Abstract
Reversible protein cysteine nitrosylation (S-nitrosylation) is a common mechanism utilized in signal transduction and other diverse cellular processes. Protein denitrosylation is largely mediated by cysteine denitrosylases, but the functional scope and significance of these enzymes are incompletely defined, in part due to limited information on their cognate substrates. Here, using Jurkat cells, we employed stable isotope labeling by amino acids in cell culture (SILAC), coupled to the biotin switch technique and mass spectrometry, to identify 46 new substrates of one denitrosylase, thioredoxin 1. These substrates are involved in a wide range of cellular functions including, cytoskeletal organization, cellular metabolism, signal transduction and redox homeostasis. We also identified multiple S-nitrosylated proteins that are not substrates of thioredoxin 1. A verification of our principal findings was made in a second cell type (RAW264.7 cells). Our results point to thioredoxin 1 as a major protein denitrosylase in mammalian cells and demonstrate the utility of quantitative proteomics for large-scale identification of denitrosylase substrates.
A multitude of studies have collectively established the importance of S-nitrosylation, the covalent attachment of a nitroso group to cysteine (Cys) thiols, in regulating the function of many proteins, thereby influencing a plethora of cellular and organismal processes, including transcription, metabolism, respiration, cell growth and survival (1). In addition to its role in physiological responses, aberrant protein S-nitrosylation that arises when cellular nitrosylation-denitrosylation equilibria are disrupted, has been implicated in a range of human pathologies, including muscle, lung and heart disease, neurodegeneration, and cancer (2, 3). Accordingly, mechanisms that regulate protein S-nitrosylation and denitrosylation represent an active area of investigation.
The level of S-nitrosylation of any cellular protein depends on the balance between nitrosylation and denitrosylation. Whereas past research has mainly focused on the addition of NO groups, analysis of denitrosylation has lagged. Elucidating mechanisms of protein denitrosylation is important in understanding both how signal transduction is regulated by protein S-nitrosylation and how cells handle nitrosative stress. Recent studies have provided new insights into cellular mechanisms of protein denitrosylation through the characterization of Cys denitrosylases, enzymes that mediate denitrosylation (4). Among those, two enzymes systems in particular, the glutathione/S-nitrosoglutathione reductase (GSH/GSNOR) and the thioredoxin/thioredoxin reductase (Trx/TrxR) systems have been established as physiologically relevant denitrosylases. Though their mechanism of action differ, both GSH/GSNOR and Trx/TrxR protect microorganisms, plants and mammalian cells from nitrosative stress, and regulate manifold NO-related cellular and systemic responses (4).
Thioredoxins (Trxs) are ubiquitous proteins that play vital roles in many processes in living cells, and are considered as key elements in the cellular response to oxidative stress (5, 6). Trxs have a conserved Cys-Gly-Pro-Cys redox active site that is essential for their function as oxidoreductases. Very recent studies have revealed a new role for Trxs in both cellular signaling and protection (4, 10), which is mediated through denitrosylation of peptides and proteins (7-13). The denitrosylase activities of Trxs are coupled to cognate Trx reductases (TrxR), flavin-containing selenoenzymes (8, 10). For example, cytosolic caspase-3 and caspase-8 are among the S-nitrosothiol (SNO)-substrates of Trx1/TrxR1 (8-11, 13), whereas mitochondria-associated caspase-3 is denitrosylated by Trx2/TrxR2 to regulate death-receptor signaling (10). In addition, both microbial and mammalian Trx systems have been shown to safeguard against nitrosative stress (14-17), which is mediated by hazardous levels of protein S-nitrosylation. Consistent with this role for Trx, NO has been shown to induce denitrosylase activity, at least in part through suppression of the Trx inhibitor, Txnip (4, 18). Thus, accumulating evidence suggests that Trx proteins, by virtue of their denitrosylase function, are important regulators of NO/SNO-dependent responses.
Recent reports have raised the idea that Trx in fact denitrosylates multiple proteins (SNO-proteins) (9, 10). Although only a few endogenous SNO-substrates (in addition to caspase-3) of Trxs have been identified (10), they may represent the tip of the iceberg. Identification of the complete set of Trx SNO-substrates will be necessary in order to better define the range of NO-related cellular processes that are regulated by Trx/TrxR as well as the molecular basis of specificity in protein denitrosylation (4). As a first step to address these questions, we devised a proteomic approach that allowed us to identify a large number of novel Trx SNO-substrates, and in situ confirmation was obtained in selected cases. The results presented here provide a proof-of-principle for the feasibility of large scale identification of protein denitrosylase substrates and underscore the large repertoire of proteins and cellular processes that may be coordinately regulated by NO and Trx.
Experimental Procedures
Materials
S-nitroso-cysteine (CysNO) was synthesized from L-cysteine using acidified nitrite. Antibodies were obtained as follows: 14-3-3θ from Santa Cruz Biotechnology, peroxiredoxin-1 from Upstate Biotechnology, and annexin-1 from BD Bioscience Laboratories. Recombinant rat TrxR1 was obtained from American Diagnostica. Recombinant human Trx1 was produced in E. coli and purified by nickel-affinity chromatography. Auranofin was purchased from BioMol. Biotin-HPDP and streptavidin agarose were purchased from Pierce. Interferon-γ was purchased from R&D systems. l-13C615N4-arginine and l-13C615N2-lysine were purchased from Sigma Isotec. Modified RPMI-1640 medium, dialyzed fetal bovine serum, and all other reagents were purchased from Sigma unless indicated.
Cell culture
Jurkat cells (human T leukemia cell line) were maintained in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C under 5% CO2. RAW264.7 cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C under 5% CO2. SILAC medium was prepared by replacing the normal arginine and lysine with the corresponding heavy isotope-labeled amino acids in RPMI. The SILAC medium also contained 10% dialyzed fetal bovine serum and antibiotics. To generate two labeling states, one population of cells was grown in “light” SILAC medium containing l-12C614N4 arginine (Arg0) and l-12C614N2-lysine (lys0), whereas the other population was grown in “heavy” SILAC medium containing l-13C615N4 arginine (Arg10) and l-13C615N2-lysine (lys8). All media were prepared at the same time and cells to be labeled were cultured in parallel for six passages before treatment.
Denitrosylation assay
Jurkat cells were homogenized in hypotonic buffer (10 mM Hepes, 3 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, pH 7.5) and then centrifuged at 100,000g for one hr at 4°C. The supernatant typically contained 5 mg/ml protein as determined by Bradford assay. Cytosolic fractions were then treated with vehicle or CysNO (final concentration, 50 μM) at room temperature for 30 min in the dark. After this step, low-molecular-weight species (including residual CysNO) were removed by repeated cycles of filtration through a 10 kDa cutoff filter (Amicon Ultra-15, Millipore) and washing with HEN buffer (25 mM Hepes, 1 mM EDTA, 10 μM neocuproine, pH 7.5). The sample was subjected to denitrosylation by incubation with GSH or a recombinant Trx system (5 μM Trx, 100 nM TrxR, 200 μM NADPH) in a total volume of 0.5 ml. To minimize spontaneous SNO decomposition, reactions were protected from light and carried out in the presence of the metal chelator EDTA. Where indicated, protein-SNOs were separated from low-molecular-mass SNO as follows: half of the sample was transferred to a separate microfuge tube. After adding three volumes of cold acetone, the sample was incubated at -20°C for 30 min, and then centrifuged at 5000 × g for 5 min at 4°C. The protein pellet was resuspended in HEN buffer and kept on ice until analysis. The other half of the sample was transferred to a 10 kDa cutoff filter (Microcon, YM-10, centrifugal filter unit, Millipore). After centrifugation, the filtrate containing low molecular weight species was stored on ice until analysis. SNO content was determined by reductive chemiluminescence with an NO analyzer (NOA 280i, Sievers Instruments) as described (19). Values were derived by comparison with GSNO standards and were normalized to protein concentration in the extract.
Detection of protein S-nitrosylation with the biotin-switch technique
Detection of endogenously S-nitrosylated proteins was performed with the biotin-switch technique (20) with some modifications. In brief, whole-cell lysates were prepared with lysis buffer (50 mM Hepes, pH 7.5, 1% NP-40, 150 mM NaCl, 0.1 mM EDTA and protease inhibitor cocktail (Roche)). An equal volume of blocking buffer (0.2% S-methyl methanethiosulfonate and 20% SDS in HEN buffer) was then added and samples were incubated at 50°C for 20 min with frequent vortexing. Proteins were then precipitated with 3 volumes of acetone at -20°C for one hr. After centrifugation, the protein pellet was washed with cold acetone to remove residual S-methyl methanethiosulfonate. Pellets were then resuspended in HENS buffer (HEN with 1% SDS), and freshly prepared biotin-HPDP and sodium ascorbate were added to give final concentrations of 0.4 mM and 20 mM, respectively. After incubation for two hr, samples were subjected to streptavidin pull-down followed by Western blot or MS analysis.
Sample preparation for proteomic analysis
Formerly-nitrosylated proteins were removed from the streptavidin resin by suspension in 3× volume of 0.3% trifluoroacetic acid (Pierce) in 70/30 v/v water/MeCN (Fisher Scientific) and incubating with shaking for 30 min. The resin was pelleted and the supernatant, containing proteins, was taken to dryness in a speed-vac. Protein was then resuspended in 20 microliters of 0.1% Rapigest SF (Waters) in 50 mM ammonium bicarbonate (VWR) with mild heating. Peptides were digested with 1% w/w sequencing-grade modified trypsin (Promega) overnight at 37°C. Rapigest was hydrolyzed with the addition of trifluoroacetic acid (TFA) and MeCN to 1% and 2% v/v, with heating (60°C) for 2 hr. The peptide mixture was then cooled to room temperature and cleaned up using C18 Zip Tips (Millipore) according to the manufacturer's recommendations. Peptides were taken to dryness and resuspended in 20 microliter 0.1% v/v TFA with 2% v/v acetonitrile. Each sample was analyzed by LC-MS/MS in duplicate.
LC-MS/MS analysis
Five microliters of the sample were injected onto a 75μm × 250 mm BEH C18 column (Waters) and separated using a gradient of 5 to 40% acetonitrile with 0.1% formic acid, with a flow rate of 0.3 mL/min, in 90 min on a nanoAcquity liquid chromatograph (Waters). The outlet of the liquid chromatography column was coupled to an LTQ-Orbitrap hybrid mass spectrometer (Thermo). The Orbitrap MS/MS method utilized CID fragmentation, with product ions also measured in the Orbitrap. Briefly, the precursor scan method utilized profile mode and 60,000 resolution, with AGC target of 1e6 and 1 microscans. MS/MS acquisition was performed on the top 3 precursor ions above a 1000 count threshold, using collisionally induced dissociation (CID) with a 3 Da isolation window, normalized collision energy of 35%, and 1 microscans. Product ion spectra were collected in profile mode with a resolution of 7500 and AGC target setting of 2e5. Dynamic exclusion settings were repeat count = 3, repeat duration = 30 sec, exclusion list = 250, and exclusion time = 120 sec.
Peptide identification and quantification
SILAC quantification and peptide identification was accomplished using Rosetta Elucidator v3.2 (Rosetta Biosoftware) and Mascot v2.2 (Matrix Sciences). Mascot searches were accomplished directly from Elucidator, against the Swissprot database v57.1, with human taxonomy (http://www.uniprot.org/). Database search parameters were 10 ppm precursor and 0.02 Da product ion tolerance, with oxidized (M), methylthio (C), Label:13C615N2 (K) and Label:13C615N4 (R) as variable modifications. Peptide identifications were accepted if they passed the 0.8 peptideteller confidence threshold in Elucidator; the peptide FDR was determined to be less than 1% using decoy database searching (21). Labeled (SILAC) pairs were located in Elucidator with a minimum m/z tolerance of 15 ppm and retention time tolerance of 0.2 min, and a maximum of 3 labeled amino acids per peptide. The SILAC ratios were calculated without intensity normalization in Elucidator, using the peak height of the highest abundance isotopomer.
Results
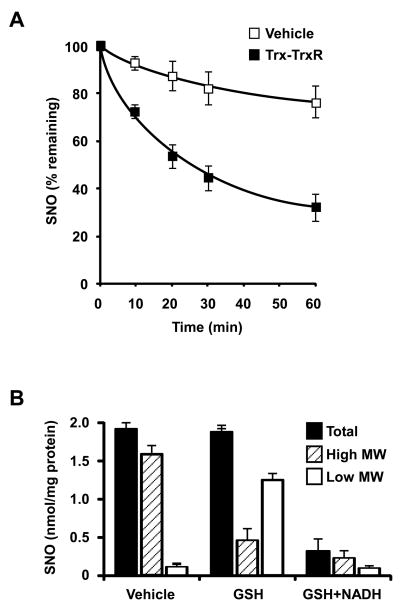
To assess Trx-mediated denitrosylation, we established an in vitro system that allowed us to monitor denitrosylation under well-controlled conditions. We prepared a cytosolic fraction from Jurkat cells and subjected it to in vitro nitrosylation by exposure to the nitrosylating agent S-nitrosocysteine (CysNO, 50 μM for 30 min, see Experimental Procedures). CysNO treatment resulted in appreciable protein S-nitrosylation (∼ 2 nmol total SNO per mg protein), as determined by reductive-chemiluminescnce (also assessed be by the biotin-switch method, below). Incubation of the nitrosylated lysate at 37°C for up to one hr was accompanied by slow denitrosylation, evident as a small progressive decrement in SNO content (Figure 1A). When a Trx system—consisting of Trx, TrxR and NADPH—was added to the lysate, the denitrosylation rate was markedly enhanced. For example, after 30 min incubation with Trx system, SNO content decreased by ∼ 46% compared to ∼ 12% decrease in vehicle-treated lysates. At 60 min, denitrosylation approached a plateau, with ∼ 30% SNO remaining, reflecting failure of Trx to denitrosylate a subset of SNOs. These data—as well as similar data obtained in other cells (8-10, 12)—suggest that Trx can catalyze the denitrosylation of multiple proteins.
Figure 1.
Biochemical analysis of denitrosylation. (A) The cytosolic fraction prepared from Jurkat cells was S-nitrosylated with CysNO (50 μM) for 30 min at room temperature. S-nitrosylated samples were then incubated with vehicle or Trx system (5 μM Trx, 100 nM TrxR, 200 μM NADPH) at 37°C for the indicated time. SNO content was assessed by reductive chemiluminescence. (B) S-Nitrosylated samples were incubated with GSH (1 mM) or with GSH and 200 μM NADH at 37°C for 10 min. SNO content in total, high- or low-MW fractions was determined by reductive chemiluminescence. Data are presented as mean ± SEM; n = 3.
We employed the same assay to assess GSH-dependent denitrosylation. Incubation of the nitrosylated sample with GSH resulted in a modest decline in total SNO content. Ten min exposure to 1 mM or 5 mM GSH produced ∼ 5% or ∼ 15% decrease in total SNO content, respectively. A major product of the reaction of GSH with protein SNO is GSNO; hence analysis of total SNO will not reveal the fate of SNOs. Separation of the reaction products according to molecular-weight (MW) revealed that 10 min treatment with 1 mM GSH resulted in ∼ 75% decrease in high-MW SNO, most of which was recovered as low-MW SNO (Figure 1B). Increasing incubation time to 60 min lead to near-complete denitrosylation of high-MW SNO (data not shown). Cellular GSNO is metabolized substantially by NADH-dependent GSNOR (22) and we observed that the addition of NADH to the reaction mixture caused a sharp decline in the level of both high- and low-MW SNO (Figure 1B). These data suggest that, under our experimental conditions (SNO production by addition of exogenous CysNO), GSH denitrosylates the majority of protein SNOs and that GSNOR-like activity in Jurkat cells efficiently metabolizes GSNO, thereby facilitating denitrosylation of SNO-proteins.
The analysis presented thus far was based on measurement of total SNO content. Next, we assessed S-nitrosylation and denitrosylation by the biotin-switch technique (BST), in which S-nitrosylated Cys are selectively biotinylated (20). Using this method, changes in S-nitrosylation of individual proteins can be visualized. Supporting and adding to the previous results, CysNO treatment resulted in robust protein S-nitrosylation and incubation with the Trx system reversed the S-nitrosylation of some but not all proteins (Figure 2A).
Figure 2.
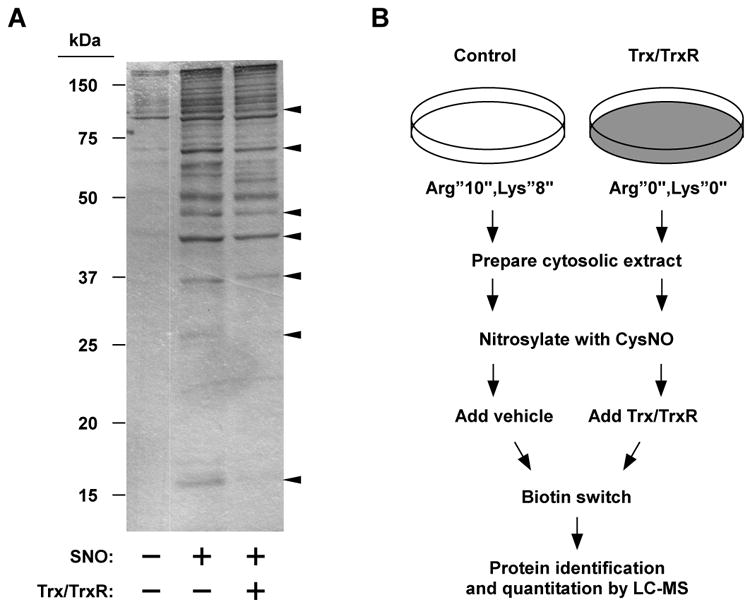
Biotin-switch based analysis of Trx-mediated protein denitrosylation. (A) The cytosolic fraction prepared from Jurkat cells was S-nitrosylated with CysNO (50 μM) for 30 min at room temperature. S-Nitrosylated samples were then incubated with vehicle or Trx system at 37°C for 20 min. Samples were subjected to BST analysis and biotin was detected with peroxidase-linked streptavidin. Arrow heads point to several proteins that appear to be denitrosylated by Trx. (B) Schematic overview of SILAC-based analysis of protein denitrosylation.
We sought to identify novel Trx SNO-substrates by coupling the BST with mass spectrometry (MS)-based proteomics. We used stable isotope labeling by amino acids in cell culture (SILAC), a proteomic strategy that metabolically labels the entire proteome, making it amenable to quantitative MS analysis (Figure 2B) (23). Two populations of cells were grown in medium containing distinct forms of arginine and lysine—the normal (“light”) l-12C614N4 arginine (Arg0) and l-12C614N2-lysine (lys0) or the isotopic variants (“heavy”) l-13C615N4 arginine (Arg10) and l-13C615N2-lysine (lys8)—until complete incorporation of the amino acids was achieved. Cytosolic fractions prepared from both light and heavy cells were S-nitrosylated in vitro as above. The S-nitrosylated “light” sample was then incubated for 20 min with the Trx system whereas the “heavy” sample served as the untreated control. The heavy and light samples were subsequently pooled and processed by BST. Biotinylated proteins were purified by avidin pull-down and analyzed by liquid chromatography-mass spectrometry (LC-MS, see Experimental Procedures).
LC-MS results obtained using high stringency parameters and compiled from two independent experiments identified a total of ∼ 280 proteins (∼ 1500 unique peptides) in the BST pull-down. To increase confidence in the quantitative accuracy, only the 145 proteins that were identified in both experiments were selected for further analysis. After annotation of heavy or light peaks (in which spectrum peaks were identified with peptide sequences), the light/heavy (L/H) pairs were located in the data set and relative peptide expression ratios were calculated.
Based on a standard power calculation (n=2, 10% RSD, 95% confidence interval) and the likelihood of being able to validate a change by alternative methodology, we considered a SILAC ratio (L/H) of 0.65 (or fold-change of 1.5) as a cutoff value for determining a significant reduction in S-nitrosylation level between Trx/TrxR and control BST pull-downs. Calculation of peptide ratios in the pull-downs revealed that 46 of 145 SNO-proteins were substantially lower in the Trx/TrxR sample compared to control (L/H ratio ≤ 0.65) (Table 1). Thus, we considered these 46 proteins to be putative Trx SNO-substrates. These proteins belong to diverse functional categories, including cytoskeletal organization, metabolism, signaling and stress responses (Table 1). The remaining 99 proteins (L/H ratio > 0.65) were considered as poor or non-Trx substrates (Supplemental Table 1, and see Discussion).
Table 1. SNO Proteins Displaying SILAC Ratio (L/H) ≤ 0.65 (Substantial Effect of Trx/TrxR Treatment).
| primary protein name | accession number | protein description | L/H ratio | function |
|---|---|---|---|---|
| LCAP_HUMAN | Q9UIQ6 | leucyl-cystinyl aminopeptidase | 0.01a | protein turnover |
| GLRX1_HUMAN | P35754 | glutaredoxin-1 | 0.28 ± 0.06 | antioxidant defense, redox signaling |
| 1433T_HUMAN | P27348 | 14-3-3 protein theta | 0.31 ± 0.01 | signal transduction |
| RANG_HUMAN | P43487 | Ran-specific GTPase-activating protein | 0.31 ± 0.08 | intracellular transport, signal transduction |
| DBNL_HUMAN | Q9UJU6 | drebrin-like protein | 0.31 ± 0.07 | cytoskeletal organization |
| NUDC2_HUMAN | Q8WVJ2 | NudC domain-containing protein 2 | 0.32 ± 0.15 | unknown |
| FUBP2_HUMAN | Q92945 | far upstream element-binding protein 2 | 0.37 ± 0.12 | mRNA metabolism |
| PAIRB_HUMAN | Q8NC51 | plasminogen activator inhibitor 1 RNA-binding protein | 0.37 ± 0.12 | mRNA metabolism |
| 2AAA_HUMAN | P30153 | serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A α isoform | 0.39 ± 0.15 | signal transduction |
| HNRPK_HUMAN | P61978 | heterogeneous nuclear ribonucleoprotein K | 0.41 ± 0.04 | mRNA metabolism |
| CHSP1_HUMAN | Q9Y2V2 | calcium-regulated heat-stable protein 1 | 0.42 ± 0.03 | signal transduction |
| PUR6_HUMAN | P22234 | multifunctional protein ADE2 | 0.43 ± 0.05 | purine biosynthesis |
| ANXA1_HUMAN | P04083 | annexin A1 | 0.44 ± 0.12 | membrane organization and trafficking, signal transduction |
| CAPZB_HUMAN | P47756 | F-actin-capping protein subunit beta | 0.45 ± 0.05 | cytoskeletal organization |
| EF1G_HUMAN | P26641 | elongation factor 1-gamma, Homo sapiens | 0.47 ± 0.04 | protein biosynthesis |
| 1433E_HUMAN | P62258 | 14-3-3 protein epsilon | 0.47 ± 0.1 | signal transduction |
| TCPB_HUMAN | P78371 | T-complex protein 1 subunit beta | 0.47 ± 0.04 | protein folding |
| DUT_HUMAN | P33316 | deoxyuridine 5′-triphosphate nucleotidohydrolase, mitochondrial precursor | 0.48 ± 0.20 | nucleotide metabolism |
| NASP_HUMAN | P49321 | nuclear autoantigenic sperm protein | 0.49 ± 0.18 | chromatin assembly |
| COF1_HUMAN | P23528 | cofilin-1 | 0.49 ± 0.04 | cytoskeletal organization |
| PSME2_HUMAN | Q9UL46 | proteasome activator complex subunit 2 | 0.49 ± 0.11 | protein turnover |
| SERA_HUMAN | O43175 | d-3-phosphoglycerate dehydrogenase | 0.50 ± 0.17 | amino acid biosynthesis |
| DPYL2_HUMAN | Q16555 | dihydropyrimidinase-related protein 2 | 0.50 ± 0.07 | signal transduction |
| GDS1_HUMAN | P52306 | Rap1 GTPase-GDP dissociation stimulator | 0.50 ± 0.014 | signal transduction |
| NDKB_HUMAN | P22392 | putative nucleoside diphosphate kinase | 0.52 ± 0.14 | nucleotide metabolism |
| CH60_HUMAN | P10809 | 60 kDa heat shock protein, mitochondrial precursor | 0.53 ± 0.02 | protein folding, stress response |
| DNJA1_HUMAN | P31689 | DnaJ homologue subfamily A member 1 | 0.53 ± 0.03 | protein folding, stress response |
| NHERF_HUMAN | O14745 | ezrin-radixin-moesin-binding phosphoprotein 50 | 0.54 ± 0.13 | membrane organization and trafficking, signal transduction |
| PUR8_HUMAN | P30566 | adenylosuccinate lyase, Homo sapiens | 0.55 ± 0.03 | purine biosynthesis |
| IMDH2_HUMAN | P12268 | inosine-5′-monophosphate dehydrogenase 2 | 0.56 ± 0.11 | purine biosynthesis |
| NUCL_HUMAN | P19338 | nucleolin | 0.58 ± 0.13 | chromatin structure, transcription, intracellular transport |
| SYG_HUMAN | P41250 | glycyl-tRNA synthetase | 0.59 ± 0.01 | protein biosynthesis |
| PPM1G_HUMAN | O15355 | protein phosphatase 1G | 0.59 ± 0.26 | signal transduction |
| PLSL_HUMAN | P13796 | plastin-2 | 0.60 ± 0.05 | cytoskeletal organization |
| SIAS_HUMAN | Q9NR45 | sialic acid synthase | 0.60 ± 0.07 | sugar metabolism |
| 1433B_HUMAN | P31946 | 14-3-3 protein beta/alpha | 0.60 ± 0.02 | signal transduction |
| NUDC_HUMAN | Q9Y266 | nuclear migration protein nudC | 0.61 ± 0.13 | cytoskeletal dynamics |
| MYL6_HUMAN | P60660 | myosin light polypeptide 6 | 0.61 ± 0.03 | cytoskeletal dynamics |
| TTL12_HUMAN | Q14166 | tubulin–tyrosine ligase-like protein 12 | 0.61 ± 0.10 | unknown |
| PPIA_HUMAN | P62937 | peptidyl-prolyl cis–trans isomerase A | 0.62 ± 0.01 | protein biosynthesis |
| HSP74_HUMAN | P34932 | heat shock 70 kDa protein 4 | 0.63 ± 0.14 | protein folding, stress response |
| ADRM1_HUMAN | Q16186 | protein ADRM1 | 0.64 ± 0.03 | protein turnover |
| LGUL_HUMAN | Q04760 | lactoylglutathione lyase | 0.64 ± 0.04 | metabolism |
| ENOA_HUMAN | P06733 | alpha-enolase | 0.65 ± 0.02 | metabolism |
| IF5A1_HUMAN | P63241 | eukaryotic translation initiation factor 5A-1 | 0.65 ± 0.04 | protein biosynthesis |
| HYES_HUMAN | P34913 | epoxide hydrolase 2 | 0.65 ± 0.14 | metabolism |
No light peptide detected.
To verify the results of the SILAC-BST analysis, we analyzed denitrosylation of individual proteins in Jurkat cell lysates. We focused on two candidate Trx-substrate proteins, annexin-1 and 14-3-3θ (Table 1), as well as a third protein, peroxiredoxin 1 (Prx1), which was unaffected by Trx (Supplemental Table 1). BST analysis revealed that incubation with Trx/TrxR lead to denitrosylation of annexin-1 and 14-3-3θ, but not of Prx1 (Figure 3A). To verify these findings in vivo, RAW264.7 macrophages were treated with lipopolysaccharide (LPS) plus interferon-γ (IFNγ), which induces iNOS, in combination with (or without) auranofin, a potent TrxR inhibitor (24). Whereas treatment with auranofin markedly increased LPS-IFNγ-dependent S-nitrosylation of annexin-1 and 14-3-3θ, it had little effect on S-nitrosylation of Prx1 (Figure 3B). These data strongly suggest that annexin-1 and 14-3-3θ are bona fide endogenous Trx SNO-substrates, and more generally, support the validity our proteomic approach.
Figure 3.
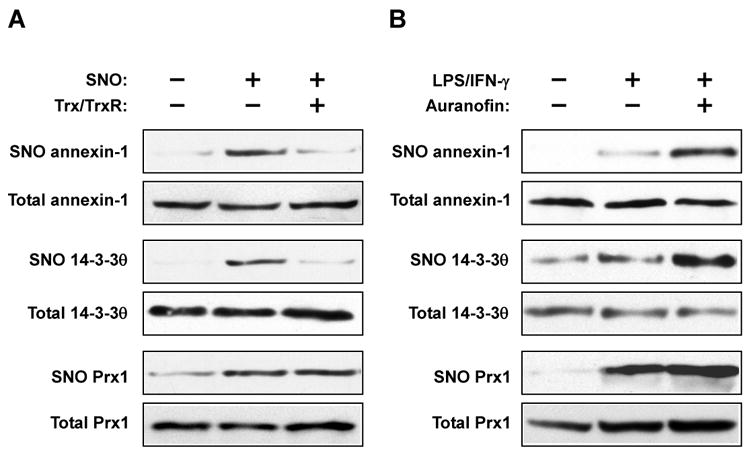
Verification of endogenous denitrosylation of selected proteins. (A) S-Nitrosylated samples were treated with the Trx system for 20 min. S-Nitrosylation of annexin-1, 14-3-3θ and Prx1 was assessed by BST. (B) RAW264.7 cells were treated with LPS (1 μg/ml)-IFNγ (10 ng/ml) for 16 hr, or with LPS-IFNγ plus auranofin (1 μM, added for the last hr). S-Nitrosylation of annexin-1, 14-3-3θ and Prx1 was assessed by BST.
Discussion
The importance of protein denitrosylation in regulating a host of cellular responses is increasingly recognized (4). Recent studies have provided important insights into mechanisms of protein denitrosylation, and in particular, pointed to two Cys denitrosylases, GSH/GSNOR and Trx/TrxR, as physiological effectors of denitrosylation (4). Notwithstanding recent progress, the pursuit of physiological denitrosylase substrates in different cell types and organisms is in its early stages, and major questions remain regarding the complement of SNO-substrates, the relative specificity of denitrosylases and their functions.
Recent proteomic studies have begun to characterize protein denitrosylation on a global scale (25, 26). Analysis of GSH-mediated denitrosylation of ∼100 proteins in brain cytosolic lysates found that the majority of SNOs are highly reactive (i.e., undergo rapid denitrosylation), whereas a smaller subset of SNOs are un-reactive towards GSH (25). Evidence was obtained that protein conformation and solvent exposure may determine susceptibility to GSH-mediated denitrosylation (25), but the overall structural features that dictate reactivity await future investigation. In a recent study, global analysis of denitrosylation (profiling hundreds of SNO sites simultaneously) revealed varying rates of SNO turnover, suggestive of multiple denitrosylation mechanisms (26). It should be noted that the conditions used in those studies as well as in the present work simulate or approximate conditions of nitrosative stress (i.e., high levels of NO/SNO). As such, the findings may be most relevant to understanding the role of denitrosylases in protection from NO-related cellular stress, as shown here and elsewhere in the context of iNOS- and endotoxin-induced nitrosylation (4, 18, 27).
Although the biochemical basis of Trx/TrxR-dependent denitrosylation has been studied is some detail (7, 8, 10), only a handful of Trx S-nitrosylated targets have been identified to date. The present study is the first to systematically identify Trx SNO-substrates. Our experimental strategy enabled the identification, among a large number of SNO-proteins, of a subset of proteins that are preferentially denitrosylated by Trx/TrxR (Table 1). These proteins are involved in a range of cellular processes, including metabolism, cytoskeletal organization, protein folding and signal transduction, supporting the idea that Trx may regulate multiple NO-related cellular functions. Notably, about a third of the candidate Trx substrates that we identified (Table 1) have been reported to undergo rapid denitrosylation in intact cells (26), consistent with the idea that Trx/TrxR is involved in rapid SNO turnover on a wide range of proteins in vivo (8-10). Denitrosylation of these substrates could be important in preserving protein and cellular function, a notion consistent with recent studies documenting a protective role of Trx/TrxR in settings of nitrosative stress (14-18, 28).
Previous SNO proteomic studies in which S-nitrosylation sites were mapped have typically identified one SNO group per protein, although examples of multiple S-nitrosylated thiols have also been documented (25, 26, 29-31). In cases of multisite protein S-nitrosylation or “hyper-S-nitrosylation”, it is conceivable that one Cys may be denitrosylated by Trx whereas others may not, or that reaction rates for different SNO-Cys may vary considerably. In such cases, our experimental approach may underestimate the extent of Trx-mediated denitrosylation. Proteomic approaches that are therefore geared towards SNO site identification (25, 26, 29, 30) may afford a more precise analysis of nitrosylation sites, and may thus be of benefit in revealing determinants of denitrosylase specificity (e.g., Trx/TrxR versus GSH/GSNOR).
Many of the SNO-proteins identified in the present study have been previously identified as S-nitrosylation targets (25, 26, 29, 30, 32), but our findings also include new substrates as well as an assignment to their cognate denitrosylase (Trx). Our results suggest, for example, that S-nitrosylated glutaredoxin-1 (Grx1) is a preferred Trx substrate (Table 1). Grx1 can be S-nitrosylated on multiple Cys residues, a process that is associated with loss of activity (33). Although it remains to be determined which Grx1 thiol(s) is denitrosylated by Trx, a likely scenario is that Trx preserves Grx1 redox activity in cells experiencing nitrosative stress. In contrast to Grx1, Trx does not appear to denitrosylate the key antioxidant enzyme, Prx1 (Supplemental Table 1, Figure 3). Several members of the Prx family were previously identified in large scale S-nitrosylation studies (32, 34-38), and S-nitrosylation of mammalian Prx2 in particular has been shown to inhibit its peroxide-reducing activity (39). The importance of stable S-nitrosylation to the function of Prxs merits further investigation. Finally, in support of our findings, several Trx SNO-substrates identified here (including 14-3-3θ) are also up-regulated in HepG2 cells treated with auranofin and CysNO (40).
Recent advances in proteomic methodologies have greatly expanded our view of the multifaceted roles that Trx and NO/SNO play in the biology of microbes, plants and metazoans (25, 26, 41-44). The current observations support the idea that mammalian Trx/TrxRs are major protein denitrosylases, which act on a broad spectrum of cellular targets. As such, Trx/TrxRs may regulate a large number of NO-related cellular processes, including receptor-mediated signaling, metabolism and stress responses. Global identification of physiological denitrosylase substrates combined with structure-function analyses will likely contribute significantly to a coherent picture of the mechanisms and roles of denitrosylases in cellular function and disease.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01HL059130 and R01HL096673.
Abbreviations
- NO
nitric oxide
- iNOS
inducible nitric oxide synthase
- GSNO
S-nitrosoglutathione
- GSNOR
S-nitrosoglutathione reductase
- SNO
S-nitrosothiol
- Trx
thioredoxin
- TrxR
thioredoxin reductase
- SILAC
stable isotope labeling by amino acids in cell culture
- LC-MS
liquid chromatography-mass spectrometry
Footnotes
Supporting Information Available: Supplementary Table 1 lists SNO-proteins displaying SILAC ratio (L/H) > 0.65. This material is available free of charge via the Internet at htttp://pubs.acs.org.
References
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura T, Lipton SA. Emerging roles of S-nitrosylation in protein misfolding and neurodegenerative diseases. Antioxid Redox Signal. 2008;10:87–101. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 3.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 5.Kondo N, Nakamura H, Masutani H, Yodoi J. Redox regulation of human thioredoxin network. Antioxid Redox Signal. 2006;8:1881–1890. doi: 10.1089/ars.2006.8.1881. [DOI] [PubMed] [Google Scholar]
- 6.Lillig CH, Holmgren A. Thioredoxin and related molecules-from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 7.Nikitovic D, Holmgren A. S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J Biol Chem. 1996;271:19180–19185. doi: 10.1074/jbc.271.32.19180. [DOI] [PubMed] [Google Scholar]
- 8.Stoyanovsky DA, Tyurina YY, Tyurin VA, Anand D, Mandavia DN, Gius D, Ivanova J, Pitt B, Billiar TR, Kagan VE. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J Am Chem Soc. 2005;127:15815–15823. doi: 10.1021/ja0529135. [DOI] [PubMed] [Google Scholar]
- 9.Sengupta R, Ryter SW, Zuckerbraun BS, Tzeng E, Billiar TR, Stoyanovsky DA. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry. 2007;46:8472–8483. doi: 10.1021/bi700449x. [DOI] [PubMed] [Google Scholar]
- 10.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta R, Billiar TR, Atkins JL, Kagan VE, Stoyanovsky DA. Nitric oxide and dihydrolipoic acid modulate the activity of caspase 3 in HepG2 cells. FEBS Lett. 2009;583:3525–3530. doi: 10.1016/j.febslet.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sengupta R, Billiar TR, Stoyanovsky DA. Studies toward the analysis of S-nitrosoproteins. Org Biomol Chem. 2009;7:232–234. doi: 10.1039/b817981f. [DOI] [PubMed] [Google Scholar]
- 13.Sengupta R, Billiar TR, Kagan VE, Stoyanovsky DA. Nitric oxide and thioredoxin type 1 modulate the activity of caspase 8 in HepG2 cells. Biochem Biophys Res Commun. 2010;391:1127–1130. doi: 10.1016/j.bbrc.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferret PJ, Soum E, Negre O, Wollman EE, Fradelizi D. Protective effect of thioredoxin upon NO-mediated cell injury in THP1 monocytic human cells. Biochem J. 2000;346:759–765. [PMC free article] [PubMed] [Google Scholar]
- 15.Comtois SL, Gidley MD, Kelly DJ. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology. 2003;149:121–129. doi: 10.1099/mic.0.25896-0. [DOI] [PubMed] [Google Scholar]
- 16.Potter AJ, Kidd SP, Edwards JL, Falsetta ML, Apicella MA, Jennings MP, McEwan AG. Thioredoxin reductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J Infect Dis. 2009;199:227–235. doi: 10.1086/595737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arai RJ, Ogata FT, Batista WL, Masutani H, Yodoi J, Debbas V, Augusto O, Stern A, Monteiro HP. Thioredoxin-1 promotes survival in cells exposed to S-nitrosoglutathione: Correlation with reduction of intracellular levels of nitrosothiols and up-regulation of the ERK1/2 MAP kinases. Toxicol Appl Pharmacol. 2008;233:227–237. doi: 10.1016/j.taap.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Forrester MT, Seth D, Hausladen A, Eyler CE, Foster MW, Matsumoto A, Benhar M, Marshall HE, Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J Biol Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang K, Ragsdale NV, Carey RM, MacDonald T, Gaston B. Reductive assays for S-nitrosothiols: implications for measurements in biological systems. Biochem Biophys Res Commun. 1998;252:535–540. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- 20.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analyt Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 23.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 24.Gromer S, Arscott LD, Williams CH, Jr, Schirmer RH, Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 25.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 28.Edes K, Cassidy P, Shami PJ, Moos PJ. JS-K, a nitric oxide prodrug, has enhanced cytotoxicity in colon cancer cells with knockdown of thioredoxin reductase 1. PLoS One. 2010;5:e8786. doi: 10.1371/journal.pone.0008786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 32.Lam YW, Yuan Y, Isaac J, Babu CV, Meller J, Ho SM. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PLoS One. 2010;5:e9075. doi: 10.1371/journal.pone.0009075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashemy SI, Johansson C, Berndt C, Lillig CH, Holmgren A. Oxidation and S-nitrosylation of cysteines in human cytosolic and mitochondrial glutaredoxins: effects on structure and activity. J Biol Chem. 2007;282:14428–14436. doi: 10.1074/jbc.M700927200. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Ruiz A, Lamas S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch Biochem Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Dall'Agnol M, Bernstein C, Bernstein H, Garewal H, Payne CM. Identification of S-nitrosylated proteins after chronic exposure of colon epithelial cells to deoxycholate. Proteomics. 2006;6:1654–1662. doi: 10.1002/pmic.200500240. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Sanchez LM, Corrales FJ, Gonzalez R, Ferrin G, Munoz-Castaneda JR, Ranchal I, Hidalgo AB, Briceno J, Lopez-Cillero P, Gomez MA, De La Mata M, Muntane J, Rodriguez-Ariza A. Alteration of S-nitrosothiol homeostasis and targets for protein S-nitrosation in human hepatocytes. Proteomics. 2008;8:4709–4720. doi: 10.1002/pmic.200700313. [DOI] [PubMed] [Google Scholar]
- 37.Romero-Puertas MC, Campostrini N, Matte A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8:1459–1469. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- 38.Han P, Chen C. Detergent-free biotin switch combined with liquid chromatography/tandem mass spectrometry in the analysis of S-nitrosylated proteins. Rapid Commun Mass Spectrom. 2008;22:1137–1145. doi: 10.1002/rcm.3476. [DOI] [PubMed] [Google Scholar]
- 39.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Sanchez LM, Corrales FJ, Lopez-Pedrera C, Aranda E, Rodriguez-Ariza A. Pharmacological impairment of S-nitrosoglutathione or thioredoxin reductases augments protein S-nitrosation in human hepatocarcinoma cells. Anticancer Res. 2010;30:415–421. [PubMed] [Google Scholar]
- 41.Fu C, Wu C, Liu T, Ago T, Zhai P, Sadoshima J, Li H. Elucidation of thioredoxin target protein networks in mouse. Mol Cell Proteomics. 2009;8:1674–1687. doi: 10.1074/mcp.M800580-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagglund P, Bunkenborg J, Maeda K, Svensson B. Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J Proteome Res. 2008;7:5270–5276. doi: 10.1021/pr800633y. [DOI] [PubMed] [Google Scholar]
- 43.Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. Thioredoxin targets in plants: the first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc Natl Acad Sci U S A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.