SUMMARY
We sequenced all protein-coding regions of the genome (the “exome”) in two family members with combined hypolipidemia, marked by extremely low plasma levels of low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, and triglycerides. These two participants were compound heterozygotes for two distinct nonsense mutations in ANGPTL3 (encoding the angiopoietin-like 3 protein). ANGPTL3 has been reported to inhibit lipoprotein lipase and endothelial lipase, thereby increasing plasma triglyceride and HDL cholesterol levels in rodents. Our finding of ANGPTL3 mutations highlights a role for the gene in LDL cholesterol metabolism in humans and shows the usefulness of exome sequencing for identification of novel genetic causes of inherited disorders. (Funded by the National Human Genome Research Institute and others.)
Familial hypobetalipoproteinemia is an inherited disorder of lipid metabolism defined by very low levels (<5th percentile of age- and sex-specific values) of plasma apolipoprotein B and LDL cholesterol. Familial hypobetalipoproteinemia is genetically heterogeneous.1,2 The best-characterized cases have been linked to mutations in the gene encoding apolipoprotein B (APOB) that lead to less apolipoprotein B synthesis and reduced secretion of very-low-density lipoprotein (VLDL) from the liver. As a consequence of impaired hepatic export of VLDL, persons with familial hypobetalipoproteinemia due to a deficiency of apolipoprotein B are prone to hepatic steatosis.3,4 Persons with hypobetalipoproteinemia may also have fat malabsorption due to impaired incorporation of dietary fats into chylomicrons in the absorptive cells of the intestine.
In a number of families with familial hypobetalipoproteinemia,5 sequencing of APOB has failed to identify mutations, and linkage analyses have pointed to other chromosomal regions.6,7 Furthermore, despite family members having plasma apolipoprotein B and LDL cholesterol levels as low as those of persons with mutations in APOB, they do not appear to have a greater susceptibility to hepatic steatosis or gastrointestinal symptoms than their unaffected relatives.8 To date, efforts to identify the gene defects in these families have not been successful.
Exome sequencing (i.e., sequencing of all protein-coding regions of the genome) in a few affected persons has proved successful at uncovering novel causal mutations in genetic disorders.9–11 This technique is considered to be unbiased — it does not focus on specific genes or loci deemed of interest on the basis of assumptions about possible causal associations. We applied next-generation sequencing (the most recent sequencing methods enabling low-cost, rapid, broad sequencing) to exomes chosen on the basis of solution hybrid selection (a method for capturing a targeted subgroup of genomic DNA) from two siblings with familial hypobetalipoproteinemia not linked to APOB, to identify the causative gene.
METHODS
PARTICIPANTS
We studied a family of European descent, some of whose members had hypobetalipoproteinemia in which linkage analysis had ruled out APOB as the causative gene (the F kindred in the study by Pulai and colleagues5) (Fig. 1). Members of this family were recruited from 1994 through 1997. Blood lipid levels were measured at multiple time points from 1994 through 2003; averages of the measurements were used in analyses. The last follow-up visit occurred in 2003; at that time, none of the family members had a history of coronary heart disease, and the oldest member was 81 years of age. Selected family members underwent lipoprotein metabolic studies and magnetic resonance spectroscopic measurements of hepatic fat.8,12
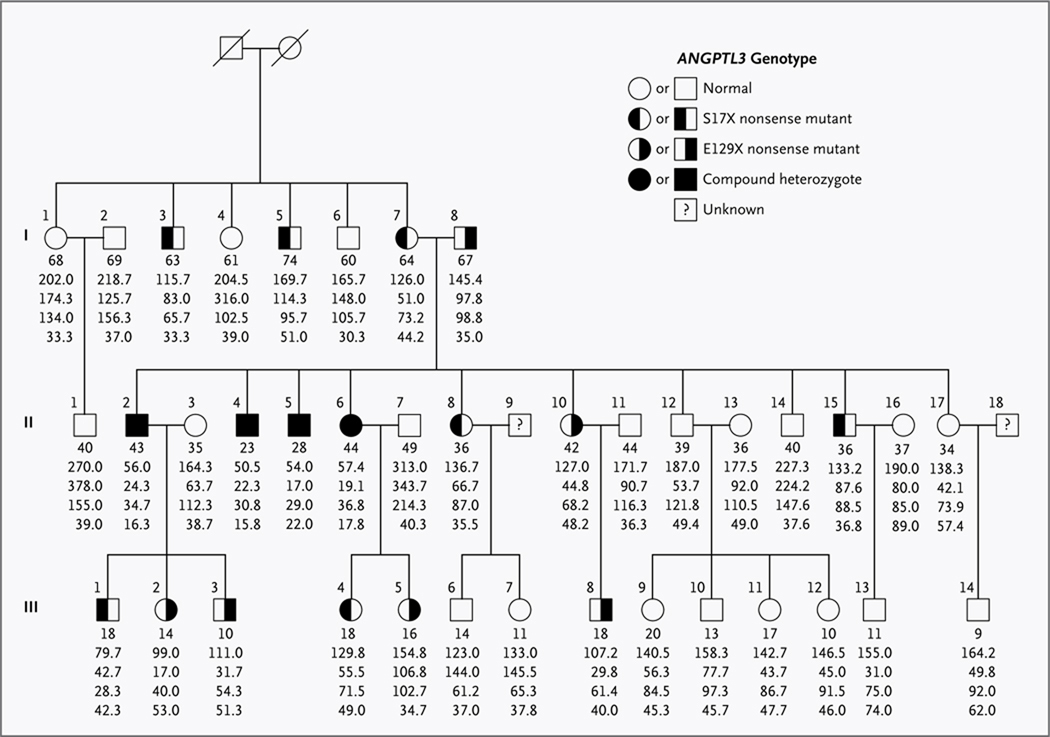
Figure 1. Pedigree of the Study Family with Familial Combined Hypolipidemia.
Squares indicate male family members, and circles female family members. Slashes indicate deceased persons. Roman numerals to the left of the pedigree indicate the generation; numerals to the upper left of each symbol indicate the individual family member. For the ANGPTL3 genotype, the X denotes a stop codon. The column of five values under each symbol indicates, from top to bottom, the age in years (as of 1997) and the total cholesterol, triglyceride, and LDL and HDL cholesterol levels (all in milligrams per deciliter). The values are the means of measurements from multiple fasting lipid profiles. To convert values for cholesterol to millimoles per liter, multiply by 0.02586. To convert values for triglycerides to millimoles per liter, multiply by 0.01129.
DNA samples were available from 38 members across three generations. Full lipid profiles, including apolipoprotein B levels, of these family members are listed in Table 1 in the Supplementary Appendix, available with the full text of this article at NEJM.org. Several family members had low plasma levels of LDL cholesterol and triglycerides. Four persons in the second generation (Participants II.2, II.4, II.5, and II.6) had combined hypolipidemia, with extremely low fasting plasma levels of LDL cholesterol, triglycerides, and HDL cholesterol. Two of the siblings with combined hypolipidemia (Participants II.4 and II.5) underwent exome sequencing.
Statistical modeling of genotypes and phenotypes (lipid and lipoprotein metabolic phenotypes) consisted of linear regression performed with the use of R software (www.r-project.org). “Residual” lipid phenotypes were ascertained after adjustment for age, age squared, and sex. The institutional review boards at the Washington University in St. Louis School of Medicine and the Broad Institute approved the study protocols. All participants provided written informed consent.
EXOME SEQUENCING, SANGER SEQUENCING, AND LINKAGE ANALYSES
A solution hybrid selection method13 was used to isolate DNA of the exome, which was subjected to sequencing (on the Genome Analyzer II platform; Illumina). Sixty exomes from persons of European descent, sequenced with the same methods, were used as controls. The full methods used for exome sequencing, Sanger sequencing, and linkage analysis are described in the Supplementary Appendix.
REPLICATION
The sequencing of ANGPTL3 in participants in the Dallas Heart Study has been described previously.14 Statistical modeling of genotypes and phenotypes was performed with the use of linear regression analysis involving adjustment for age, sex, race or ethnic group, and treatment for hypercholesterolemia.
RESULTS
EXOME SEQUENCING
A total of 28,646,006 bases in 164,688 exons from 15,994 genes were targeted for sequencing (Table 1). For each of the two participants, we generated approximately 6 billion bases of sequence, allowing each targeted base to be read approximately 200 times. For each participant, approximately 270 new single-nucleotide variants were found, of which approximately 150 were missense mutations and approximately 6 were nonsense (premature termination) mutations (Table 1).
Table 1.
Details of Solution Hybrid Selection, and Results of Exome Sequencing, in Two Study Participants.*
| Sample | Participant II.4 | Participant II.5 |
|---|---|---|
| No. of bait DNA bases | 37,640,396 | 37,640,396 |
| No. of target DNA bases | 28,646,006 | 28,646,006 |
| Total bases sequenced — no. (%) | 10,529,696,342 | 9,902,941,465 |
| Outside bait region | 2,062,882,735 (19.6) | 1,905,438,918 (19.2) |
| Near bait region | 1,090,792,362 (10.4) | 943,516,981 (9.5) |
| Within bait region | 7,376,021,245 (70.0) | 7,053,985,566 (71.2) |
| Within target region | 6,007,508,537 (57.1) | 5,793,926,100 (58.5) |
| Coverage† | ||
| Within bait region | 196 | 187 |
| Within target region | 210 | 202 |
| Base covered ≥10 times — % of region | 93.4 | 92.1 |
| Base covered ≥20 times — % of region | 90.1 | 88.0 |
| Identified variants‡ | ||
| Total no. | 18,259 | 18,123 |
| Transition-to-transversion ratio | 2.93 | 2.98 |
| No. of variants meeting quality-control standard | 16,426 | 16,246 |
| No. in dbSNP | 15,680 | 15,512 |
| Concordance with dbSNP — % | 99.50 | 99.52 |
| No. not in dbSNP | 746 | 734 |
| No. not in 1000 Genomes Project database | 481 | 481 |
| No. not in the 60 control exomes | 266 | 277 |
| Coding single-nucleotide variants | ||
| Known | ||
| Total — no. | 16,160 | 15,969 |
| Transition-to-transversion ratio | 3.20 | 3.26 |
| Homozygous — no. | 6284 | 6419 |
| Heterozygous — no. | 9876 | 9550 |
| Synonymous — no. | 9260 | 9179 |
| Missense — no. | 6622 | 6525 |
| Premature termination — no. | 40 | 41 |
| Splice site — no. | 238 | 224 |
| Novel | ||
| Total — no. | 266 | 277 |
| Transition-to-transversion ratio | 2.86 | 2.85 |
| Homozygous — no. | 6 | 7 |
| Heterozygous — no. | 260 | 270 |
| Synonymous — no. | 107 | 116 |
| Missense — no. | 151 | 149 |
| Premature termination — no. | 5 | 7 |
| Splice site — no. | 3 | 4 |
| No. of genes with novel homozygous or compound heterozygous variants shared between participants§ | 1 | 1 |
The bait DNA is the region of the genome preferentially retained during the hybrid selection step; it is therefore vastly enriched during sequencing, as compared with unselected regions. The target DNA is the subgroup of the bait DNA that is of special interest — in this study, the 164,688 exon sequences with additional two-nucleotide flanking sequences, to capture splice sites.
“Coverage” is the average number of times each base is represented in the sequence reads, as a measure of the degree to which the region of interest is captured by the sequencing method.
The term dbSNP refers to the Single Nucleotide Polymorphism Database (build 130) of the National Center for Biotechnology Information.
The one shared gene is ANGPTL3.
NONSENSE VARIANTS IN ANGPTL3
To identify the causal variants in the two participants who underwent sequencing (Participants II.4 and II.5), we hypothesized that the phenotype of combined hypolipidemia (i.e., the concurrence of low levels of plasma HDL cholesterol, LDL cholesterol, and triglycerides) was inherited in an autosomal recessive fashion. Therefore, we sought genes harboring novel variants in both alleles of both participants. We eliminated variants whose features did not meet quality-control standards (see the Supplementary Appendix) and variants that were not shared between the two probands. We also eliminated any variant sites that were present in the Single Nucleotide Polymorphism Database (dbSNP, build 130) of the National Center for Biotechnology Information, in the genomes sequenced by the 1000 Genomes Project, or in the 60 control exomes (Table 1).
Only one gene harbored novel variants in both alleles in both siblings: ANGPTL3. ANGPTL3 harbored two nonsense variants: a single-nucleotide variant (GAA→TAA, at nucleotide 62,836,210 on chromosome 1) that introduces a nonsense mutation at position 129, resulting in the amino acid mutation E129X; and a double nucleotide variant (TCC→TGA, at nucleotides 62,835,875 to 62,835,876 on chromosome 1) that introduces a nonsense mutation at position 17, resulting in the amino acid mutation S17X. (The X in both amino acid mutations denotes a stop codon.) Both mutations are located in the first exon of ANGPTL3 (Fig. 1 in the Supplementary Appendix).
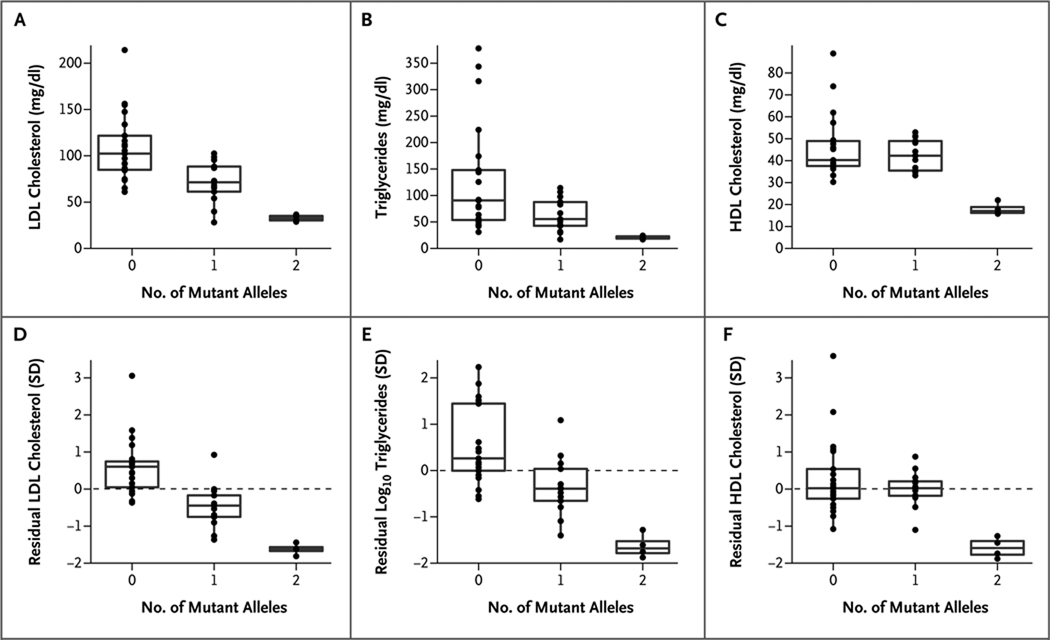
Sanger sequencing of the first ANGPTL3 exon of all 38 persons in the study family who were available confirmed that the two nonsense mutations segregated independently (Fig. 1). The 13 family members heterozygous for just one of the two nonsense mutations had significantly lower plasma levels of LDL cholesterol (mean, 72 mg per deciliter [1.9 mmol per liter]) and triglycerides (mean, 64 mg per deciliter [0.7 mmol per liter]) than did the 21 family members with neither mutation (mean for LDL cholesterol, 109 mg per deciliter; P<0.001; mean for triglycerides, 130 mg per deciliter; P = 0.01). The remaining 4 participants were compound heterozygotes and had even lower plasma LDL cholesterol and triglyceride levels (means, 33 mg per deciliter [0.9 mmol per liter] and 21 mg per deciliter [0.2 mmol per liter], respectively). Thus, the nonsense mutations in ANGPTL3 affected phenotypes in a manner dependent on the gene dosage — the number of alleles present in a given person (P<0.001 for LDL cholesterol and P = 0.004 for triglycerides, from additive models) (Fig. 2).
Figure 2. Phenotypes Associated with Familial Combined Hypolipidemia in the Study Family, According to Number of Mutant ANGPTL3 Alleles.
Shown are the levels of low-density lipoprotein (LDL) cholesterol (Panel A), triglycerides (Panel B), and high-density lipoprotein (HDL) cholesterol (Panel C) in the family members, grouped according to ANGPTL3 genotype. The corresponding “residual” phenotypes are shown (in Panels D, E, and F, respectively), in units normalized to the standard deviation, after adjustment for age, age squared, and sex. The box plots give the median levels (middle horizontal line in each box), the interquartile ranges (delineated by the top and bottom of each box), and outliers falling below the 5th percentile or above the 95th percentile (points below or above the vertical lines, respectively). To convert values for cholesterol to millimoles per liter, multiply by 0.02586. To convert values for triglycerides to millimoles per liter, multiply by 0.01129.
In contrast, the nonsense mutations were recessive with respect to HDL cholesterol: family members heterozygous for a single nonsense mutation had plasma HDL cholesterol levels (mean, 43 mg per deciliter [1.1 mmol per liter]) that did not differ significantly from those of family members with no ANGPTL3 mutation (mean, 46 mg per deciliter [1.2 mmol per liter]; P = 0.34), whereas compound heterozygotes had a very low level of HDL cholesterol (mean, 18 mg per deciliter [0.5 mmol per liter]; P<0.001 from a recessive model). These relations were maintained even after adjustment of lipid levels for age and sex (Fig. 2).
LINKAGE ANALYSES
To determine whether mutations at another locus could account for the hypolipidemia in this family, we performed linkage analyses, using LDL cholesterol and HDL cholesterol as quantitative traits. The highest lod scores for both traits occurred between the DNA markers GATA72H07 and GATA109 on chromosome 1p33–31.1, the region harboring ANGPTL3. The peak lod scores for LDL cholesterol were 3.07 (at GATA165C03) and 3.89 (at GATA61A06) (Table 2 in the Supplementary Appendix). Significant lod scores were not found for regions on chromosomes 3 and 10 previously reported to be associated with familial hypobetalipoproteinemia,6,7 nor for any other regions in the genome. These results provide additional evidence that the nonsense mutations in ANGPTL3 are causal for the combined hypolipidemia in this family. Additional details regarding the linkage analyses are given in the Supplementary Appendix.
REPLICATION IN A POPULATION-BASED COHORT
Sequencing of ANGPTL3 in participants in the Dallas Heart Study14 showed that carriers of frameshift mutations had significantly lower LDL cholesterol levels than noncarriers. Details regarding these analyses are given in the Supplementary Appendix.
STUDIES OF LIPOPROTEIN METABOLISM
Physiological studies of selected members of the study family indicated that carriers of ANGPTL3 mutations had decreased rates of VLDL production and increased rates of LDL fractional catabolism. Details regarding these analyses are given in the Supplementary Appendix.
DISCUSSION
Whole-exome sequencing in two siblings with a clinical syndrome of combined hypolipidemia revealed two independent nonsense mutations in ANGPTL3. One, S17X, is predicted to terminate translation near the N-terminal of the protein; the other, E129X, is predicted to prematurely terminate the protein at residue 129. Family members who were heterozygous for either mutation had plasma levels of LDL cholesterol and triglycerides that were intermediate between the levels in persons with neither mutation and those with both mutations, findings consistent with a codominant mode of inheritance for the LDL cholesterol and triglyceride phenotypes. In contrast, the level of HDL cholesterol appears to segregate as a recessive trait. Family members who had both nonsense alleles, but not members with just one, had significantly lower plasma HDL cholesterol levels than members with neither mutation. Thus, persons with the two nonsense ANGPTL3 alleles had low plasma levels of LDL cholesterol, triglycerides, and HDL cholesterol, a phenotype we propose to call familial combined hypolipidemia.
The protein ANGPTL3 is secreted and expressed primarily in the liver. The underlying gene was originally associated with lipid levels on the basis of positional cloning of a causal mutation in a hypolipidemic mouse strain.15 Expression of the recombinant protein in the livers of these mice increased plasma triglyceride and cholesterol levels. Subsequent studies showed that targeted inactivation of Angptl3 in mice lowered plasma cholesterol and triglyceride levels — both in wild-type mice,16 in which cholesterol is transported primarily in HDL particles, and in mice in which Apoe or Ldlr (the LDL receptor gene) had been knocked out,17,18 in which cholesterol is carried primarily by VLDL, intermediate-density lipoprotein, and LDL. Taken together, these data suggest that ANGPTL3 normally acts to increase plasma levels of triglycerides, LDL cholesterol, and HDL cholesterol.
This conclusion is consistent with the finding of combined hypolipidemia in persons with two defective ANGPTL3 alleles. The activities of lipoprotein lipase and endothelial lipase, key enzymes in the metabolism of circulating triglycerides and HDL cholesterol, respectively, are elevated in Angptl3-knockout mice,19–21 suggesting that increased activity of these enzymes may explain the low levels of triglycerides and HDL cholesterol in humans with ANGPTL3 nonsense mutations. The mechanism by which deficiency of ANGPTL3 lowers LDL cholesterol remains to be determined.
Confirming the link between ANGPTL3 and plasma LDL cholesterol levels is the finding that Dallas Heart Study participants with frameshift mutations had significantly lower LDL cholesterol levels than the rest of the cohort. Furthermore, in genomewide association screening of common variants in more than 100,000 participants, we found that polymorphisms near ANGPTL3 are highly significantly associated with lipid traits — most strongly with the triglyceride level (e.g., the polymorphism rs2131925, associated with a change of 4.9 mg per deciliter [0.05 mmol per liter] per allele [P = 9×10−43 for the association]), but also with the LDL cholesterol level (e.g., rs2131925, associated with a change of 1.6 mg per deciliter [0.04 mmol per liter] per allele [P = 2×10−17 for the association]). These two associations were consistent with the lipid patterns observed in the current family under study.22
This contrasts with studies of common variants near LPL (the gene encoding lipoprotein lipase), which are associated with triglycerides and HDL cholesterol, and those near LIPG (encoding endothelial lipase), which are associated with HDL cholesterol; neither the LPL variants nor the LIPG variants are associated with LDL cholesterol.22 These studies are consistent with observations in genetically modified mice that effects of ANGPTL3 on triglycerides and HDL cholesterol are mediated by lipoprotein lipase and endothelial lipase, respectively, and that the mechanism by which ANGPTL3 modulates plasma LDL cholesterol levels is distinct.
The apparent discrepancy between the modes of inheritance of reduced LDL cholesterol and triglyceride levels and reduced HDL cholesterol levels also argues for distinct mechanisms of action on different lipoproteins. We found that carriers of ANGPTL3 nonsense mutations had decreased rates of VLDL apolipoprotein B production and increased fractional catabolic rates for LDL apolipoprotein B. This suggests that ANGPTL3 acts directly in the liver to regulate hepatocellular lipoprotein secretion and clearance, which is distinct from the role of the protein in the inhibition of lipoprotein lipase and endothelial lipase in the bloodstream.
In conclusion, we found that nonsense mutations in ANGPTL3 resulted in decreased plasma LDL cholesterol levels and, in persons harboring two nonsense alleles, a phenotype of familial combined hypolipidemia. This finding highlights the promise of exome sequencing in medical genetics. It also underscores a new mechanism for the lowering of LDL cholesterol in humans.
Supplementary Material
Acknowledgments
Supported by grants from the National Human Genome Research Institute of the National Institutes of Health (NIH) (Medical Sequencing Program grant U54 HG003067, to the Broad Institute for exome sequencing), the National Heart, Lung, and Blood Institute (NHLBI) of the NIH (RC1 HL099793, RC1 HL099634, and RC2 HL101864, to Dr. Kathiresan; K99 HL098364, to Dr. Musunuru; and R01 HL082896, to Drs. Cohen and Hobbs for analysis of ANGPTL3 in the Dallas Heart Study), the Donovan Family Foundation (to Dr. Kathiresan), the Sarnoff Cardiovascular Research Foundation (a research fellowship, to Mr. Pirruccello), and the Canadian Institutes of Health Research (a Canada Graduate Doctoral Scholarship, to Mr. Do).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Schonfeld G, Lin X, Yue P. Familial hypobetalipoproteinemia: genetics and metabolism. Cell Mol Life Sci. 2005;62:1372–1378. doi: 10.1007/s00018-005-4473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper AJ, van Bockxmeer FM, Burnett JR. Monogenic hypocholesterolaemic lipid disorders and apolipoprotein B metabolism. Crit Rev Clin Lab Sci. 2005;42:515–545. doi: 10.1080/10408360500295113. [DOI] [PubMed] [Google Scholar]
- 3.Tarugi P, Lonardo A, Ballarini G, et al. Fatty liver in heterozygous hypobetalipoproteinemia caused by a novel truncated form of apolipoprotein B. Gastroenterology. 1996;111:1125–1133. doi: 10.1016/s0016-5085(96)70082-3. [DOI] [PubMed] [Google Scholar]
- 4.Schonfeld G, Patterson BW, Yablonskiy DA, et al. Fatty liver in familial hypobetalipoproteinemia: triglyceride assembly into VLDL particles is affected by the extent of hepatic steatosis. J Lipid Res. 2003;44:470–478. doi: 10.1194/jlr.M200342-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Pulai JI, Neuman RJ, Groenewegen AW, Wu J, Schonfeld G. Genetic heterogeneity in familial hypobetalipoproteinemia: linkage and non-linkage to the apoB gene in Caucasian families. Am J Med Genet. 1998;76:79–86. [PubMed] [Google Scholar]
- 6.Yuan B, Neuman R, Duan SH, et al. Linkage of a gene for familial hypobetalipoproteinemia to chromosome 3p21.1-22. Am J Hum Genet. 2000;66:1699–1704. doi: 10.1086/302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherva R, Yue P, Schonfeld G, Neuman RJ. Evidence for a quantitative trait locus affecting low levels of apolipoprotein B and low density lipoprotein on chromosome 10 in Caucasian families. J Lipid Res. 2007;48:2632–2639. doi: 10.1194/jlr.M700078-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Yue P, Tanoli T, Wilhelm O, Patterson B, Yablonskiy D, Schonfeld G. Absence of fatty liver in familial hypobetalipoproteinemia linked to chromosome 3p21. Metabolism. 2005;54:682–688. doi: 10.1016/j.metabol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias N, Patterson BW, Schonfeld G. In vivo metabolism of ApoB, ApoA-I, and VLDL triglycerides in a form of hypobetalipoproteinemia not linked to the ApoB gene. Arterioscler Thromb Vasc Biol. 2000;20:1309–1315. doi: 10.1161/01.atv.20.5.1309. [DOI] [PubMed] [Google Scholar]
- 13.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romeo S, Yin W, Kozlitina J, et al. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koishi R, Ando Y, Ono M, et al. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 16.Köster A, Chao YB, Mosior M, et al. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;146:4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- 17.Ando Y, Shimizugawa T, Takeshita S, et al. A decreased expression of angiopoietin-like 3 is protective against atherosclerosis in apoE-deficient mice. J Lipid Res. 2003;44:1216–1223. doi: 10.1194/jlr.M300031-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Lee EC, Desai U, Gololobov G, et al. Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL) J Biol Chem. 2009;284:13735–13745. doi: 10.1074/jbc.M807899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizugawa T, Ono M, Shimamura M, et al. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 20.Shimamura M, Matsuda M, Yasumo H, et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 21.Jin W, Wang X, Millar JS, et al. Hepatic proprotein convertases modulate HDL metabolism. Cell Metab. 2007;6:129–136. doi: 10.1016/j.cmet.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.