SUMMARY
Advanced stage papillary serous tumors of the ovary are responsible for the majority of ovarian cancer deaths, yet the molecular determinants modulating patient survival are poorly characterized. Here, we identify and validate a prognostic gene expression signature correlating with survival in a series of microdissected serous ovarian tumors. Independent evaluation confirmed the association of a prognostic gene microfibril-associated glycoprotein 2 (MAGP2) with poor prognosis, whereas in vitro mechanistic analyses demonstrated its ability to prolong tumor cell survival and stimulate endothelial cell motility and survival via the αVβ3 integrin receptor. Increased MAGP2 expression correlated with microvessel density suggesting a proangiogenic role in vivo. Thus, MAGP2 may serve as a survival-associated target.
INTRODUCTION
Ovarian cancer is the fifth most common form of cancer in women in the United States, accounting for 3% of the total number of cancer cases and for 26% of those cases occurring in the female genital tract. Epithelial ovarian cancer is highly lethal. In 2009, there will be an estimated 21,550 new cases and 14,660 deaths from ovarian caner in the United States (Jemal et al., 2009). Of these deaths, the vast majority will be attributed to papillary serous tumors, which account for 60% of all cases (Boring et al., 1994). Most of these tumors are detected at an advanced stage with metastases present beyond the ovaries precluding curative treatment (Boente et al., 1996). Clinical management of this disease involves tumor debulking followed by administration of carboplatin chemotherapy. Despite the fact that 80% of advanced ovarian cancers (stages III and IV) respond to primary treatment with surgery and chemotherapy, the disease usually recurs and is ultimately fatal. A subset of these patients will develop a more chronic form of ovarian cancer and may survive 5 years or more with treatment. Consequently, there is a pressing need for diagnostic classifiers that can reliably stratify patients for therapy, as well as targets for therapeutic intervention.
Previous studies have shown that large-scale transcription profiling can identify differentially expressed genes and molecular signatures in numerous biological systems including ovarian cancer (Alizadeh et al., 2000; Bonome et al., 2005; DeRisi et al., 1997; Golub et al., 1999). More recent efforts to derive clinical predictors for survival in ovarian cancer from gene expression data have focused on discrete patient groups clustered at either end of the survival spectrum (Berchuck et al., 2005; Lancaster et al., 2004). Yet, expression patterns identified in this manner may not adequately differentiate the majority of patients who will succumb at an intermediate endpoint. In addition, the evaluation of undissected tumor isolates may introduce erroneous data attributable to varying amounts of intervening stroma and lymphocytic infiltrate. In this study, we describe the use of a whole-genome oligonucleotide array to perform expression profiling on a series of microdissected late-stage, high-grade papillary serous ovarian adenocarcinomas to identify aprognostic gene signature correlating with survival as a continuous variable.
RESULTS
Derivation and Validation of a Gene Signature Predictive for Survival in Patients with Advanced Papillary Serous Ovarian Cancer
We identified 53 advanced stage, high-grade primary tumor specimens from patients with papillary serous adenocarcinomas of the ovary whose survival spanned a spectrum of 145 months (see Table S1, available online). The average age for the cohort was 61.9 years (SD = 12.7), with an average survival time of 40.5 months following surgery (SD = 41.3 months). Among these patients, 12 were still alive when we analyzed the data and 11 patients were suboptimally debulked. All specimens were subjected to laser-based microdissection and analyzed as pure, microdissected epithelial cell populations on whole-genome Affymetrix U133 Plus 2.0 GeneChip microarrays.
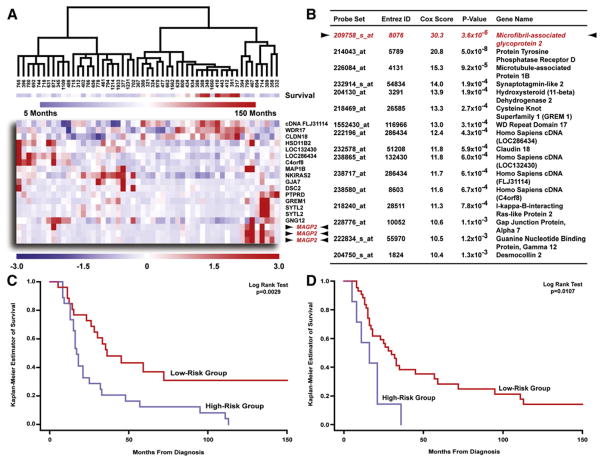
To derive the predictor, we used a two-step “semi-supervised” approach using Cox regression analysis and leave-one-out cross-validation to identify and validate the survival signature. The performance of the prediction analysis was visualized by hierarchical clustering, which demonstrated the ability of the top scoring genes (Cox hazard ratio >10) to cluster the 53 specimens according to survival (Figure 1A). As detailed in Figure 1B, the gene possessing the highest hazard ratio was MAGP2. Finally, the validity of the entire 200 probe set classifier (see Table S2) was evaluated by a non-parametric log rank test using median hazard to stratify the patients. The test was highly significant, with the high-risk group, defined by predicted hazard greater than median hazard, having a significantly shorter survival than the low-risk group (Figure 1C). This result confirmed that our model was able to predict a patient’s hazard accurately. We also investigated other predictor compositions varying the number (100 or 300) of probe sets and principal components (PCs) (four or six) that were assessed and found our results remained nearly identical (data not shown).
Figure 1. Identification and Validation of a Prognostic Gene Expression Signature Correlating with Survival in 53 Microdissected Late-Stage High-Grade Papillary Serous Ovarian Tumors.
(A) Hierarchical clustering of 53 advanced stage, high-grade serous adenocarcinomas using expression values for genes possessing a Cox score >10 (gene expression: red, upregulated; blue, downregulated; survival: blue, short survival; red, long survival).
(B) Genes presented in this table possessed a large Cox score (>10). Only the probe set with the highest Cox score is presented for MAGP2.
(C) Kaplan Meier analysis of the predictor demonstrated a significant difference in survival time (p = 0.0029).
(D) Kaplan Meier survival analysis of 49 patients using qRT-PCR validation data obtained for the top 11 survival-signature genes confirmed the two groups retained significantly different survival endpoints (p = 0.0107).
In order to validate the survival signature, the 11 genes possessing the highest Cox hazard ratios were selected. Quantitative RT-PCR (qRT-PCR) was performed on all 53 RNA samples, which were included in the microarray analysis, using primers specific for each gene. Assayable expression levels for each gene were obtained in 49/53 samples and used to generate a Kaplan Meier plot in a fashion analogous to the microarray analysis. A nonparametric log rank test showed the two groups retained a significant survival difference (Figure 1D), confirming the microarray data.
Identification of Signaling Events Affecting Patient Survival
To ascertain whether subsets of the survival-associated genes might be participating in coordinated signaling pathway(s) contributing to patient outcome, we compared the 53 advanced stage ovarian adenocarcinoma specimens to 10 normal ovarian surface epithelium (OSE) brushings. A total of 5022 probe sets were differentially regulated. Both differential gene expression data and identifiers for the top 150 survival-associated probe sets (Cox Hazard >8.5) were overlaid onto a biological association network for identification of coregulated pathways.
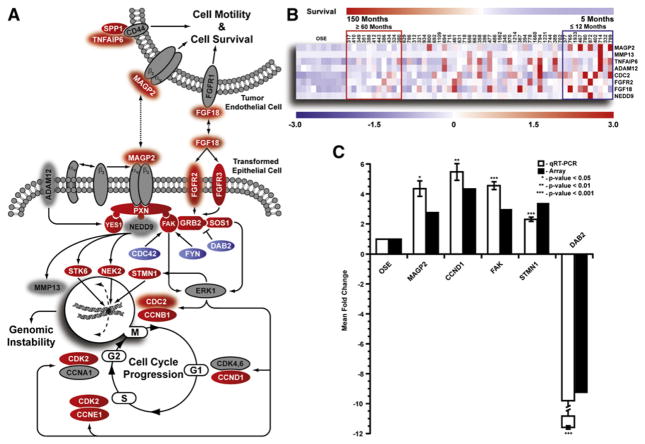
Integrin-mediated signaling stimulated by MAGP2 engagement of the αVβ3 receptor featured prominently in the analysis (Figure 2A). Although the receptor subunits were not differentially regulated in the tumor specimens, a number of downstream effectors were overexpressed versus OSE including PXN, FAK, GRB2, and SOS1. Contributing to this pathway were a number of genes implicated in patient survival including MAGP2, FGF18, FGFR2, ADAM12, NEDD9, MMP13, and CDC2. Of these, MAGP2, FGF18, FGFR2, and CDC2 were also significantly upregulated, when compared to normal OSE. A unique feature of MAGP2, FGF18, and TNFAIP6 is their ability to modulate endothelial cell behavior. The cognate receptors of each protein are also expressed in tumor endothelial cells, suggesting that patient survival may consist of signaling events specific to the transformed cell, as well as induced endothelial cell changes. Furthermore, it is possible that pronounced expression of one or more survival-associated genes may dramatically enhance the aggressiveness of the disease negatively impacting patient outcome (Figure 2B).
Figure 2. Assessment of Putative Signaling Events Contributing to Patient Survival.
(A) Pathway analysis of select differentially regulated genes identified in the 53 microdissected serous tumors, as compared to 10 normal OSE brushings (red, upregulated; blue, downregulated; gray, no change; red halo, survival-associated gene).
(B) Heat map demonstrating association between survival signature genes identified in the pathway and overall patient survival. Increased expression of one or more genes greatly reduced patient survival.
(C) qRT-PCR validation of select genes identified in the pathway analysis for all 53 tumor samples and 10 normal OSE. Error bar represents mean fold change ± SD.
To substantiate the pathway analysis, we completed qRT-PCR for five differentially regulated genes including MAGP2, CCND1, FAK, STMN1, and DAB2 for all 53 tumor specimens, as well as the ten normal OSE isolates. Student’s t test confirmed all five genes were differentially regulated relative to normal OSE at levels comparable to the array data (Figure 2C).
Characterization of Clinical Correlates Associated with the Survival Signature Gene MAGP2
Although the probe sets identified in the analysis predict for patient survival as a group, each gene was selected according to its individual Cox hazard ratio. Thus, genes possessing a high hazard ratio may independently predict for patient survival. MAGP2 was identified by three separate probe sets and scored the highest hazard ratio. In addition, pathway analysis suggested it may participate in coregulated signaling events contributing to enhanced tumor cell survival and prolonged endothelial cell survival and motility distinguishing MAGP2 as a suitable candidate for further characterization.
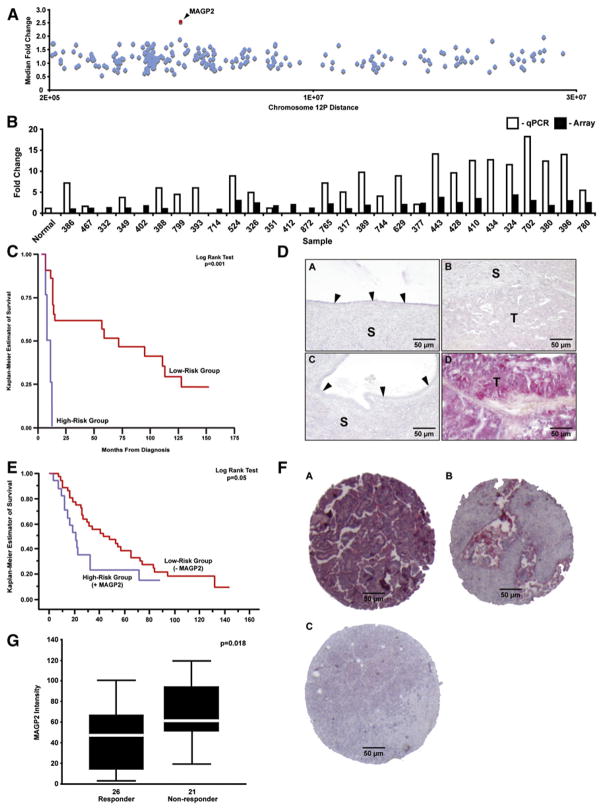
To extend the analysis, we evaluated MAGP2 as an independent prognostic factor. Comparative genome hybridization (CGH) with a 20,000 element oligonucleotide microarray demonstrated a median MAGP2 copy number approaching 2.5 in a subset of the tumor specimens (Figure 3A). This observation was supported by qPCR analysis correlating amplification of the locus with CGH copy number (Figure 3B). qRT-PCR analysis using all 53 tumor isolates confirmed the association between MAGP2 mRNA expression and patient survival. Stratifying the expression values according to the mean showed a significantly shorter survival time for patients expressing MAGP2 mRNA above the mean (Figure 3C). A significant correlation between MAGP2 DNA and mRNA copy number was not observed, suggesting that amplification may only be one of the mechanisms leading to upregulation of MAGP2 in epithelial ovarian cancer cells.
Figure 3. The MAGP2 Locus Is Amplified in Serous Ovarian Tumors and Its Expression Products Are Significantly Correlated with Overall Survival and Chemoresponse.
(A) Oligonucleotide microarray CGH for 42 of the serous tumors established the median MAGP2 copy number (2.5) on chromosome 12p. MAGP2 is highlighted in red.
(B) qPCR correlated with the CGH microarray data for 29 of the serous tumors (r = 0.61, p = 0.004).
(C) Kaplan Meier survival analysis of MAGP2 mRNA expression using the 53 patients with stage III/IV high-grade serous ovarian cancer demonstrated a significant difference in patient outcome (p = 0.001).
(D) Low-level MAGP2 protein staining was observed in normal surface epithelia (A), epithelial and stromal components of benign ovarian cysts (C), and in some high-grade serous tumors (B). Strong MAGP2 staining was observed in a small proportion of high-grade serous ovarian tumor tissues (D). Arrowheads indicate the epithelial layer of the normal ovary and benign ovarian cyst (S, stroma; T, tumor cells).
(E) Kaplan Meier survival analysis of MAGP2 protein expression using 53 patients with stage III/IV high-grade serous ovarian cancer. A statistically significant difference was observed between the outcome groups (p = 0.05).
(F) Independent TMA validation of MAGP2 expression in 64 serous ovarian cancers. A, high-level staining; B, moderate staining; C, low-level staining.
(G) Box plot analysis of MAGP2 protein expression levels among chemotherapy responders and non-responders. A significant correlation was found between poor response and increased MAGP2 protein levels (p = 0.018).
Immunolocalization of MAGP2 protein in all 53 optimally debulked stage III grade 3 serous adenocarcinoma, as well as normal ovarian epithelium and benign cysts, demonstrated low-level expression of MAGP2 in normal ovarian epithelial cells (Figure 3D, subpanel A) and benign cysts (Figure 3D, subpanel C), but elevated levels in a proportion of malignant tumors (Figure 3D, subpanels B and D). The intensity of MAGP2 staining was analyzed with the survival data and examined by Kaplan Meier survival analysis. The results indicated that patients positive for MAGP2 possessed a poor prognosis (Figure 3E). Independent validation of this association was completed with a 64 element tissue microarray (TMA) consisting of advanced stage high-grade serous adenocarcinoma specimens (Figure 3F and Table S1). MAGP2 staining intensity was correlated with survival and adjusted for debulking status by multivariate Cox regression analysis. A significant association was found between the degree of MAGP2 staining and survival (hazard ratio: 1.857; p = 0.004; 95% confidence interval [1.253 and 2.752]).
Resistance to chemotherapy has been linked to αVβ3 signaling in a number of tumor systems including ovarian cancer (Maubant et al., 2002). Consequently, stimulation of the receptor by MAGP2 may attenuate chemoresponse, ultimately affecting patient survival. A subset (47) of the 64 tumors from the validation set analyzed was evaluable for chemotherapeutic response status. Tumors were stratified into two groups, resistant and responsive to chemotherapy, as defined by objective evidence of complete or partial remission. MAGP2 expression levels were significantly lower in patients who responded to chemotherapy (Figure 3G).
Recombinant MAGP2 Stimulates Serous Ovarian Cancer Cell Adhesion and Survival
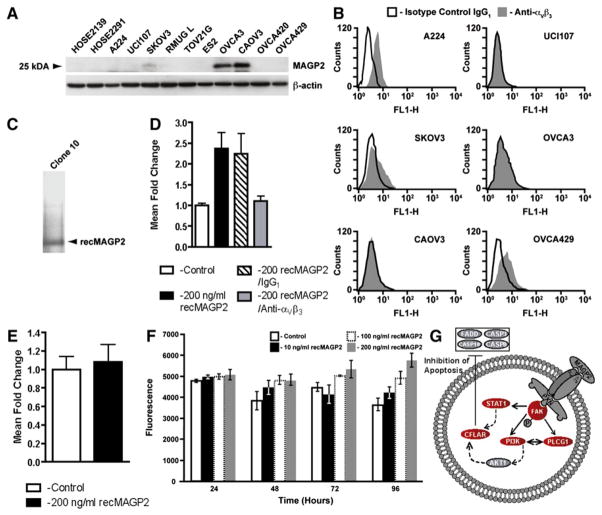
To determine potential mechanisms by which MAGP2 negatively impacts patient survival, we conducted in vitro assays on a panel of serous ovarian cancer cell lines. Screening the panel for MAGP2 expression by western blotting (Figure 4A), as well as αVβ3 receptor status through flow cytometry analysis (Figure 4B, select cell lines only), facilitated the selection of cell lines amenable to biological characterization. Among cell lines expressing MAGP2, only SKOV3 coexpressed the αVβ3 receptor. More importantly, two cell lines, A224 and OVCA429, were positive for αVβ3, yet lacked measurable levels of MAGP2. These cell lines were chosen for subsequent analyses involving the recombinant protein along with UCI107, which was negative for both the receptor and MAGP2.
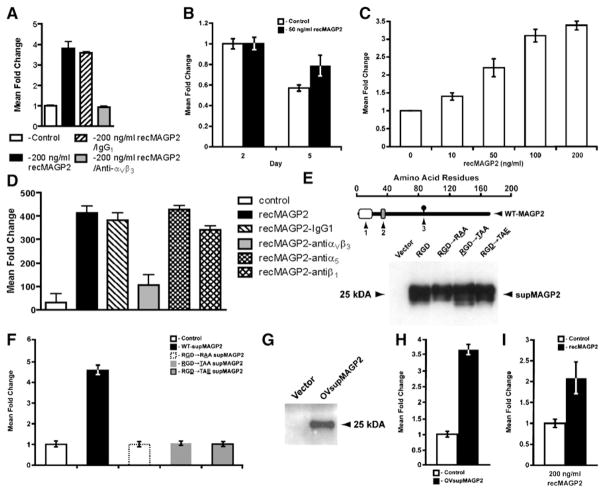
Figure 4. MAGP2 Is Expressed in Ovarian Cancer Cells and Enhances Adhesion and Survival In Vitro.
(A) Western blot analysis of MAGP2 protein isolated from 2 normal HOSE cultures and 12 ovarian cancer cell lines.
(B) Flow cytometry analysis of αVβ3 cell surface receptor levels in select high and low MAGP2-expressing cell lines using monoclonal antibodies against αVβ3 or IgG1 isotype control.
(C) Silver staining for total protein obtained from yeast clone 10 supernatant identifies recMAGP2 as the principal secreted product following purification.
(D) Increased adhesion of A224 ovarian cells in the presence of purified recMAGP2 (p < 0.02), with reduced adhesion following anti-αVβ3 integrin antibody treatment (p < 0.01).
(E) αVβ3 integrin receptor-negative UCI107 cells displayed no significant increase in adhesion in the presence of recMAGP2.
(F) OVCA429 cell viability was evaluated with increasing concentrations of recMAGP2. Increased cell survival was seen at higher concentrations of recMAGP2 reaching a maximum at 200 ng/ml (p = 0.004). For (E) and (F), data are the mean ± SD of three independent experiments.
(G) Survival signaling in OVCA429 cells in response to treatment with recMAGP2. Differentially regulated genes were identified (red, upregulated; blue, downregulated; gray, no change).
To test whether MAGP2 can modulate tumor cell biology, we synthesized purified recombinant MAGP2 (recMAGP2) using a yeast expression system (Figure 4C). Given that MAGP2 has been shown to induce adhesion in a number of different cell types via the αVβ3 integrin receptor (Gibson et al., 1999), we selected this endpoint to verify the biological activity of the construct. The effect of recMAGP2 on cellular adhesion was evaluated for A224 and UCI107 cell lines. When plated on recMAGP2-coated wells, A224 cells displayed an increase in adhesion (Figure 4D), relative to control wells. Preincubation of A224 cells with anti-αVβ3 integrin-blocking antibody resulted in a decrease in adhesion on recMAGP2-coated wells (Figure 4D), whereas pretreatment with control IgG1 antibody had no effect. In contrast, UCI107 showed no increase in adhesion at high concentrations of recMAGP2, further demonstrating the suitability of recMAGP2 for biological evaluation (Figure 4E).
Based on the pathway analysis, αVβ3-mediated signaling was implicated in ovarian tumor cell survival through the stimulation of critical cell-cycle checkpoint regulators. To assess this observation, we cultured OVCA429 cells under increasing concentrations of purified recMAGP2. At 96 hr, a significant difference in survival was observed for cells treated with 200 ng/ml of recMAGP2 in serum-free media (Figure 4F). This observation substantiated the pathway, suggesting that differential patient survival may be attributed in part to enhanced tumor cell survival through MAGP2-induced signaling events.
RecMAGP2 Stimulates the Adhesion, Survival, Migration, and Invasion of Human Umbilical Vein Endothelial Cells
As indicated in Figure 2A, we hypothesized that secreted MAGP2 may modulate the biology of surrounding endothelial cells ultimately promoting neovascularization. αVβ3 integrin is expressed on endothelial cells and has been associated with tumor angiogenesis (Ellis et al., 2003). To probe these biological effects, we cultured human umbilical vein endothelial cells (HUVECs) in the presence of recMAGP2.
As observed for A224 ovarian cancer cells, HUVECs plated on recMAGP2 showed a 3.5-fold increase in cellular adhesion (Figure 5A). This effect was blocked by pretreatment with an anti-αVβ3 integrin antibody, whereas control IgG1 antibody had no effect (Figure 5A). This suggests that recMAGP2 also exerts its biological activity through the αVβ3 integrin receptor in HUVECs. To determine the effect of recMAGP2 on survival, HUVECs were grown under low serum conditions. Cells grown in the absence of recMAGP2 for 5 days displayed a reduction (25%) in survival, which was partially restored in the presence of recombinant protein (Figure 5B).
Figure 5. RecMAGP2 Stimulates Adhesion, Survival, Motility, and Invasion of HUVECs In Vitro.
(A) HUVECs cultured on recMAGP2 showed increased adhesion (p < 0.0002) when compared to anti-αVβ3 integrin antibody preincubation.
(B) Enhanced survival of HUVECs treated with recMAGP2 (p < 0.05).
(C) Dose-dependent HUVEC motility in response to recMAGP2 protein was significant at all concentrations evaluated (p ≤ 0.01).
(D) Anti-αVβ3 integrin antibody attenuated re-cMAGP2-stimulated HUVEC motility through αVβ3 receptor inhibition (p < 0.002), whereas anti-α5 and anti-β1 integrin antibody treatments did not.
(E) Successful processing and secretion of wild-type supMAGP2 and RGD motif mutant proteins by 293T cells (1, signal sequence; 2, RGD motif; 3, N-linked carbohydrate consensus sequence; the underlined letter refers to the mutated RGD motif amino acid).
(F) supMAGP2 stimulated HUVEC motility (p = 0.002), whereas the three mutant constructs had no significant effect.
(G) Proper processing and secretion of wild-type OVsupMAGP2 protein by UCI107 ovarian cancer cells.
(H) Ovarian cancer cell-derived OVsupMAGP2 induced HUVEC motility (p < 0.001).
(I) Increased invasion of recMAGP2-treated HU-VECs into matrigel, as compared to untreated cells (p < 0.05). For (A)–(D), (F), (H), and (I), the data represents the mean ± SD of three independent experiments.
The effect of recMAGP2 on HUVEC motility and invasion was also evaluated. Application of recMAGP2 increased cell motility in a dose-dependent manner, as compared with cells incubated with medium alone, for all of the concentrations evaluated (Figure 5C). The addition of anti-αVβ3 antibody in the presence of recMAGP2 attenuated cell motility (Figure 5D). Given that other integrin protein family members also possess the ability to bind proteins that possess an RGD motif, including the α5 and β1 receptors, their contribution to re-cMAGP2-induced motility was determined (Takada et al., 2007). Relative to IgG1, neither anti-α5 nor anti-β1 antibodies had a significant impact on recMAGP2-stimulated HUVEC motility, suggesting that signaling is specific to the αVβ3 receptor (Figure 5D). To confirm that the RGD motif of MAGP2 is required for its biological activity, we evaluated supernatants containing secreted MAGP2 (supMAGP2) from 293T cells transfected with wild-type MAGP2 or one of three RGD motif mutant constructs (Figure 5E). Only wild-type supernatant stimulated HUVEC migration (Figure 5F). Finally, wild-type MAGP2 was transfected into ovarian cancer cell line UCI107, which does not express MAGP2, so that the ability of an ovarian tumor cell to successfully process and secrete functional protein could be verified (Figure 5G). Application of MAGP2-enriched tumor cell-derived supernatant (OVsupMAGP2) induced HUVEC motility compared to the vector control (Figure 5H).
In addition to survival and motility, exposing HUVECs seeded onto Matrigel matrix to reMAGP2 stimulated a 2-fold increase in invasion (Figure 5I). Summation of these results implicates the RGD motif of MAGP2 as a potent modulator of endothelial cell behavior acting through the αVβ3 receptor, which may contribute to angiogenesis, affecting patient survival.
Identification of Signaling Events Contributing to the Effect of Recombinant MAGP2 on HUVECs
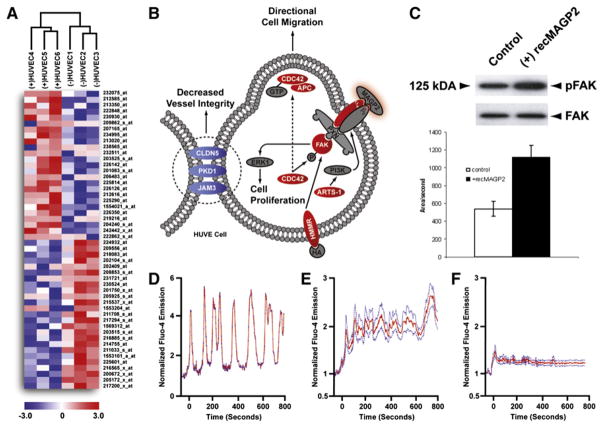
The marked effect of recMAGP2 on HUVEC behavior implied a change in cellular regulation had taken place, affecting gene expression (Figure 6A). To explore the consequences of recMAGP2-induced signaling, we completed microarrays for a series of recMAGP2-treated (n = 3) and untreated (n = 3) HUVEC isolates. A total of 274 probe sets were identified for pathway analysis in cells exposed to recMAGP2 (Figure 6B). Among them were ITGAV, FAK, and CDC42, suggesting that MAGP2-stimulated signaling may induce the expression of key effectors enhancing its activity in HUVECs. In addition, a number of genes involved in the formation of endothelial cell tight junctions including CLDN5, PKD1, and JAM3 were downregulated, whereas motility genes CDC42 and APC were upregulated. Deregulation of these genes may lead to a reduction in vessel integrity, permitting endothelial cell migration and neovascularization.
Figure 6. Identification of Putative Signaling Events Mediating the Effects of MAGP2 on HUVECs.
(A) Hierarchical clustering of recMAGP2-treated (n = 3) and untreated (n = 3) HUVECs following supervised class comparison. The top 50 differentially regulated probe sets are displayed (red, upregulated; blue, downregulated).
(B) Signaling events stimulated in HUVECs treated with recMAGP2 as compared to untreated cells. Differentially regulated genes were identified (red, upregulated; blue, downregulated; gray, no change).
(C) recMAGP2 treatment of HUVECs increased the proportion of phosphorylated FAK, as compared to untreated cells. Error bar represents the area/second mean ± SD of three independent experiments.
(D) Time course changes in [Ca2+] levels in a single recMAGP2 induced in a single HUVEC.
(E) Mean normalized time course changes in [Ca2+] induced by recMAGP2 in HUVECs.
(F) Mean normalized time course of changes in [Ca2+] induced by recMAGP2 in HUVECs pre-treated with GRGDSP competitor peptide.
Given that FAK is a critical mediator of αVβ3 signaling, we confirmed its phosphorylation status in recMAGP2-treated cells. With a phosphorylation-specific antibody recognizing tyrosine 397, a western blot comparing phospho-FAK to total FAK was completed for both treated and untreated HUVEC isolates (Figure 6C). In cells treated with the recombinant protein, there was a 2-fold increase in phosphorylated FAK, indicating elevated levels of activated protein, whereas no change was seen in untreated cells. Furthermore, treatment of UCI107 ovarian cancer cells, which are null for αVβ3, and HUVEC cells in the presence of blocking antibody to αVβ3 with recMAGP2 resulted in no activation of FAK (Figure S1).
In order to identify the mechanism underlying the stimulation of FAK by recMAGP2, we assessed Ca2+ oscillation in treated HUVECs. Ca2+ mobilization and oscillation has been associated with the activation of FAK in motile cells (Giannone et al., 2004). recMAGP2 induced an immediate increase in [Ca2+]I levels (Figures 6D and 6E). When cells were pretreated with a synthetic RGD peptide, the effect of recMAGP2 on [Ca2+]I levels was severely diminished (Figure 6F). Furthermore, the addition of BAPTA/AM to HUVEC cells treated with recMAGP2 inhibits the activation of FAK (Figure S2). These data imply that FAK is activated through Ca2+ mobilization in response to recMAGP2 in HUVECs. This may account in part for the enhanced motility and survival of HUVECs described in the pathway analysis.
In parallel with these experiments, we sought to determine the mechanism(s) of MAGP2-induced survival of ovarian cancer cells. To explore the consequences of recMAGP2-induced survival, we completed microarrays for a series of recMAGP2-treated (n = 3) and untreated (n = 3) OVCA429 cells. A total of 389 probe sets were identified as differentially expressed under recMAGP2 treatment (Table S3). As one might expect, there was overlap between this gene list and that obtained by exposure of HUVEC cells to recMAGP2. A pathway was identified that again centered on fak but included other important survival-related genes such as cflar (Figure 4G). Cflar is upregulated by recMAGP2 and in turn this gene inhibits and regulates multiple important mediators of apoptosis.
MAGP2 Expression Is Necessary for In Vivo Tumor Size Growth
To investigate the effects of decreased MAGP2 expression on ovarian tumor growth and microvessel density in vivo, we “knocked down” MAGP2 expression in SKOV3 clones by stably transfecting the cells with MAGP2 shRNA (Figure 7A). Four clones with decreased MAGP2 along with two vector control clones were injected into nude mice. After tumor development, the animals were sacrificed and tumors were removed and weighed. Tumors with reduced levels of MAGP2 were significantly smaller than those that developed from control cells (Figure 7B). To validate these results, we used lentiviral particles expressing shRNA to knock down MAGP2 expression in both SKOV3 and OVCAR3 cell lines. Pools of clones were isolated, each pool expressing a MAGP2 shRNA targeting a different region of the MAGP2 gene sequence. Reduced expression of MAGP2 was seen for all pools compared with controls (nontargeting shRNA) and this resulted in decreased tumor size (Figure S3).
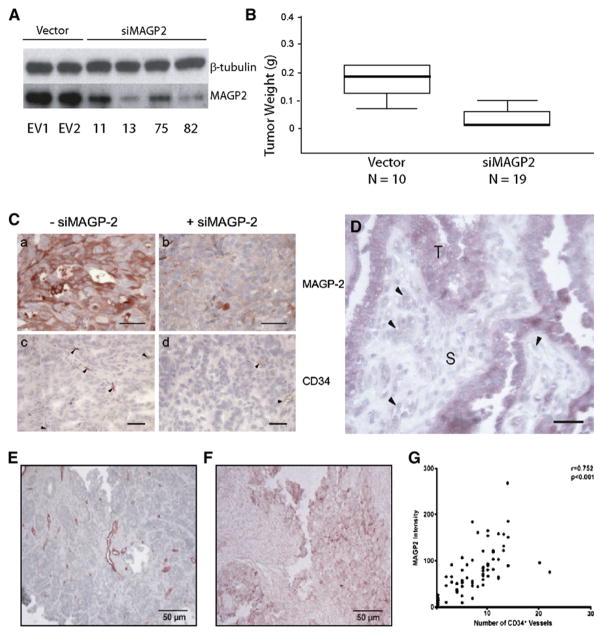
Figure 7. MAGP2 Expression Correlates with Tumor Size and CD34+ Microvessel Density in Serous Ovarian Cancer Tumors and Tissue Specimens.
(A) Western blot demonstrated knockdown of MAGP2 in five clones isolated from stably transfected SKOV3 cells, as well as three empty vector clones.
(B) MAGP2 knockdown resulted in significantly decreased tumor size in mice. The median weight of tumors from SKOV3 cells is 0.18 g; knockdown of MAGP2 results in a significant decrease in tumor weight (0.02 g; p < 0.01).
(C) MAGP2 knockdown resulted in decreased MAGP2 expression and CD34+ microvessel density in tumors developed in mice. Scale bars represent 50 μm.
(D) A section of a high-grade serous adenocarcinoma showing strong MAGP2 expression in tumor cells (T) but not in the stromal component (S) or the endothelial cells (arrowheads). The scale bar represents 100 μm.
(E) Immunolocalization of CD34+ microvessels in a human serous ovarian adenocarcinoma tumor section.
(F) MAGP2 expression in a late-stage high-grade human serous ovarian adenocarcinoma tumor section.
(G) Correlation between CD34+ and MAGP2 expression.
Microvessel density in the tumors was measured by immunolocalization of CD34+. A decrease in MAGP2 expression resulted in decreased CD34+ microvessel density (Figure 7C). It is important to note that these ovarian tumors express MAGP2 specifically in the tumor epithelial cells and not the stromal or endothelial cells (Figure 7D).
MAGP2 Expression Is Significantly Correlated with CD34 Expression in Serous Ovarian Cancer
Given the pronounced biologic effect of MAGP2 on endothelial cells, we hypothesized that tumors with elevated MAGP2 expression would display features indicative of a proangiogenic microenvironment. To evaluate whether MAGP2 may induce angiogenesis in ovarian tumors, MAGP2 expression was correlated with microvessel density in 30 human advanced serous cancers. MAGP2 expression was determined by immunolocalization of the MAGP2 protein with an anti-MAGP2 polyclonal antibody, whereas microvessel density was assayed for by immunolocalization of CD34+ microvasculature within the tissue with an anti-CD34 monoclonal antibody (Figures 7E and 7G). A significant correlation between MAGP2 expression and CD34+ microvessel density was detected (Figure 7F, p = 0.009). These data suggest that MAGP2 may play a role in ovarian tumor neovascularization in vivo.
DISCUSSION
Patients suffering from advanced stage ovarian cancer often display a heterogeneous clinical course with overall survival spanning less than 5 months to beyond 150. The development of genomic classifiers will aid in the clinical management of the disease, as well as identify new opportunities for therapeutic intervention. The work reported here describes a validated gene expression signature correlating with survival in patients with advanced serous ovarian cancer. To biologically validate the signature, we have characterized the gene possessing the highest Cox score, a secreted factor, MAGP2, and demonstrated that the protein promotes ovarian tumor cell survival as well as endothelial cell survival, motility, and invasion. Furthermore, expression of MAGP2 correlates with microvessel density in ovarian cancer.
Transcription profiling is widely used to derive molecular signatures associated with ovarian cancer (Bonome et al., 2005; Wamunyokoli et al., 2006). These signatures have been correlated with histology, grade, and stage (Berchuck et al., 2005; Bonome et al., 2005; Wamunyokoli et al., 2006). However, only a few methods have been proposed to analyze survival outcome using microarray data. The first approach uses unsupervised hierarchical clustering followed by an assessment of survival distributions among clusters using traditional approaches, e.g., log rank test; however, our samples were all of the same histological subtype (Sorlie et al., 2001). Furthermore, subgroups in this cancer type have not been reported precluding use of a cluster-based strategy. The second genre of analysis reduces data dimensionality by clustering gene expression including partial least-squares or support vector machines (Nguyen and Rocke, 2002). These methods use a combination of all the genes to predict survival by adding noise to the model, since the vast majority of the genes should be unrelated to survival. Indeed, studies have shown that using only a subset of genes generally performs better than using all genes (Bair and Tibshirani, 2004). Recent work by Berchuck et al. (2005), as well as other groups, has used this type of approach to identify genes associated with long-term survival in serous ovarian cancer (Lancaster et al., 2004; Spentzos et al., 2004). However, it remains unclear whether these predictors are applicable to the majority of patients who will eventually die at an intermediate time point. In addition, their algorithms did not account for censoring age and/or debulking status and were derived from undissected isolates containing intervening stroma-associated cells.
To overcome these limitations, our approach relied on a Cox proportional hazards model evaluating survival as a continuous variable relative to gene expression for a cohort of serous epithelial cell tumor isolates (Cox, 1972). This statistical method incorporated censoring age and debulking variables returning a hazard ratio measuring each gene’s effect on survival, rather than scoring a multigene classifier. In this way, top survival-associated genes were easily selected for PC analysis and evaluated as a complex multigene predictor applicable to a broad range of survival endpoints.
Despite clear delineation and validation of the 200 probe set classifier, the broad range of genes that were identified precluded unaided identification of coregulated signaling pathways. By using pathway analysis software, we identified αVβ3 activation as a prominent survival-associated event. On the basis of the expression data, the pathway was activated on average across the panel of tumors that were evaluated versus OSE, including a number of survival-associated genes. However, in patients surviving less than 1 year, there was a pronounced overexpression of multiple prognostic genes, as compared with individuals surviving beyond 5 years. This implies that these genes may function as critical regulators suitable for further evaluation as therapeutic targets.
Motivated by these findings, we used the data to characterize the top survival-associated signature gene MAGP2. This approach not only provided a biological validation of the prognostic list but also demonstrated a link between protein expression and a number of tumor-associated processes. MAGP2 is a 25 kDa matrix glycoprotein, which was originally identified by its coextraction from developing fetal nuchal ligament tissue (Gibson et al., 1996, 1989). The C-terminal half of MAGP2 interacts with fibrillin-1 and -2, as well as fibulin-1, another component of elastic fibers, suggesting that MAGP2 is an important component in the assembly of microfibrils (Penner et al., 2002). The N-terminal half of MAGP2 contains an RGD motif that has been shown to mediate adhesion and spreading of fetal bovine aortic smooth-muscle cells, ear cartilage chondrocytes, human skin fibroblasts, and osteoblasts through the αVβ3 integrin receptor (Gibson et al., 1999). However, it has never been identified or evaluated in the context of ovarian cancer.
Increased MAGP2 copy number supports the notion that MAGP2 may play a causative role in the development of the disease, and qRT-PCR and IHC both validated its prognostic value. Interestingly, MAGP2 levels were also associated with chemosensitivity. There is evidence that αVβ3 signaling may contribute to chemoresistance in ovarian cancer (Maubant et al., 2002). In addition, chromosomal instability has been linked to poor response to therapy (Salisbury et al., 2004). Survival-associated NEDD9, which acts downstream of the αVβ3 integrin receptor, can activate both STK6/Aurora-A and NEK2, possibly leading to aberrant chromosomal separation (Pugacheva and Golemis, 2005). This suggests a mechanism whereby MAGP2 may be stimulating chemoresistance through a second prognostic gene.
Beyond its clinical correlation, pathway analysis also suggested a role for MAGP2 in ovarian tumor cell survival and endothelial motility and survival. Indeed, treatment of both cell types with recombinant and secreted supernatant proteins substantiated these hypotheses. In endothelial cells, dissolution of tight junctions was evidenced by downregulation of PDK1, CLDN5, and JAM3. These events may ultimately lead to decreased vessel integrity and enhanced endothelial cell motility (Ebnet et al., 2003; Kim et al., 2000; Morita et al., 1999). Increased FAK phosphorylation, as well as calcium oscillation, also supports the notion that MAGP2 can actuate αVβ3 integrin receptor signaling and promote motility.
Finally, correlation of MAGP2 expression with microvessel density provided an in vivo indication that the cell line data is of clinical relevance. This raises the potential that MAGP2 is a proangiogenic factor in ovarian cancer. In addition, localization to the extracellular matrix makes it an attractive target for therapeutic intervention. Without sufficient blood supply, tumor expansion is severely limited. Furthermore, targeting a factor affecting an endothelial cell with presumed genetic stability may increase response. Ultimately, the development and expansion of serous ovarian cancer involves a host of complex biological processes. Nevertheless, this work identified at number of prognostic genes and a coregulated regulated pathway that may directly contribute to patient outcome. Among these genes, MAGP2 featured prominently as a mediator of survival. Subsequent analyses will be necessary to confirm its proangiogenic role as well as to assess the contribution of other survival-associated genes.
EXPERIMENTAL PROCEDURES
Tissue Samples
A total of 53 snap-frozen tissue specimens and a validation set of 64 paraffin-embedded tumor samples were obtained from previously untreated ovarian cancer patients, who were hospitalized at the Brigham and Women’s Hospital between 1990 and 2000. All patients had advanced stage, high-grade serous ovarian cancer according to the International Federation of Gynecology and Obstetrics standards. All specimens and their corresponding clinical information were collected under protocols approved by the institutional review boards of the corresponding institutions (for details, see tumor specimens section in Supplemental Experimental Procedures) and informed consent was obtained from all subjects.
Microdissection, Total RNA Extraction, Affymetrix GeneChip Hybridization, and Image Acquisition
RNA was isolated and amplified from microdissected specimens as described previously (Bonome et al., 2005). Hybridization, processing, and image acquisition was done as described previously (Bonome et al., 2005).
To demonstrate that pure populations of epithelial cells were obtained, we checked our arrays for the expression of endothelial cell markers (TIE-2 and VEGFR2) and T cell markers (CD8 and CD45). None of the markers we examined were expressed on our arrays (see Figure S4).
Microarray Survival Analysis
Low-level analysis included array normalization and estimation of expression level using an invariant set of probe sets to adjust the overall signal level of the arrays to the same level (Sorlie et al., 2001). Next, a model-based PM-only approach established gene expression levels using dChip software (Li and Wong, 2001).
A modified “semi-supervised” method was applied (Bair and Tibshirani, 2004) in two stages: (1) Supervised dimension reduction was done by fitting a univariate Cox model that included a jackknifing step to each gene, so only those genes with a consistently large Cox score were included in the signature. Among 200, 100, or 300 probe sets, 200 yielded an optimally sized predictor. (2) In stage 2, the dimensions of the data set were reduced from 200 to 5 by PC analysis. The number of PCs was set at five, capturing 90% of the expression data. Four or six PCs were investigated to see whether this parameter affected the results. A prediction model was then built using multivariate Cox regression, where independent variables included the first five PCs and debulking status.
Standard leave-one-out validation evaluated the prediction model reserving one sample for testing and using the remaining 52 patients to establish the prediction model following the method described above. The reserved patient had no contribution to the prediction model, yielding 53 separate predicted hazards. Patients were equally divided into low- and high-risk groups according to whether their hazard was less or greater than the sample median and were compared by Kaplan Meier analysis with a nonparametric log rank test.
Microarray Pathway Analysis
The dChip PM-only algorithm was applied to the normal OSE and HUVEC line data. Biometric Research Branch ArrayTools version 3.2.2 software developed by R. Simon and A. Peng Lam of the Biometrics Research Branch of the National Cancer Institute was used to filter and complete the statistical analysis. Only those probe sets present in greater than 50% of the arrays and displaying a variance in the top 50th percentile were evaluated.
Differentially expressed genes were identified for tumor and OSE specimens using a multivariate permutation t test (p < 0.001). A total of 2000 permutations were completed to identify a list of probe sets containing fewer than 5% false positives at a confidence interval of 90%. For the HUVEC isolates, differentially regulated genes were identified by a univariate t test (p < 0.01 and fold change >1.5). A random-variance model was selected to permit information sharing among probe sets.
Coregulated pathways underlying the survival-associated gene expression signature and HUVEC signaling were analyzed using PathwayStudio version 4.0 software (Ariadne Genomics).
qPCR Analyses
Real-time PCR was performed as previously described (Wamunyokoli et al., 2006). To calculate the relative expression for each gene, we used the 2−ΔΔCT method to average the CT values for the three housekeeping genes for a single reference gene value (Livak and Schmittgen, 2001).
Comparative Genome Hybridization and qPCR Validation
DNA from tumor cells from 42 of the late-stage, high-grade serous adenocarcinomas evaluated by microarray gene expression analysis were analyzed for copy number alterations as previously described (Birrer et al., 2007). qPCR validation of MAGP2 copy number was completed as previously described (Birrer et al., 2007).
MAGP2 Survival and Chemoresponse Immunohistochemical Analyses
The 53 stage III/IV grade 3 serous cases evaluated by gene expression analysis, five normal ovaries, and five serous cystadenoma cases were used. Immunolocalization and quantification of MAGP2 was performed using a polyclonal anti-MAGP2 antibody at a dilution of 1:1000 and the ImagePro Plus 5.1 software (Media Cybermetics) was used as previously described (Birrer et al., 2007). The results were compared with matched survival data and examined by Kaplan Meier survival analysis. Statistical significance was determined by log rank test.
An independent validation set of 64 advanced stage high-grade papillary serous tumors comprising a TMA were quantified for MAGP2 expression using the antibody, instrumentation, and scoring method described above. Protein expression levels were compared to overall survival by multivariate Cox regression analysis adjusting for debulking status. p < 0.05 was considered significant.
Synthesis of Recombinant and Mutant MAGP2 Constructs
Full-length human MAGP2 cDNA was amplified from Image Clone 377692 (primers in Supplemental Experimental Procedures) and cloned into the EcoRI/NotI sites of yeast vector pPICZα(A) of the EasySelect Pichia expression kit according to the manufacturer’s protocol (Invitrogen). For large-scale growth, individual clones were induced with 0.5% methanol in BMMY media. Recombinant protein in the supernatant was purified with the Ni+-NTA purification system (Invitrogen) under native binding conditions. Isolation of re-cMAGP2 was verified by western blotting after elution in 250 mM imidazole and was quantified by silver staining (Invitrogen).
MAGP2 RGD motif mutants were constructed using the GeneEditor in vitro site-directed mutagenesis system (Promega), according to the manufacturer’s protocol. Three mutants were synthesized, including RGD→RAA, RGD→TGD, and RGD→RGE (primers in Supplemental Experimental Procedures). pcDna3.1(−):MAGP2 was used as the template.
Cell Lines and Culture Conditions
Human ovarian adenocarcinoma cell lines A224, OVCAR3, SKOV3, and UCI107, OVCA429, and 293T cells were grown in RPMI-1640 or DMEM, supplemented with 10% FBS, 2 mM glutamine, and penicillin/streptomycin (Invitrogen). HUVECs (Cambrex) were grown in EBM supplemented with 2% FBS and EGM-2 MV SingleQuots (Cambrex). All cells were maintained in exponential growth phase at 37°C in 5% CO2.
Western Blot Analysis
Protein lysates from ovarian cancer and OSE cultures were separated on 4%–12% SDS gels (Amersham Biosciences) and incubated with an anti-MAGP2 antibody (Rockland Inc.). For assessing FAK phosphorylation in HUVECs, cells were plated on 60 mm2 dishes and treated with 100 ng/ml recMAGP2 protein for 30 min or with elute buffer control. For testing Ca2+ dependence, 50 μM of the calcium signal blocking reagent BAPTA/AM (Molecular Probes) was added to the HUVECs for 30 min before treatment with recMAGP2. Western blotting was then performed on cell lysates under denaturing conditions on a 4%–20% NuPAGE Tris-Glycine gel with a 1:1000 dilution of anti-FAK[pY397] mouse monoclonal primary antibody (BD Biosciences) followed by a 1:20,000 dilution of goat anti-mouse HRP-conjugated secondary antibody (Amersham Biosciences). Bands were detected via ECL (Amersham Biosciences).
Flow Cytometry
Flow cytometry analysis was performed with anti-αVβ3 integrin (23C6) FITC conjugate and negative control IgG1 FITC conjugate (Santa Cruz Biotechnology). Cells were analyzed on a FACSAria Cell Sorting System (BD Biosciences).
In Vitro Cell Line Assays
A224, UCI107, and HUVEC adhesion was assayed in 96-well plates were treated with 30 μl recMAGP2 at the desired concentration for 18 hr at 4°C. The cells were suspended in serum-free media (1 mg/ml BSA) and plated in quadruplicate in the pretreated 96-well plates and observed every 5 min. Once adhesion/spreading was observed in treated wells, the plate was shaken for 30 s on an automatic plate shaker to remove unattached cells. The medium was removed, washed once with PBS, and replaced with fresh medium. Cell number was determined by MTT (Promega). Blocking of adhesion was performed by treating cells with antibody for 30 min in serum-free media with 1 mg/ml BSA before plating [anti-integrin αVβ3 (LM609) and IgG1 (MABC002) (Chemicon)].
For HUVEC motility assays, cells were plated in triplicate in 350 μl in the upper chamber of a 8.0 μM pore size PET membrane 24-well cell culture insert (BD Biosciences). The bottom chamber contained 500 μl of 0.2% FBS media with 100 ng/ml recMAGP2 as a chemoattractant. After cells attached for 3 hr, the insert was removed and the cells on the underside were fixed and stained with Diff-Quick Stain Set (Dade Behring). Cell number was quantified by counting five 200× fields of view per membrane.
Blocking of motility was performed as above, with a 30 min antibody pretreatment using anti-αVβ3 integrin (LM609), anti-α5 integrin (IIA1), anti-β1 integrin (4B4) (BD Biosciences), or IgG1 (MABC002) antibodies.
HUVEC motility was also assessed using supernatant from 293T or UCI107 cells transfected with pcDNA3.1(−):MAGP2 or RGD motif mutants. Each cell line was plated in 100 mm2 dishes O/N in DMEM or RPMI, respectively, transfected with 3 μg DNA O/N, and cultured in new media containing 0.2% EBM for 36 hr.
For HUVEC invasion, inserts were pretreated with 100 μl Matrigel (BD Biosciences) at 0.5 mg/ml in the dark for 18 hr at RT. The bottom chamber contained 200 ng/ml recMAGP-2. Cells were allowed to invade for 24 hr before staining.
HUVEC survival was evaluated by plating cells in quadruplicate in 96-well plates at 25% confluence in 100 μl of 0.2% EBM. recMAGP2 was added on day 0 and 2 at 50 ng/ml. Cell number was quantified by MTT on day 5.
Measurement of HUVEC Ca2+ Oscillation
The effect of recMAGP2 on [Ca2+]i of cultured ovarian HUVECs were monitored with confocal fluorescence microscopy using Fluo-4/AM (a cell-permeable calcium-sensitive fluorescence dye). Cultured cells on coverslips were loaded with Fluo-4/AM (5 M) in phenol-red-free culture medium for 20 min, washed, and incubated for another 20 min for deesterification. Coverslips were then positioned in a temperature-controlled perfusion chamber mounted on a Zeiss inverted microscope. Confocal fluorescence images were collected with a Bio-Rad MRC-1000 confocal scanning unit at 0.5 HZ through a 40× objective (NA 1.2). recMAGP2 (100 ng/ml) was introduced into the perfusion chamber through the inlet port. Changes in emission intensity of Fluo-4 from individual cell induced by recMAGP2 were measured retrospectively from each image.
In Vivo Studies
Ovarian cancer cells, SKOV3, were stably transfected with MAGP2 shRNA or the vector. Five stable clones were selected for each group. For lentivirus experiments, four different viral transduction particles were used: three expressing shRNA targeting different regions of MAGP2 and one expressing a nontargeting shRNA. SKOV3 and OVCAR 3 were transduction as per the vendor’s instruction (Sigma-Aldrich). Pools of resistant clones were isolated.
The animal protocol is approved by the Institutional Animal Care and Use Committee at the University of Texas M.D. Anderson Cancer Center. A total of 5 × 105 cells from each clone or pool were injected subcutaneously into the posterior neck region of five nude mice (6- to 8-week-old female nude mice; Charles River). After 4 weeks, the mice were sacrificed and the tumors developed from each mouse were removed and weighed. The clones were subsequently fixed in formalin and processed for histological evaluation. MAGP2 expression in tumor tissues was evaluated using a rabbit anti-human MAGP-2 antibody (Rockland Inc.) and the microvessel density was determined by immunolocalization of CD34+ blood vessels using a goat anti-mouse polyclonal antibody. Significant differences in the weight of the tumors between the MAGP2 shRNA and the control group were determined by Mann-Whitney U test. A p-value < 0.05 was considered significant.
Evaluation of CD34+ Microvessel Density
Immunolocalization of CD34+ endothelial cells was performed on 30 of the tumors included in the microarray analysis using formalin-fixed paraffin-embedded tissue with a commercially available anti-CD34 monoclonal antibody (Dako Cytomation) and the ABC system (Vector Laboratories). Grading of CD34-positive blood vessels was performed as previously described (Birrer et al., 2007). A Spearmen’s rho tested the correlation between MAGP2 expression with CD34+ microvessel density.
SIGNIFICANCE.
Ovarian cancer is the most lethal gynecologic cancer in the United States, yet little is known about the molecular events impacting survival. Thus, efforts to stratify patients for therapy and identify therapeutic targets have been severely hampered. We have now identified and confirmed a gene expression signature correlating with poor survival in microdissected advanced serous ovarian tumors. In addition, we have completed an initial characterization of a proangiogenic factor associated with prognosis, MAGP2, which can promote tumor epithelial cell survival and stimulate endothelial cell motility and survival. Correlation of MAGP2 with microvessel density suggests a role in neovascularization in vivo. This work has developed a prognostic gene signature of biological significance and identified a putative ovarian cancer target.
Supplementary Material
Acknowledgments
This study was supported in part by Dana-Farber Ovarian Cancer SPORE grant P50CA105009, R33CA103595, and RO1CA133057 from the National Institutes of Health, Department of Health and Human Services, the Ovarian Cancer Research Fund, Inc., and the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
ACCESSION NUMBERS
Microarray and CGH data described herein are available at NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE18521 and GSE18154.
Supplemental Data include Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online at http://www.cell.com/cancer-cell/supplemental/S1535-6108(09)00386-9.
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11:3686–3696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, Welch WR, Berkowitz RS, Mok SC. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- Boente MP, Hamilton TC, Godwin AK, Buetow K, Kohler MF, Hogan WM, Berchuck A, Young RC. Early ovarian cancer: A review of its genetic and biologic factors, detection, and treatment. Curr Probl Cancer. 1996;20:83–137. [PubMed] [Google Scholar]
- Bonome T, Lee JY, Park DC, Radonovich M, Pise-Masison C, Brady J, Gardner GJ, Hao K, Wong WH, Barrett JC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- Boring CC, Squires TS, Tong T, Montgomery S. Cancer statistics, 1994. CA Cancer J Clin. 1994;44:7–26. doi: 10.3322/canjclin.44.1.7. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life tables (with discussion) Journal of the Royal Statistical Society, Series B (Statistical Methodology) 1972;34:187–220. [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Ebnet K, Aurrand-Lions M, Kuhn A, Kiefer F, Butz S, Zander K, Meyer zu Brickwedde MK, Suzuki A, Imhof BA, Vestweber D. The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: A possible role for JAMs in endothelial cell polarity. J Cell Sci. 2003;116:3879–3891. doi: 10.1242/jcs.00704. [DOI] [PubMed] [Google Scholar]
- Ellis PD, Metcalfe JC, Hyvonen M, Kemp PR. Adhesion of endothelial cells to NOV is mediated by the integrins alphavbeta3 and alpha5-beta1. J Vasc Res. 2003;40:234–243. doi: 10.1159/000071887. [DOI] [PubMed] [Google Scholar]
- Giannone G, Ronde P, Gaire M, Beaudouin J, Haiech J, Ellenberg J, Takeda K. Calcium rises locally trigger focal adhesion disassembly and enhance residency of focal adhesion kinase at focal adhesions. J Biol Chem. 2004;279:28715–28723. doi: 10.1074/jbc.M404054200. [DOI] [PubMed] [Google Scholar]
- Gibson MA, Kumaratilake JS, Cleary EG. The protein components of the 12-nanometer microfibrils of elastic and nonelastic tissues. J Biol Chem. 1989;264:4590–4598. [PubMed] [Google Scholar]
- Gibson MA, Hatzinikolas G, Kumaratilake JS, Sandberg LB, Nicholl JK, Sutherland GR, Cleary EG. Further characterization of proteins associated with elastic fiber microfibrils including the molecular cloning of MAGP-2 (MP25) J Biol Chem. 1996;271:1096–1103. doi: 10.1074/jbc.271.2.1096. [DOI] [PubMed] [Google Scholar]
- Gibson MA, Leavesley DI, Ashman LK. Microfibril-associated glycoprotein-2 specifically interacts with a range of bovine and human cell types via alphaVbeta3 integrin. J Biol Chem. 1999;274:13060–13065. doi: 10.1074/jbc.274.19.13060. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Kim K, Drummond I, Ibraghimov-Beskrovnaya O, Klinger K, Arnaout MA. Polycystin 1 is required for the structural integrity of blood vessels. Proc Natl Acad Sci USA. 2000;97:1731–1736. doi: 10.1073/pnas.040550097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JM, Dressman HK, Whitaker RS, Havrilesky L, Gray J, Marks JR, Nevins JR, Berchuck A. Gene expression patterns that characterize advanced stage serous ovarian cancers. J Soc Gynecol Investig. 2004;11:51–59. doi: 10.1016/j.jsgi.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maubant S, Cruet-Hennequart S, Poulain L, Carreiras F, Sichel F, Luis J, Staedel C, Gauduchon P. Altered adhesion properties and alphav integrin expression in a cisplatin-resistant human ovarian carcinoma cell line. Int J Cancer. 2002;97:186–194. doi: 10.1002/ijc.1600. [DOI] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: Claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DV, Rocke DM. Partial least squares proportional hazard regression for application to DNA microarray survival data. Bioinformatics. 2002;18:1625–1632. doi: 10.1093/bioinformatics/18.12.1625. [DOI] [PubMed] [Google Scholar]
- Penner AS, Rock MJ, Kielty CM, Shipley JM. Microfibril-associated glycoprotein-2 interacts with fibrillin-1 and fibrillin-2 suggesting a role for MAGP-2 in elastic fiber assembly. J Biol Chem. 2002;277:35044–35049. doi: 10.1074/jbc.M206363200. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–946. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, D’Assoro AB, Lingle WL. Centrosome amplification and the origin of chromosomal instability in breast cancer. J Mammary Gland Biol Neoplasia. 2004;9:275–283. doi: 10.1023/B:JOMG.0000048774.27697.30. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spentzos D, Levine DA, Ramoni MF, Joseph M, Gu X, Boyd J, Libermann TA, Cannistra SA. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J Clin Oncol. 2004;22:4700–4710. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamunyokoli FW, Bonome T, Lee JY, Feltmate CM, Welch WR, Radonovich M, Pise-Masison C, Brady J, Hao K, Berkowitz RS, et al. Expression profiling of mucinous tumors of the ovary identifies genes of clinicopathologic importance. Clin Cancer Res. 2006;12:690–700. doi: 10.1158/1078-0432.CCR-05-1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.