which encodes the initiating repeat unit glycosyltransferase (8). An insertion in cps2E also turned up in a transposon screen of genes that affect pneumococcal biofilm formation (35). For these reasons, we amplified and sequenced the 3.7-kb region containing cps2CDET′ from nine independently isolated ΔackA medium-size colony suppressors (Table 1). These strains contained mutations in csp2E (7/9) or cps2C (2/9), five of which were frameshift mutations. One mutation, cps2E (ΔA), consisting of a deletion of 1 A from a run of 7 A bases, arose three times independently and was used in subsequent experiments.
TABLE 1.
Spontaneous suppressor mutations that accumulated in ΔackA mutants
| Phenotype group and mutated gene | Mutation in independent isolates |
|---|---|
| Mucoid large colony suppressors | |
| spxB | Phe269Leu (TTT to CTT) |
| spxB | Frameshift mutation ΔC in codon 264 |
| spxB | Gly472Asp (GGC to GAC) |
| spxB | Change in mapped transcription start from G to T |
| Rough medium-sized colony suppressors | |
| spxB | Trp339STOP (TGG to TAG) |
| cps2Ea | Frameshift mutation ΔA in codon 326 (in run of 7 As) |
| spxB | Gly423Cys (GGT to TGT) |
| cps2Ea | Frameshift mutation ΔA in codon 326 (in run of 7 As) |
| spxR | Frameshift mutation ΔG in codon 115 (in run of 5 Gs) |
| cps2Ea | Frameshift mutation ΔA in codon 326 (in run of 7 As) |
| Additional capsule mutations identified in rough medium-sized colony suppressors | |
| cps2E | Frameshift mutation ΔG in codon 111 |
| cps2E | Arg347Cys (CGT to TGT) |
| cps2E | Frameshift mutation ΔA in codon 419 |
| cps2E | Insertion of T after codon 204 |
| cps2C | Insertion of A after codon 45 |
| cps2C | Insertion of T in codon 113 |
Isolated three times from separate independent transformations.
Spontaneous mutations leading to loss of capsule were not associated with reduced H2O2 production in ΔackA mutants.
Medium-size ΔackA mutants containing the cps2E (ΔA) mutation produced low levels of H2O2 (Fig. 2A, bars 7 and 10). To determine if this phenotype was caused by the combination of ΔackA and cps2E (ΔA) mutations, we repaired the ΔackA allele alone and then both the ΔackA and cps2E (ΔA) mutations back to their wild-type alleles and assayed H2O2 production (Fig. 2A, bars 8, 9, 11, and 12). We also constructed and assayed isogenic strains containing a Δcps2E deletion/insertion or the cps2E (ΔA) mutation (Fig. 2A, bars 3 and 4, and B, tube 4). H2O2 production was comparable in the cps2E mutants and the cps2E+ parent (Fig. 2A, bar 1), whereas all strains in which the ΔackA or cps2E (ΔA) alleles were repaired still produced relatively low levels of H2O2 (Fig. 2A, bars 7 to 12), indicating that the medium-size ΔackA mutants contained a second suppressor mutation that decreased H2O2 production.
Mutations in spxB and spxR suppress the deleterious effects of ΔackA deletion.
The ΔackA mutants that formed larger, mucoid colonies retained capsule and showed a decrease in H2O2 production similar to that of the medium-size, rough ΔackA mutants (Fig. 2A, bars 5, 6, and 10). Since both medium- and large-size ΔackA colonies were defective in H2O2 production (Fig. 2A), we sequenced the spxB and spxR genes (Fig. 1) (42) of seven independent ΔackA suppressor strains (Table 1). Five isolates had spontaneous lesions in the spxB reading frame, including one frameshift and one nonsense mutation, one isolate had a change in the mapped transcription start site of spxB from G to T, and one had a frameshift mutation in spxR. All of the mutations were different and likely reduced SpxB activity or expression as reflected by reduced H2O2 production (data not shown).
These results suggested that inactivation of spxB or spxR stabilized ΔackA deletion strains. This conclusion was corroborated in two ways. First, transformation of the ΔackA deletion into ΔspxB or spxR::Mariner null mutants (IU2173 or IU2072, respectively [see Table S1 in the supplemental material]) resulted in uniform, large, mucoid colonies, similar to the appearance of one class of the spontaneous suppressors (data not shown). Second, ΔackA transformants with uniform colony size and morphology were obtained in the D39 rpsL1 parent strain under anaerobic conditions, in which SpxB is not functional (Fig. 1). When ΔackA transformants obtained under anaerobic conditions were streaked onto aerobic plates, the same mixture of colony sizes described above was observed. We also confirmed that a constructed cps2E (ΔA) mutation alone did not stabilize ΔackA mutants (data not shown). Finally, various strain constructions confirmed that spxB cps2 or spxB mutants alone formed medium-sized rough or large mucoid colony morphologies, respectively, similar to those observed for the spontaneous ΔackA suppressor mutants. Together, these findings suggested that the medium-sized rough colonies resulted from tandem mutation, starting with inactivation of capsule biosynthesis followed by inactivation of spxB or spxR.
Δpta deletion is not deleterious to cells.
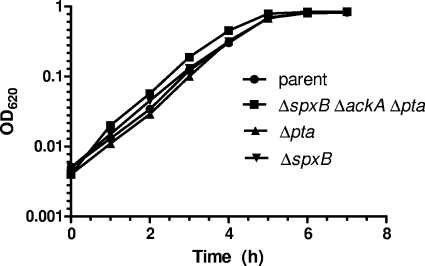
In contrast to ΔackA mutants, deletion of pta did not cause overt accumulation of spontaneous suppressor mutations. The S. pneumoniae serovar 2 D39 genome contains only a single ortholog of pta (spd_0985) (28). Similar to Pta from other low-GC Gram-positive bacteria, PtaSpn lacks the extra amino-terminal domains present in Pta from E. coli and related species (data not shown). ptaSpn is preceded by rluD (putative pseudouridine synthase) and followed by a 270-bp intercistronic region preceding an oppositely oriented IS1167 element (data not shown). Because of PCR amplification issues, the Δpta::Kanr rpsL+ insertion/deletion mutation retained only the last 200 bp of pta (see Tables S1 and S2 in the supplemental material). The only phenotypic change for Δpta mutants was formation of slightly smaller colonies than the parent strain on BA plates incubated in an atmosphere of 5% CO2 (data not shown). This slight growth defect was complemented by expression of pta+ under the control of a fucose-inducible promoter from the ectopic CEP site (strain IU4497 [see Table S1]) (data not shown). However, there was no defect in relative growth rate or yield of the Δpta mutant grown in BHI broth under similar incubation conditions (Fig. 3). The Δpta mutant formed normal-sized colonies on anaerobic BA plates, and Δpta ΔspxB mutants formed large, mucoid colonies indistinguishable from ΔspxB mutants on plates incubated aerobically. These observations suggest that Δpta mutants may be slightly inhibited by aerobic growth on plates but not in certain complex broths grown with limited aeration. Consistent with its similar growth in BHI broth, relative transcript amounts of the exponentially growing Δpta mutant showed relatively few differences compared to its isogenic pta+ parent (see Table S3 in the supplemental material). Notably, there were moderate increases in the relative transcript amounts of the prtA gene (cell wall-associated serine protease) and psa regulon (manganese transport), but little else, in the Δpta mutant compared to its parent. Finally, unlike deletion of spxB, deletion of pta did not stabilize a ΔackA mutant, and the three sizes of colonies described above appeared for the Δpta ΔackA mutants (data not shown). Together, these results indicated that we could construct mutants containing single ΔspxB or Δpta mutations, both ΔspxB and Δpta mutations, or combinations of ΔackA, ΔspxB, and Δpta mutations, provided that ΔspxB mutations were introduced before ΔackA mutations to stabilize strains (see Table S1 in the supplemental material).
FIG. 3.
Typical growth curves of encapsulated parent strain IU1781 (D39 rpsL1) and isogenic mutants IU2837 (D39 rpsL1 ΔspxB Δpta ΔackA), IU2687 (D39 rpsL1 Δpta), and IU2173 (D39 rpsL1 ΔspxB) in BHI broth at 37°C with limited aeration (static) in an atmosphere of 5% CO2 (see Materials and Methods). See Table S1 of the supplemental material for full genotypes. The experiment was repeated numerous times.
Characterization of cellular ATP and AcP amounts in ΔackA, ΔspxB, and Δpta mutant combinations.
ΔackA mutants are blocked in the conversion of AcP to acetate and ATP (Fig. 1) (53). Therefore, the immediate effect of blocking the AckA step would be a decrease in the cellular ATP amount and a possible accumulation of AcP. In contrast, decreased activity or expression of SpxB decreases AcP amount (40) and H2O2 production. To better understand the effects of these mutations on the SpxB-Pta-AckA pathway (Fig. 1), we determined the relative amounts of ATP and AcP in these mutants and their parent strain growing exponentially in BHI broth (see Materials and Methods). All samples were taken in early exponential phase, when the bacteria are primarily excreting lactate and little acetate and before significant depletion of the glucose contained in BHI broth (47).
In contrast to a preliminary result suggesting much greater amounts of AcP than ATP (40), we found that the amount of AcP was only ≈1.5-fold greater than that of ATP in S. pneumoniae D39 (Table 2). We estimated that the cellular concentrations of extractable ATP and AcP were ≈2 mM and ≈3 mM, respectively, under these culture conditions (Table 2; row 1). Validation experiments in which E. coli cells were extracted in parallel experiments gave cellular ATP concentrations of ≈3.0 mM, which is comparable to values reported previously (6, 25, 36). Different extraction formats which increased the yield of ATP from E. coli (6) did not appreciably affect the yield of ATP from S. pneumoniae (data not shown). We conclude that the ATP pool in S. pneumoniae is comparable to that of E. coli during exponential growth in rich media.
TABLE 2.
Relative cellular amounts of ATP and AcP in the parent D39 strain and SpxB-Pta-AckA pathway mutants growing exponentially in BHI broth
| Straina | Presence of gene |
Additionb | ||||
|---|---|---|---|---|---|---|
| spxB | pta | ackA | Relative amt (n)c |
|||
| ATPd | AcPe | |||||
| IU1781 | + | + | + | None | ≡1 (9) | ≡1 (8) |
| IU1781 | + | + | + | 15 mM potassium acetate | 1.0 ± 0.05 (3) | 1.6 ± 0.10 (2) |
| IU2173 | − | + | + | None | 1.0 ± 0.08 (3) | 0.18 ± 0.02 (3) |
| IU2687 | + | − | + | None | 1.1 ± 0.06 (3) | 1.0 ± 0.10 (3) |
| IU2687 | + | − | + | 15 mM potassium acetate | 0.90 ± 0.08 (2) | 1.5 ± 0.10 (2) |
| IU3590 | − | + | − | None | 0.53 ± 0.04 (3) | 1.9 ± 0.20 (2) |
| IU2689 | − | − | + | None | 0.50 ± 0.05 (3) | 0.08 ± 0.04 (3) |
| IU2837 | − | − | − | None | 0.54 ± 0.09 (3) | 0.07 ± 0.01 (3) |
Further information on the indicated strains is provided in Table S1 of the supplemental material. The presence of capsule was confirmed by smooth colony formation on BA plates and Quellung reactions (see Materials and Methods).
Potassium acetate (CH3CO2K) was added to some cultures at a final concentration of 15 mM at the beginning of culture growth.
Bacteria were grown to an OD620 of 0.2, and relative cellular amounts of ATP and AcP were determined as described in Materials and Methods. The number of biological replicates (n) is indicated, and determinations were performed in duplicate within each replicate experiment.
The amount of ATP recovered from 20 ml of encapsulated strain IU1781 (D39 cps+ rpsL1) cells at an OD620 of 0.2 was 0.0067 μmol/mg of protein extract. This amount corresponds to an estimated cellular concentration of ≈2 mM ATP (see Materials and Methods). The experimental error of the ATP determinations was ≈8%.
The amount of AcP recovered from 20 ml of IU1781 (D39 cps+ rpsL1) cells at an OD620 of 0.2 was 0.011 μmol/mg of protein extract. This amount corresponds to an estimated cellular concentration of ≈3 mM AcP. The experimental error of the AcP determinations was ≈5%.
We noted strong homeostasis to maintain cellular ATP amounts in S. pneumoniae by the parallel SpxB and Pta routes (Fig. 1). Addition of 15 mM potassium acetate to the medium increased the relative cellular amount of AcP by ≈1.6-fold, which was the maximum that could be obtained by acetate addition, but the relative amount of ATP remained unchanged (Table 2, row 2). ΔspxB mutants contained only about 20% of the AcP of the parent strain, but again ATP amount was maintained. This finding supports the previous report that the relative amount of AcP is reduced to 20% in spxB mutants (40). Δpta mutants had the same amounts of ATP and AcP as the parent strain (Table 2, row 4), and addition of 15 mM potassium acetate to the Δpta mutant increased the relative AcP amount by ≈1.5-fold but left the relative ATP amount unchanged (Table 2, row 5).
In contrast, ΔspxB ΔackA mutants contained only about half the ATP amount of the parent strain and accumulated about twice as much AcP in the dead-end pathway created by the absence of SpxB and AckA (Fig. 1; Table 2, row 6). Consistent with this result, the ΔspxB Δpta ackA+ mutant showed the same 50% reduction in relative ATP amount and contained negligible AcP, indicating little conversion of acetate to AcP by AckA under the growth conditions sampled here. The ΔspxB Δpta ΔackA triple mutant could be readily constructed starting with the ΔspxB mutation to suppress the ΔackA mutation (see above; see also Table S1 in the supplemental material). Like the ΔspxB Δpta mutant, the triple mutant grew comparably to the parent strain (Fig. 3 and data not shown), even though it produced 50% of the normal ATP amount and contained negligible AcP (Table 1, row 8).
Consideration of these amounts of ATP and AcP suggests that a ΔackA deletion initially decreases the cellular ATP amount by ≈50% and increases AcP amounts. Therefore, the reason ΔackA mutants need to accumulate spxB or spxR suppressor mutations may be to reduce the H2O2 level in response to the lower cellular ATP amount, or to reduce high AcP amounts that are somehow deleterious, or both. Addition of acetate to the parent and the Δpta mutant increased the cellular AcP amount by ≈1.5-fold without changing the ATP level (Table 2, rows 2 and 5). Plating the parent and Δpta mutant on BA plates containing acetate at 15 mM and higher concentrations did not cause colony heterogeneity indicative of suppressor accumulation (data not shown). Therefore, modest increases in AcP amounts did not appear deleterious to cells, although we were unable to increase cellular AcP amounts above this range by adding more potassium acetate (data not shown).
In addition, we attempted to deplete AckA expression and then follow the effects on the ATP and AcP pools. As described above, expression of an ectopic copy of ackA+ under the control of a fucose-inducible promoter complemented the ΔackA mutation on BA plates containing fucose (strain IU4493 [ΔackA CEP::PfcsK-ackA+]) (see Table S1 in the supplemental material). We tested whether we could also deplete AckA sufficiently in strain IU4493 in BHI broth. Cells were grown in relatively low amounts of fucose (0.2% [wt/vol]), washed, and resuspended at a low density (OD620, 0.005) in BHI broth lacking or containing fucose. However, cultures lacking or containing fucose after the shift grew similarly (data not shown). Either it was not possible to sufficiently deplete the accumulated AckA amount and activity within the time frame of these growth experiments, or more likely, the PfcsK promoter was leakier in cells grown in BHI broth than those on BA plates. Regardless, we were unable to obtain conclusive data about the extent of AcP accumulation following depletion of AckA before suppressor accumulation.
Effects of the SpxB-Pta-AckA pathway on TCS expression.
Previously, we reported that ΔspxB mutants growing exponentially caused a limited number of modest changes in relative transcript amounts compared to the spxB+ parent, even though relative AcP amount and H2O2 production were reduced by 80% and 90%, respectively (Table 2, row 3) (40, 42). In particular, there were no large changes in the transcription patterns of known TCS regulons (42). However, we could not rule out a role for AcP in TCS regulation, because the cellular concentration of AcP was unknown, and we had not reduced it to a negligible level (42). We used the strains described above to perform an epistasis analysis, comparing the global transcription patterns of the ΔspxB, ΔspxB ΔackA, ΔspxB Δpta, and ΔspxB ΔackA Δpta mutants (Table 3; see also Tables S3 to S6 in the supplemental material and the information in the GEO database under accession number GSE23404) (a rationale for such work was reported in reference 54). These results confirmed our previous data, showing that the ΔspxB mutation does not strongly affect relative transcript amounts in exponentially growing cells (Table 3, IU2173). Unexpectedly, the changes in relative transcript amounts of the ΔspxB ΔackA Δpta mutant, which produced 50% ATP and negligible AcP compared to its parent (Table 2, row 8), were largely confined to the WalRKSpn, CiaRHSpn, and LiaSRSpn TCS regulons (see Table S6 in the supplemental material). Relative transcript levels of some of the more strongly responsive regulon genes changed the most, especially in the CiaRHSpn regulon (Table 3, IU2837). This result shows that the function of the SpxB-Pta-AckA pathway influences the expression of only a limited number (3/13) of the complete TCSs in S. pneumoniae.
TABLE 3.
Microarray analysis of transcripts strongly regulated by the WalRKSpn, CiaRHSpn, or LiaSRSpn TCS in SpxB-Pta-AckA pathway mutants, compared to the D39 spxB+ pta+ ackA+ parent strain
| Regulon and gene tag (spd no./ spr no.)a | Metabolite or gene description | Relative metabolite or transcript fold changeb in strain (genotype) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| IU2687 (Δpta) | IU2173 (ΔspxB) | IU3590 (ΔspxB ΔackA) | IU2689 (ΔspxB Δpta) | IU2837 (ΔspxB ΔackA Δpta) | IU1885 (ΔwalKSpn) | IU4319 (ΔspxB ΔackA ΔwalKSpn) | IU3107 (ΔspxB Δpta ΔackA ΔwalKSpn) | ||
| ATP | 1.1 ± 0.1 | 1.0 ± 0.1 | 0.53 ± 0.04 | 0.50 ± 0.05 | 0.54 ± 0.09 | ND | ND | ND | |
| AcP | 1.0 ± 0.1 | 0.18 ± 0.02 | 1.9 ± 0.2 | 0.08 ± 0.04 | 0.07 ± 0.01 | ND | ND | ND | |
| WalRKSpn regulonc | |||||||||
| 0104/0096 | LysM domain protein | 1.0 | 1.2 | 1.1 | −2.2 ± 0.5 | −2.5 ± 0.2 | −3.2 ± 0.1 | −2.0 | −4.4 ± 0.9 |
| 1874/1875 | LysM domain protein (putative lysozyme) | −1.1 | −1.2 | 1.1 | −3.4 ± 0.7 | −2.9 ± 0.3 | −7.8 ± 1.5 | −7.2 | −10.0 ± 1.4 |
| 2043/2021 | pcsB; cell division protein | 1.0 | 1.1 | 1.1 | −1.3 | −1.9 ± 0.1 | −4.1 ± 0.7 | −2.1 | −3.9 ± 0.7 |
| CiaRHSpn regulonc | |||||||||
| 0775/0782 | Conserved hypothetical protein | −1.2 | −1.9 ± 0.4 | −2.2 ± 0.8 | −2.0 ± 0.7 | −4.8 ± 0.4 | 1.7 | −1.1 | −1.6 |
| 0913/0931 | Conserved hypothetical protein | −1.0 | −2.0 ± 0.4 | −3.7 ± 1.2 | −3.1 ± 0.1 | −7.8 ± 1.5 | 1.4 | −1.1 | −2.0 ± 0.8 |
| 2068/2045 | htrA; serine protease | −1.0 | −2.2 ± 0.5 | −3.5 ± 1.6 | −3.1 ± 0.6 | −9.4 ± 1.9 | −1.0 | −1.5 | −2.5 ± 0.5 |
| LiaSRSpn regulonc | |||||||||
| 0178/0173 | Conserved hypothetical protein | 1.2 | 1.1 | −1.1 | −1.2 | −1.6 | 1.1 | 1.2 | 1.1 |
| 0351/0343 | liaS; sensor histidine kinase | −1.0 | −1.4 | −1.2 | −1.2 | −2.2 ± 0.3 | −1.2 | −1.1 | −1.1 |
| 0803/0810 | Conserved hypothetical protein | 1.1 | −1.2 | −1.3 | −1.6 | −2.5 ± 0.3 | 1.2 | 1.0 | 1.0 |
Gene tags are shown for both the D39 serovar 2 strain (spd no.) used in the study and laboratory strain R6 (spr no.) used in previous studies by various laboratories.
All amounts are relative to those in parent strain IU1781 (D39 cps+ rpsL1). ATP and AcP values are restated from Table 2 for ease of comparison. ND, not determined.
Microarray analyses were performed as described in Materials and Methods. At least three biological replicates were performed for IU2687 (Δpta) and IU2837 (ΔspxB ΔackA Δpta). Two biological replicates were performed for the other strains, except IU4319 (ΔspxB ΔackA ΔwalKSpn), which was evaluated once for comparison. Standard errors are indicated for fold changes above ±1.8, which was used as the cutoff. Relative transcript amounts of other WalRKSpn, CiaRHSpn, and LiaSRSpn regulon members also changed significantly but are not shown here for the sake of simplicity. Lists of all significant changes in relative transcript amounts are provided in the supplemental material and in the GEO database (accession number GSE23404).
Of these three TCSs affected, only the WalRKSpn TCS regulon had changes in expression correlated with the AcP amount. The ΔspxB Δpta mutant produced similar amounts of AcP and ATP as the triple ΔspxB ΔackA Δpta mutant (Table 3). The changes in the relative transcript amounts of genes encoding the two LysM domain proteins in the WalRKSpn regulon were similar in the double and triple mutants. Notably, the relative transcript amounts of the WalRKSpn regulon genes were restored to the parent level in the ΔspxB ΔackA mutant, which still produced a lower ATP amount, but had a modestly increased amount of AcP compared to the parent (Table 3, IU3590). We compared the effects of eliminating the WalKSpn histidine kinase with the modest changes in WalRKSpn regulon expression observed in the SpxB-Pta-AckA pathway mutants. Decreases in relative amounts of WalRKSpn regulon transcripts were greater in the ΔwalKSpn mutant (Table 3, IU1885) than in the SpxB-Pta-AckA pathway mutants (Table 3, IU2837). Elimination of both WalKSpn and the SpxB-Pta-AckA pathway resulted in an approximately additive decrease (Table 3, IU3107), consistent with the interpretation that WalKSpn kinase activity and AcP can contribute to WalRSpn phosphorylation. However, this effect of AcP was only detected when AcP amounts were reduced to negligible levels, since increasing AcP amount when WalKSpn was absent did not significantly increase WalRKSpn regulon expression (Table 3, IU4319), despite the absence of the WalKSpn phosphatase activity for WalRSpn∼P (16).
Similar to the WalRKSpn TCS regulon, expression levels of the CiaRHSpn and LiaSRSpn TCS regulons were reduced strongly or weakly, respectively, by elimination of the SpxB-Pta-AckA pathway (Table 3, IU2837). In contrast to the WalRKSpn regulon, expression of the CiaRHSpn regulon was marginally reduced in the ΔspxB mutant, which synthesizes only 20% of the AcP of the parent strain (Table 3, IU2173). However, the epistasis analysis described above failed to show a correlation between AcP amount and CiaRHSpn regulon expression (Table 3). Absence of the WalKSpn histidine kinase did not significantly affect expression of the CiaRHSpn and LiaSRSpn regulons (Table 3, IU1885), consistent with minimal or no cross talk. But elimination of both WalKSpn and the SpxB-Pta-AckA pathway produced less reduction in CiaRHSpn regulon expression than elimination of the SpxB-Pta-AckA pathway alone (Table 3, IU2837 and IU3107). Together, these results imply that transcription levels of the CiaRHSpn and LiaSRSpn TCS regulons are influenced in complex ways by perturbations of the SpxB-Pta-AckA pathway.
DISCUSSION
In this paper, we report the first characterizations of the Pta-AckA pathway in S. pneumoniae and its metabolic relationship to SpxB. Both SpxB pyruvate oxidase and Pta phosphotransacetylase synthesize AcP, which can be used as an additional source of ATP in these bacteria that otherwise must synthesize ATP by substrate-level phosphorylation during glycolysis (Fig. 1). Homologs of Pta are present in most bacterial species (although Pta from Gram-positive bacteria lacks the additional amino-terminal domains present in E. coli Pta), but relatively few bacterial species contain SpxB, which also synthesizes H2O2 as a product (Fig. 1) (42). In contrast to mutations in pta and spxB, strains deleted for ackA, which encodes acetate kinase (Fig. 1), were unstable and accumulated suppressors at a high frequency that reduced SpxB function or expression (Table 1), including inactivation of spxR, which encodes a positive regulator of spxB and strH (glycoprotein exoglycosidase) transcription that we identified previously (42).
This collection of mutants defective in different parts of the SpxB-Pta-AckA pathway revealed some unanticipated properties of ATP and AcP biosynthesis in S. pneumoniae. Cellular ATP amounts were maintained at constant levels in ΔspxB and Δpta mutants, and also upon acetate addition to medium, which increased the AcP amount by ≈1.6-fold (Table 2). Notably, ΔspxB mutants contained the same amount of ATP as the parent strain. This comparison was not made in a previous study (40), which did show an 80% reduction in AcP amount in spxB mutants similar to that reported here (Table 2). Our results suggest that in the absence of SpxB, AcP synthesized by Pta is consumed, but not replenished, to maintain the cellular ATP pool.
In suppressed ΔspxB ΔackA mutants or in a ΔspxB Δpta mutant, the cellular ATP amount dropped to 50% of the wild-type level, and in the ΔspxB Δpta and ΔspxB Δpta ΔackA mutants, the AcP amount dropped to a negligible level, although a small (≈7%) residual amount was always detected (Table 2). Surprisingly, these drastic drops in the ATP and AcP energy pools did not detectably impede the growth of mutants in BHI broth or on BA plates (Fig. 3). The cellular concentrations of ATP and AcP were estimated at ≈2 mM and ≈3 mM, respectively, in exponentially growing serotype 2 strain D39 (Table 2). Several estimates of the pneumococcal ATP pool, ranging from ≈0.3 mM to ≈6 mM, have been reported, although most reports did not provide details of how the cellular amounts were calculated. Our estimate of 2 mM most closely matches the 1 mM estimate reported in reference 49. The cellular ATP pools of S. pneumoniae D39 (≈2 mM [Table 2]) and E. coli K-12 (≈3 mM [6, 25]) were comparable in exponentially growing bacteria. Although E. coli has a greater capacity to synthesize ATP, it also contains nearly twice as many genes and forms cells at least twice as large as S. pneumoniae (3, 28). Likewise, the steady-state cellular pool of AcP in S. pneumoniae (≈3 mM) (Table 2) was comparable to that of E. coli under some growth conditions (≈3 mM) (25, 54). Thus, during exponential growth, S. pneumoniae reserves a remarkable amount of phosphate bond energy in AcP, which is synthesized by a pyruvate oxidase activity not present in E. coli and many other bacterial species (Fig. 1). An important implication from the combined results in this paper is that AcP seems to be a major energy reservoir for ATP synthesis in pneumococcus, rather than a signaling conduit between metabolic state and TCS regulation.
The accumulation of suppressors was high in ΔackA mutants, which formed very small, unstable colonies under aerobic growth conditions. Every time a small, single ΔackA colony was streaked, multiple large mucoid and medium-sized rough colony types were obtained, and all suppressor mutants contained mutations in spxB or spxR that reduced SpxB function or expression (Table 1; Fig. 2). In addition, the medium-sized rough colonies contained mutations in the cps2E or cps2C genes that halted capsule biosynthesis (Table 1). Genetic reconstruction experiments demonstrated that ΔspxB or spxR::Mariner mutations were necessary and sufficient to stabilize ΔackA mutants, whereas Δcps2A′-Δcps2H′ and cps2E (ΔA) mutations were not (see Results). Therefore, the cps2E and cps2C mutations likely occurred first in the medium-sized rough ΔackA mutants, perhaps to reduce the considerable ATP load of capsule biosynthesis, but loss of capsule was not sufficient to relieve the metabolic stress caused by ΔackA deletion, which further requires reduced SpxB function or expression.
The mutations that accumulated in cps2E, cps2C, spxB, and spxR in independently isolated, spontaneous ΔackA suppressor strains fell into a limited set of changes that perhaps is indicative of a mutational stress response. Except for one C→T change, the eight other sequenced cps2E and cps2C mutants contained frameshift mutations involving an insertion or deletion of a single base. Of these eight frameshift mutations, three were caused by a ΔA deletion in a single run of 7 A residues in cps2E (Table 1). However, the other five frameshift mutations were not in runs of bases characteristic of a phase variation mechanism (34). On the other hand, we found that the cps2E (ΔA) could shift back to the in-frame 7-A run if bacteria were subjected to strong selection for capsule production in an animal model of infection (K. J. Wayne, unpublished results). In addition, one spxB and one spxR frameshift mutation were recovered among the seven spxB and spxR suppressor mutants (Table 1). The other five spxB mutations were single nucleotide changes, four of which were changes of G to other bases, including two G→T mutations, consistent with oxidative damage of G to 8-oxo-G (32). Finally, preliminary experiments using a newly constructed frameshift indicator strain suggested that the ΔackA mutants accumulated most of these suppressor mutations on the selection plates following transformation (data not shown), which is again consistent with a highly stressed state that would increase frameshifts and base damage.
We considered two nonexclusive explanations as to why ΔackA mutants need to reduce or eliminate SpxB function. AckASpn is homologous to canonical AckAEco over its whole length (data not shown), and the instability of ΔackASpn mutants is likely caused by a metabolic pathway block, rather than some unprecedented essential interaction between AckA and another protein that does not occur in E. coli. The ΔackA block should decrease ATP production and increase AcP amounts (Fig. 1). Reduced SpxB expression decreases H2O2 and AcP production (Table 2; Fig. 2). Therefore, according to one scenario, the reduced spxB expression simply decreases highly toxic levels of AcP back to levels that are tolerated. According to the other scenario, resistance to the high endogenous levels of H2O2 produced by SpxB is an energy-dependent process that requires wild-type ATP levels.
Several observations tend to support the energy dependence hypothesis, but none is definitive. The reaction catalyzed by Pta is usually reversible (Fig. 1), so high levels of AcP could be reduced by funneling it back toward acetyl-CoA and other metabolites in a ΔackA mutant. But disruptions in the acetyl-CoA pathway itself could be deleterious, because of its essential role in the biosynthesis of fatty acids and other key metabolites (Fig. 1). Acetate addition led to modest increases of AcP levels in spxB+ strains (Table 2) and did not result in spxB suppressor accumulation, as was observed for ΔackA mutants. But, we were limited in how much we could increase the AcP amount by this approach. Attempts to downshift AckA expression were not successful in liquid medium, probably because of leakiness of the fucose-regulated PfcsK promoter. Elsewhere, we have demonstrated that several essential genes are required for resistance to the endogenous H2O2 synthesized by SpxB, and different genes are used to cope with endogenously produced and exogenously added H2O2 (unpublished data). The protein encoded by one of these genes consumes energy as part of its metabolic activity. Moreover, recent work has implicated the pentose phosphate shunt, which consumes NADPH, in resistance mechanisms against H2O2 (39). Together, these observations are consistent with the idea that a sufficient ATP level needs to be maintained for resistance to the endogenous H2O2 synthesized by SpxB.
Finally, this set of mutants (Table 2) allowed us to analyze effects of the SpxB-Pta-AckA pathway and AcP amount on TCS regulation patterns in S. pneumoniae (Table 3; see also the supplemental material). AcP phosphorylation of some response regulators plays important roles in the signaling pathways of several TCSs in E. coli and other bacterial species (reviewed in references 39, 53, and 54). In S. pneumoniae, the WalRSpn (VicR) response regulator is essential, whereas the WalKSpn (VicK) histidine kinase is not essential under standard growth conditions (16, 37, 52). One hypothesis proposed to explain this nonessentiality of WalKSpn is cross talk phosphorylation of WalRSpn by AcP, which occurs at high (≈10 mM) AcP concentrations in purified biochemical reactions over extended times (16, 38). In addition, it has been proposed that the AcP produced by SpxB generally phosphorylates response regulators (27, 45).
Previously, we reported that the 80% reduction in AcP that occurs in ΔspxB mutants (Table 2) (40) did not lead to large changes in transcription patterns indicative of altered TCS regulation (42). The most conspicuous change in the ΔspxB mutant was a small decrease near the microarray cutoff value in the relative transcript amount of the CiaRHSpn TCS regulon (Table 3). We also did not detect small decreases in the relative amounts of fab gene transcripts, as recently reported in reference 4. Complete disruption of the SpxB-Pta-AckA pathway in a triple mutant led to a negligible amount of AcP and 50% of the cellular ATP amount (Table 2). Surprisingly, these reductions did not significantly affect growth under the condition tested (Fig. 3) and resulted in a limited global transcription response that strongly decreased expression of the CiaRH TCS regulon and to a lesser extent that of the WalRKSpn and LiaSRSpn regulons (Table 3; see also Table S6 in the supplemental material).
Epistasis analysis using combinations of SpxB-Pta-AckA pathway mutations showed that expression of the WalRKSpn regulon was correlated with AcP amounts, but the decreases in expression levels were small (Table 3) and approximately additive, with the larger decreases in WalRKSpn regulon expression caused by the absence of the WalKSpn histidine kinase (Table 3). However, these effects only became detectable at negligible AcP amounts, which are unlikely to occur under normal physiological conditions, especially given the central role described here for AcP as an energy reservoir in S. pneumoniae. Likewise, at least an 80% reduction of cellular AcP amount was required to detect a marginal decrease in CiaRHSpn TCS regulon expression (Table 3) (42). These combined results do not support phosphorylation of pneumococcal response regulators by AcP as a general signaling mechanism, despite a relatively high (≈3 mM) estimated cellular AcP concentration (Table 2). On the other hand, normal function of the SpxB-Pta-AckA pathway quite specifically influences the expression of the three TCS regulons that mediate cell wall homeostasis and stress (reviewed in references 10 and 22). In particular, the response of the CiaRHSpn TCS regulon is complex (Table 3) and may reflect changes in cell wall biosynthesis that occur when the SpxB-Pta-AckA pathway is perturbed or changes in other key metabolic processes, such as protein acetylation, that are linked to this pathway (53, 54).
Supplementary Material
Acknowledgments
We thank Tiffany Tsui, Alina Gutu, and Kyle Wayne for comments, discussions, and some bacterial strains and PCR amplicons used in this paper and Linda Kenney (UIC) for critical comments.
This project was supported by grant number AI060744 (to M.E.W.) from the National Institute of Allergy and Infectious Diseases. S.R.-M. was a predoctoral trainee on NIH training grant T32GM007757.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 15 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allegrucci, M., and K. Sauer. 2008. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J. Bacteriol. 190:6330-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auzat, I., S. Chapuy-Regaud, G. Le Bras, D. Dos Santos, A. D. Ogunniyi, I. Le Thomas, J. R. Garel, J. C. Paton, and M. C. Trombe. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018-1028. [DOI] [PubMed] [Google Scholar]
- 3.Barendt, S. M., A. D. Land, L. T. Sham, W. L. Ng, H. C. Tsui, R. J. Arnold, and M. E. Winkler. 2009. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191:3024-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benisty, R., A. Y. Cohen, A. Feldman, Z. Cohen, and N. Porat. 2010. Endogenous H(2)O(2) produced by Streptococcus pneumoniae controls FabF activity. Biochim. Biophys. Acta 1801:1098-1104. [DOI] [PubMed] [Google Scholar]
- 5.Bortoni, M. E., V. S. Terra, J. Hinds, P. W. Andrew, and H. Yesilkaya. 2009. The pneumococcal response to oxidative stress includes a role for Rgg. Microbiology 155:4123-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckstein, M. H., J. He, and H. Rubin. 2008. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190:718-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabre, M. 2009. Pneumonia in the elderly. Curr. Opin. Pulm. Med. 15:223-229. [DOI] [PubMed] [Google Scholar]
- 8.Cartee, R. T., W. T. Forsee, M. H. Bender, K. D. Ambrose, and J. Yother. 2005. CpsE from type 2 Streptococcus pneumoniae catalyzes the reversible addition of glucose-1-phosphate to a polyprenyl phosphate acceptor, initiating type 2 capsule repeat unit formation. J. Bacteriol. 187:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crook, D. W., A. B. Brueggemann, K. L. Sleeman, and T. E. A. Peto. 2004. Pneumococcal carriage, p. 136-147. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, DC.
- 10.Dubrac, S., P. Bisicchia, K. M. Devine, and T. Msadek. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307-1322. [DOI] [PubMed] [Google Scholar]
- 11.El-Mansi, M., A. J. Cozzone, J. Shiloach, and B. J. Eikmanns. 2006. Control of carbon flux through enzymes of central and intermediary metabolism during growth of Escherichia coli on acetate. Curr. Opin. Microbiol. 9:173-179. [DOI] [PubMed] [Google Scholar]
- 12.Felmingham, D., R. Canton, and S. G. Jenkins. 2007. Regional trends in beta-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J. Infect. 55:111-118. [DOI] [PubMed] [Google Scholar]
- 13.Fredericks, C. E., S. Shibata, S. Aizawa, S. A. Reimann, and A. J. Wolfe. 2006. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61:734-747. [DOI] [PubMed] [Google Scholar]
- 14.Gottshalk, G. 1986. Bacterial metabolism, 2nd ed. Springer-Verlag, New York, NY.
- 15.Gueriri, I., S. Bay, S. Dubrac, C. Cyncynatus, and T. Msadek. 2008. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol. Microbiol. 70:1342-1357. [DOI] [PubMed] [Google Scholar]
- 16.Gutu, A. D., K. J. Wayne, L. T. Sham, and M. E. Winkler. 2010. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 192:2346-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendriksen, W. T., H. J. Bootsma, S. Estevao, T. Hoogenboezem, A. de Jong, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190:590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendriksen, W. T., T. G. Kloosterman, H. J. Bootsma, S. Estevao, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. Site-specific contributions of glutamine-dependent regulator GlnR and GlnR-regulated genes to virulence of Streptococcus pneumoniae. Infect. Immun. 76:1230-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriques-Normark, B., and S. Normark. 2010. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp. Cell Res. 316:1408-1414. [DOI] [PubMed] [Google Scholar]
- 20.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 23.Kadioglu, A., J. N. Weiser, J. C. Paton, and P. W. Andrew. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288-301. [DOI] [PubMed] [Google Scholar]
- 24.Kazmierczak, K. M., K. J. Wayne, A. Rechtsteiner, and M. E. Winkler. 2009. Roles of rel in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72:590-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, A. H., A. Shulla, S. A. Reimann, D. H. Keating, and A. J. Wolfe. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 189:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreth, J., H. Vu, Y. Zhang, and M. C. Herzberg. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J. Bacteriol. 191:6281-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuboniwa, M., G. D. Tribble, C. E. James, A. O. Kilic, L. Tao, M. C. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60:121-139. [DOI] [PubMed] [Google Scholar]
- 28.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch, J. P., III, and G. G. Zhanel. 2010. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr. Opin. Pulm. Med. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 30.Madeddu, G., M. Laura Fiori, and M. Stella Mura. 2010. Bacterial community-acquired pneumonia in HIV-infected patients. Curr. Opin. Pulm. Med. 16:201-207. [DOI] [PubMed] [Google Scholar]
- 31.Merritt, J., F. Qi, and W. Shi. 2005. A unique nine-gene comY operon in Streptococcus mutans. Microbiology 151:157-166. [DOI] [PubMed] [Google Scholar]
- 32.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moscoso, M., E. Garcia, and R. Lopez. 2009. Pneumococcal biofilms. Int. Microbiol. 12:77-85. [PubMed] [Google Scholar]
- 34.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 35.Munoz-Elias, E. J., J. Marcano, and A. Camilli. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 76:5049-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p. 445-473. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 37.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 38.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogunniyi, A. D., L. K. Mahdi, M. P. Jennings, A. G. McEwan, C. A. McDevitt, M. B. Van der Hoek, C. J. Bagley, P. Hoffmann, K. A. Gould, and J. C. Paton. 2010. Central role of manganese in regulation of stress responses, physiology and metabolism in Streptococcus pneumoniae. J. Bacteriol. 192:4489-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pericone, C. D., S. Park, J. A. Imlay, and J. N. Weiser. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 185:6815-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruss, B. M., and A. J. Wolfe. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973-984. [DOI] [PubMed] [Google Scholar]
- 42.Ramos-Montanez, S., H. C. Tsui, K. J. Wayne, J. L. Morris, L. E. Peters, F. Zhang, K. M. Kazmierczak, L. T. Sham, and M. E. Winkler. 2008. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol. Microbiol. 67:729-746. [DOI] [PubMed] [Google Scholar]
- 43.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somerville, G. A., and R. A. Proctor. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803-813. [DOI] [PubMed] [Google Scholar]
- 46.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taniai, H., K. Iida, M. Seki, M. Saito, S. Shiota, H. Nakayama, and S. Yoshida. 2008. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J. Bacteriol. 190:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 49.Trombe, M. C., G. Laneelle, and A. M. Sicard. 1984. Characterization of a Streptococcus pneumoniae mutant with altered electric transmembrane potential. J. Bacteriol. 158:1109-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsui, H. C., D. Mukherjee, V. A. Ray, L. T. Sham, A. L. Feig, and M. E. Winkler. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192:264-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Poll, T., and S. M. Opal. 2009. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374:1543-1556. [DOI] [PubMed] [Google Scholar]
- 52.Winkler, M. E., and J. A. Hoch. 2008. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190:2645-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfe, A. J. 2010. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr. Opin. Microbiol. 13:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xayarath, B., and J. Yother. 2007. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J. Bacteriol. 189:3369-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yesilkaya, H., F. Spissu, S. M. Carvalho, V. S. Terra, K. A. Homer, R. Benisty, N. Porat, A. R. Neves, and P. W. Andrew. 2009. Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence. Infect. Immun. 77:5418-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yother, J. 2004. Capsules, p. 30-48. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, DC.
- 58.Zapun, A., T. Vernet, and M. G. Pinho. 2008. The different shapes of cocci. FEMS Microbiol. Rev. 32:345-360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.