Abstract
Background:
Defective DNA repair has a causal role in hereditary colorectal cancer (CRC). Defects in the base excision repair gene MUTYH are responsible for MUTYH-associated polyposis and CRC predisposition as an autosomal recessive trait. Numerous reports have suggested MUTYH mono-allelic variants to be low penetrance risk alleles. We report a large collaborative meta-analysis to assess and refine CRC risk estimates associated with bi-allelic and mono-allelic MUTYH variants and investigate age and sex influence on risk.
Methods:
MUTYH genotype data were included from 20 565 cases and 15 524 controls. Three logistic regression models were tested: a crude model; adjusted for age and sex; adjusted for age, sex and study.
Results:
All three models produced very similar results. MUTYH bi-allelic carriers demonstrated a 28-fold increase in risk (95% confidence interval (CI): 6.95–115). Significant bi-allelic effects were also observed for G396D and Y179C/G396D compound heterozygotes and a marginal mono-allelic effect for variant Y179C (odds ratio (OR)=1.34; 95% CI: 1.00–1.80). A pooled meta-analysis of all published and unpublished datasets submitted showed bi-allelic effects for MUTYH, G396D and Y179C (OR=10.8, 95% CI: 5.02–23.2; OR=6.47, 95% CI: 2.33–18.0; OR=3.35, 95% CI: 1.14–9.89) and marginal mono-allelic effect for variants MUTYH (OR=1.16, 95% CI: 1.00–1.34) and Y179C alone (OR=1.34, 95% CI: 1.01–1.77).
Conclusions:
Overall, this large study refines estimates of disease risk associated with mono-allelic and bi-allelic MUTYH carriers.
Keywords: colorectal cancer, base excision repair, MUTYH , carrier risk estimates, meta-analysis
Oxidative damage to DNA occurs with cell proliferation and increases with age. Certain organs such as the gut are heavily exposed to oxidising agents, which impacts on carcinogenic potential. Dysfunction of base excision repair, the major pathway for repairing oxidative damage, has been implicated as a risk factor for the development of multiple colorectal adenomas and colorectal cancer (CRC; Al-Tassan et al, 2002; Croitoru et al, 2004; Farrington et al, 2005). Bi-allelic mutations of the MUTYH gene seem to be responsible for a high proportion of the multiple adenoma phenotype families (termed MUTYH-associated polyposis (MAP)) unaccounted for by germline APC mutations (Al-Tassan et al, 2002; Sampson et al, 2003; Sieber et al, 2003; Gismondi et al, 2004; Venesio et al, 2004; Nielsen et al, 2009) and predispose to CRC per se (Enholm et al, 2003; Croitoru et al, 2004; Fleischmann et al, 2004; Kambara et al, 2004; Wang et al, 2004; Farrington et al, 2005; Peterlongo et al, 2005; Zhou et al, 2005; Moreno et al, 2006; Tenesa et al, 2006; Webb et al, 2006; Küry et al, 2007; Cleary et al, 2009; Lubbe et al, 2009). Although an increased CRC risk associated with bi-allelic MUTYH mutations is incontrovertible, the risk associated with one MUTYH mutant allele is controversial (Croitoru et al, 2004; Farrington et al, 2005; Jenkins et al, 2006; Tenesa et al, 2006; Webb et al, 2006; Cleary et al, 2009; Jones et al, 2009; Lubbe et al, 2009). A statistically significant or close to significant effect, for a MUTYH mono-allelic effect, has been reported in different studies with possible age specific effects present, but the rarity of the alleles associated with the small increased risk for CRC have made it difficult to replicate study findings. A recent risk analysis of MAP family members agreed with previous family based findings (Jenkins et al, 2006) that mono-allelic carriers are at a two-fold increase in risk of CRC (Jones et al, 2009) providing further evidence of a mono-allelic effect of the gene. However, family based studies can be subject to ascertainment bias and any mono-allelic effect could potentially be modified by other inherited factors, including alleles at other loci. Furthermore, environmental risk factors also show familial aggregation and hence, studies in which there has been selection of cases based on family history may be confounded. Bi-allelic carriers may develop CRC because of the predominant effect of MUTYH, whereas the environmental effect is greater in affected siblings with mono-allelic mutations but the risk is ascribed to the MUTYH allele. Thus further work is required to resolve the mono-allelic carrier risk question.
To clarify the role of MUTYH in disease risk, we initiated a multi-centre collaboration allowing large-scale meta-analysis of the individual MUTYH variants, with special interest in determining if there were age and sex-specific effects on CRC association with MUTYH variants (Farrington et al, 2006). In this study, we present the results of this collaborative meta-analysis.
Subjects and methods
Participating studies
Relevant case–control studies to be invited for inclusion in the meta-analysis of the effect of MUTYH on CRC risk were identified by a literature search in the ISI Web of Science (http://wok.mimas.ac.uk) and PUBMED bibliographic databases (http://www.ncbi.nlm.nih.gov/pubmed/), using the search terms ‘MYH or MUTYH and CRC’. In the initial search 55 studies were identified and eight of these were considered for our study (Enholm et al, 2003; Croitoru et al, 2004; Fleischmann et al, 2004; Kambara et al, 2004; Wang et al, 2004; Farrington et al, 2005; Peterlongo et al, 2005; Zhou et al, 2005). The inclusion criteria were as follows: the patients had to be diagnosed with CRC and the studies had to have genotype data for both cases and controls. Ten additional studies were identified during the progress of the project – Webb et al (2006), Moreno et al (2006), Küry et al (2007), Cleary et al (2009), Lubbe et al (2009); and unpublished data from Koessler T and Pharoah PD; and Tomlinson – personal communication. Colebatch et al (2006); Balaguer et al (2007); Avezzù et al (2008) were used in the pooled meta-analysis of all available published and unpublished datasets.
The principal investigators (PIs) of the selected studies were contacted and were asked to participate by providing a minimum dataset including variables necessary for the analysis (Supplementary Box 1: Study questionnaire; Supplementary Table 1: Data extraction table). In cases, in whom PIs failed to respond to our invitation to participate, reminder letters were despatched. It was not possible to include data from the following studies in the logistic regression analyses because (i) data was only available for cases that were heterozygous or homozygous for a MUTYH mutation (Enholm et al, 2003); (ii) co-variate data were only available for cases, as controls were anonymous blood donors (Zhou et al, 2005; Tomlinson, unpublished data); (iii) failure to communicate with us (Kambara et al, 2004 and Wang et al, 2004). The study by Fleischmann et al (2004) and Webb et al (2006) were not used because they had been superseded by a later study (Lubbe et al, 2009), which was included.
Statistical analysis
Data from all collaborating centres were checked for completeness, coded and merged to form a core database. MUTYH defects were considered pathogenic only if there was published evidence of their pathogenicity. Individuals reported to have two defects of MUTYH in the original report were classified as mutated/mutated (MM), those with one defect as wild type/mutant (WM) and those with no mutation as wild type/wild type (WW). Descriptive statistics were produced on all subject characteristics, risk factors and event data. All populations described in the case–control studies were tested for Hardy–Weinberg equilibrium in controls and the genotype distributions between all groups were compared by χ2-test.
Three logistic regression models were applied to address confounding co-variates (model I: crude, model II: including co-variates for age and sex, model III: including co-variates for age, sex and study) on the combined datasets investigating the effect of MUTYH defects (WM vs WW and MM vs WW) as well as of the individual mutations Y179C (c.536A>G/p.Tyr179Cys; AA=WW, GG=MM) and G396D (c.1187G>A/p.Gly396Asp; GG=WW, AA=MM; previously known as Y165C and G382D), to identify any variant specific associations. The three logistic regression models were applied after sex and age (over 55 years and under or equal 55 years) stratification as previously described (Farrington et al, 2005), to assess the effect of age and sex on the association of the variants with disease risk. In all the studies, interaction associations between the MUTYH variants and study code (for each individual study) were estimated and similarly between MUTYH variants and hormone replacement therapy (HRT) among female participants in three studies (the Scottish SOCCS studies and the studies – Croitoru et al, 2004; Cleary et al, 2009). Association between both genetic (i.e., one of the MUTYH mutations) and the study code or environmental factor (i.e., HRT) and disease was assessed and interaction was tested by fitting interactive and nested multiplicative models. To assess for any small study effects, we performed Funnel plot analysis and tested for significance using the Harbord test.
Finally, the relationship between the genotype and CRC was analysed by meta-analysis, combining the effect estimates of all published and unpublished datasets.
All statistic analyses were conducted using Intercooled STATA version 10.0 (Stata Corp, College Station, TX, USA). For the logistic regression analyses, it is necessary to add a whole number to any fields containing 0 (see model Ia in Table 2), which reduces the final OR value, however, by using the META command in the STATA meta-analysis programme, a lower value can be added (0.5 as indicated by model Ib in Table 2) thereby giving a more accurate assessment of risk. However, this is a grouped analysis and therefore cannot be adjusted for confounding co-variates, such as age/sex and study as in models II and III. To account for multiple testing we applied the Bonferroni correction method, and the P-value threshold for significance was estimated to be 0.003.
Results
Table 1 details summary data from the studies included in our combined analysis (comprising a total of 20 565 cases and 15 524 controls). The two variant alleles are rare with G396D variant allele having a frequency of 0.007 in controls and the Y179C variant allele a frequency of 0.002. Tests for deviation from Hardy–Weinberg equilibrium in controls were P=0.99 and P<0.00005 for G396D and Y179C variants, respectively.
Table 1. Summary tablea.
|
G396D
|
Y179C
|
Genotype
|
Age |
Sex
|
Ethnicity
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Number | GG | GA | AA | AA | AG | GG | WW | WM | MM | n (%) | Mean (SD) | n (%) | Males (%) | n (%) | Caucasian/White (%) |
| All studies | ||||||||||||||||
| Cases | 20 565 | 19 966 | 324 | 33 | 20 260 | 154 | 11 | 19 783 | 417 | 76 | 20 555 (99.9) | 59.5 (10.1) | 20 555 (99.9) | 11 668 (56.8) | 5374 (26.1) | 5030 (93.6) |
| Controls | 15 524 | 15 145 | 211 | 0 | 15 074 | 68 | 2 | 14 723 | 280 | 2 | 15 287 (98.5) | 59.0 (10.9) | 15 521 (99.9) | 6845 (44.1) | 5597 (36.1) | 4796 (85.7) |
| Croitoru et al (2004) | ||||||||||||||||
| Cases | 1238 | 1209 | 25 | 4 | 1223 | 13 | 2 | 1197 | 29 | 12 | 1236 (99.8) | 60.1 (8.7) | 1236 (99.8) | 544 (44.0) | Missing | Missing |
| Controls | 1255 | 1238 | 17 | 0 | 1251 | 4 | 0 | 1234 | 21 | 0 | 1244 (99.1) | 63.6 (8.6) | 1252 (99.8) | 702 (56.1) | Missing | Missing |
| Peterlongo et al (2005) | ||||||||||||||||
| Cases | 585 | 567 | 3 | 1 | 569 | 3 | 0 | 552 | 4 | 2 | 584 (99.8) | 62.1 (13.1) | 584 (99.8) | 319 (54.6) | 585 (100) | 251 (42.9) |
| Controls | 1158 | 1040 | 5 | 0 | 1039 | 2 | 0 | 923 | 7 | 0 | 1148 (99.1) | 53.9 (11.5) | 1156 (99.8) | 240 (20.8) | 1158 (100) | 363 (31.3) |
| SOCCS (includes data from Farrington et al (2005), Tenesa et al (2006) and unpublished SOCCS prospective samples) | ||||||||||||||||
| Cases | 3278 | 3086 | 55 | 8 | 3192 | 23 | 0 | 3038 | 71 | 12 | 3277 (100) | 59.5 (11.3) | 3278 (100) | 1883 (57.4) | 3278 (100) | 3268 (99.7) |
| Controls | 3318 | 3239 | 43 | 0 | 3061 | 15 | 0 | 2993 | 57 | 0 | 3313 (99.8) | 61.4 (10.9) | 3318 (100) | 1861 (56.1) | 3318 (100) | 3312 (99.8) |
| Moreno et al (2006) | ||||||||||||||||
| Cases | 356 | 343 | 8 | 1 | 348 | 0 | 0 | 336 | 8 | 1 | 356 (100) | 66.5 (11.7) | 356 (100) | 217 (61.0) | Missing | Missing |
| Controls | 297 | 285 | 7 | 0 | 291 | 0 | 0 | 283 | 7 | 0 | 297 (100) | 65.3 (12.5) | 297 (100) | 158 (53.2) | Missing | Missing |
| Koessler T, (unpublished data obtained in 2007) | ||||||||||||||||
| Cases | 2262 | 2215 | 31 | 2 | 2227 | 19 | 1 | 2198 | 37 | 9 | 2253 (99.6) | 59.1 (8.1) | 2262 (100) | 1287 (57.1) | Missing | Missing |
| Controls | 2253 | 2217 | 31 | 0 | 2236 | 11 | 0 | 2204 | 42 | 0 | 2053 (91.1) | 53.4 (7.6) | 2253 (100) | 949 (42.0) | Missing | Missing |
| Küry et al (2007) | ||||||||||||||||
| Cases | 1025 | 1003 | 22 | 0 | 1021 | 4 | 0 | 999 | 25 | 1 | 1023 (99.8) | 68.7 (9.9) | 1025 (100) | 632 (61.7) | 1025 (100) | 1025 (100) |
| Controls | 1121 | 1105 | 16 | 0 | 1117 | 4 | 0 | 1100 | 21 | 0 | 1121 (100) | 61.9 (10.0) | 1121 (100) | 609 (54.3) | 1121 (100) | 1121 (100) |
| SOCCS retrospective cases (unpublished data obtained in 2008) | ||||||||||||||||
| Cases | 486 | 403 | 6 | 2 | 446 | 2 | 3 | 391 | 6 | 4 | 486 (100) | 54.7 (17.2) | 479 (98.6) | 240 (50.1) | 486 (100) | 486 (100) |
| Controls | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Cleary et al (2009) | ||||||||||||||||
| Cases | 2076 | 2023 | 33 | 5 | 2053 | 7 | 1 | 2029 | 38 | 9 | 2070 (99.7) | 56.6 (10.8) | 2076 (100) | 1114 (53.7) | Missing | Missing |
| Controls | 1049 | 1032 | 17 | 0 | 1042 | 6 | 1 | 1024 | 24 | 1 | 1047 (99.8) | 56.4 (11.3) | 1049 (100) | 444 (42.3) | Missing | Missing |
| Lubbe et al (2009) b | ||||||||||||||||
| Cases | 9268 | 9117 | 141 | 10 | 9181 | 83 | 4 | 9043 | 198 | 27 | 9268 (100) | 59.0 (8.5) | 9268 (100) | 5432 (58.6) | Missing | Missingc |
| Controls | 5064 | 4989 | 75 | 0 | 5037 | 26 | 1 | 4962 | 101 | 1 | 5064 (100) | 59.2 (10.8) | 5064 (100) | 1882 (37.2) | Missing | Missingc |
Abbreviations: MM=mutated/mutated; WM=wild type/mutant; WW=wild type/wild type.
This table presents the raw data sent to us by each group that was then included in the analyses. G396D and Y179C are looked at independently and any compound bi-allelic carriers are presented as heterozygotes for each variant; MUTYH genotype data also includes other pathogenic mutations; unpublished data are included in Peterlonogo P and Moreno V, SOCCS prospective and Küry S.
Includes data from Webb et al (2006) and Fleischmann et al (2004).
All the cases and controls were UK residents and of European ancestry (self-reported).
Bi-allelic effect of MUTYH
All three models of the logistic regression analysis gave consistent results and so the results of the crude analysis (model I) are described below and presented in Table 2; results of the other two models can be found in Supplementary Table 2. Bi-allelic carriers for the MM genotype of the combined MUTYH defects, G396D and Y179C/G396D compound heterozygotes were associated with a significant increase in CRC risk (odds ratio (OR)=28.3, 95% confidence limits (CIs): 6.95–115; 23.1 (95% CI: 3.15–169) and 21.6 (95% CI: 2.94–159), respectively). These risks are conservative, concentrating on the significant logistic regression results – model Ib results presented in Table 2 are likely a better reflection of risk and tend to be two-fold higher. There was a greater CRC risk for the MM genotype for the earlier age individuals when compared with the older age group (OR=36.2 (95% CI: 4.98–263) for ⩽55 years compared with 11.6 (95% CI: 2.77–48.2) for >55 years). However, their CIs overlapped and the results were not statistically significantly different. ANOVA analysis of mean age of carriers demonstrated that there are significant age differences between cases and controls when considering MM genotype carriers and Y179C bi-allelic carriers but not for G396D carriers (P<0.0005, P<0.0005 and P=0.27, respectively – Supplementary Table 3). Indeed there is a significant age difference between mean age of bi-allelic Y179C and G396D carriers (48.9 vs 56.7, respectively, P=0.003 based on t-test – Supplementary Table 4).
Table 2. Logistic regression analysis of the combined datasets; G396D analysis was conducted for individuals that were Y179C AA; Y179C analysis was conducted for individuals that were G396D GG; combined genotype analysis was conducted for individuals with data for both Y179C and G396D.
|
Model I
*
,
a
|
Model I
b
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Cases | Controls | OR | 95% CI | P-value | OR | 95% CI | P-value |
| G396D c | ||||||||
| Whole sample | ||||||||
| GG | 19 767 | 14 723 | 1.00 | |||||
| GA | 292 | 210 | 1.04 | 0.87, 1.24 | 0.70 | |||
| AA | 31 | 0 | 23.09 | 3.15, 169.15 | 0.002 | 46.93 | 2.87, 766.89 | 0.007 |
| ⩽55 Years old | ||||||||
| GG | 6269 | 5270 | 1.00 | |||||
| GA | 88 | 77 | 0.96 | 0.71, 1.31 | 0.80 | |||
| AA | 13 | 0 | 10.93 | 1.43, 83.57 | 0.02 | 22.70 | 1.35, 381.91 | 0.03 |
| >55 Years old | ||||||||
| GG | 13 498 | 9453 | 1.00 | |||||
| GA | 204 | 133 | 1.07 | 0.86, 1.34 | 0.52 | |||
| AA | 18 | 0 | 12.60 | 1.68, 94.37 | 0.01 | 25.91 | 1.56, 430.03 | 0.02 |
| Males | ||||||||
| GG | 11 229 | 6460 | 1.00 | |||||
| GA | 161 | 95 | 0.98 | 0.76, 1.26 | 0.85 | |||
| AA | 15 | 0 | 8.62 | 1.14, 65.25 | 0.04 | 17.84 | 1.07, 298.11 | 0.04 |
| Females | ||||||||
| GG | 8528 | 8259 | 1.00 | |||||
| GA | 131 | 115 | 1.10 | 0.86, 1.42 | 0.45 | |||
| AA | 16 | 0 | 15.49 | 2.05, 116.86 | 0.008 | 31.96 | 1.92, 532.72 | 0.02 |
| Y179C d | ||||||||
| Whole sample | ||||||||
| AA | 19 767 | 14 723 | 1.00 | |||||
| AG | 122 | 68 | 1.34 | 1.00, 1.80 | 0.05 | |||
| GG | 11 | 2 | 4.10 | 0.91, 18.48 | 0.07 | NA | ||
| ⩽55 Years old | ||||||||
| AA | 6269 | 5270 | 1.00 | |||||
| AG | 35 | 25 | 1.18 | 0.70, 1.97 | 0.54 | |||
| GG | 9 | 0 | 7.57 | 0.96, 59.74 | 0.06 | 15.97 | 0.93, 274.491 | 0.06 |
| >55 Years old | ||||||||
| AA | 13 498 | 9453 | 1.00 | |||||
| AG | 87 | 43 | 1.42 | 0.98, 2.04 | 0.06 | |||
| GG | 2 | 2 | 0.70 | 0.10, 4.97 | 0.72 | NA | ||
| Males | ||||||||
| AA | 11 229 | 6460 | 1.00 | |||||
| AG | 68 | 23 | 1.70 | 1.06, 2.73 | 0.03 | |||
| GG | 8 | 0 | 4.60 | 0.58, 36.80 | 0.15 | 9.78 | 0.56, 169.48 | 0.12 |
| Females | ||||||||
| AA | 8528 | 8259 | 1.00 | |||||
| AG | 54 | 45 | 1.16 | 0.78, 1.73 | 0.46 | |||
| GG | 3 | 2 | 1.45 | 0.24, 8.70 | 0.68 | NA | ||
| Genotype e | ||||||||
| Whole sample | ||||||||
| WW | 19 767 | 14 723 | 1.00 | |||||
| WM | 418 | 280 | 1.11 | 0.95, 1.29 | 0.17 | |||
| MM | 76 | 2 | 28.30 | 6.95, 115.26 | 3.1 × 10−6 | NA | ||
| G396D AA | 31 | 0 | 23.09 | 3.15, 169.15 | 0.002 | 46.93 | 2.87, 766.89 | 0.007 |
| Y179C GG | 11 | 2 | 4.10 | 0.91, 18.48 | 0.07 | NA | ||
| Compound heterozygousf | 29 | 0 | 21.60 | 2.94, 158.58 | 0.003 | 43.95 | 2.69, 719.26 | 0.008 |
| ⩽55 Years old | ||||||||
| WW | 6269 | 5270 | 1.00 | |||||
| WM | 124 | 104 | 1.00 | 0.77, 1.30 | 0.99 | |||
| MM | 43 | 0 | 36.15 | 4.98, 262.57 | 0.0004 | 73.14 | 4.50, 1188.3 | 0.003 |
| G396D AA | 13 | 0 | 10.93 | 1.43, 83.57 | 0.02 | 22.70 | 1.35, 381.91 | 0.03 |
| Y179C GG | 9 | 0 | 7.57 | 0.96, 59.74 | 0.06 | 15.97 | 0.93, 274.49 | 0.06 |
| Compound heterozygous | 17 | 0 | 14.29 | 1.90, 107.42 | 0.01 | 29.42 | 1.77, 489.38 | 0.02 |
| >55 Years old | ||||||||
| WW | 13 498 | 9453 | 1.00 | |||||
| WM | 294 | 176 | 1.17 | 0.97, 1.41 | 0.10 | |||
| MM | 33 | 2 | 11.56 | 2.77, 48.17 | 0.001 | NA | ||
| G396D AA | 18 | 0 | 12.60 | 1.68, 94.37 | 0.01 | 25.91 | 1.56, 430.03 | 0.02 |
| Y179C GG | 2 | 2 | 0.70 | 0.10, 4.97 | 0.72 | NA | ||
| Compound heterozygous | 12 | 0 | 8.40 | 1.09, 64.64 | 0.04 | 17.51 | 1.04, 295.75 | 0.05 |
| Males | ||||||||
| WW | 11 229 | 6460 | 1.00 | |||||
| WM | 232 | 119 | 1.12 | 0.90, 1.40 | 0.31 | |||
| MM | 36 | 0 | 20.71 | 2.84, 151.09 | 0.003 | 42.00 | 2.58, 684.38 | 0.009 |
| G396D AA | 15 | 0 | 8.62 | 1.14, 65.25 | 0.04 | 17.84 | 1.07, 298.11 | 0.04 |
| Y179C GG | 8 | 0 | 4.60 | 0.58, 36.80 | 0.15 | 9.78 | 0.56, 169.48 | 0.12 |
| Compound heterozygous | 12 | 0 | 6.90 | 0.90, 53.10 | 0.06 | 14.38 | 0.85, 242.96 | 0.06 |
| Females | ||||||||
| WW | 8528 | 8259 | 1.00 | |||||
| WM | 186 | 161 | 1.12 | 0.90, 1.38 | 0.30 | |||
| MM | 40 | 2 | 19.37 | 4.68, 80.17 | 4.3 × 10−5 | NA | ||
| G396D AA | 16 | 0 | 15.49 | 2.05, 116.86 | 0.008 | 31.96 | 1.92, 532.72 | 0.02 |
| Y179C GG | 3 | 2 | 1.45 | 0.24, 8.70 | 0.68 | NA | ||
| Compound heterozygous | 17 | 0 | 16.46 | 2.19, 123.70 | 0.006 | 33.90 | 2.04, 563.74 | 0.01 |
Abbreviations: CI=confidence interval; MM=mutated/mutated; NA=not available; OR=odds ratio; WM=wild type/mutant; WW=wild type/wild type.
*Crude analysis.
Estimated by adding one control with the variant genotype.
Estimated using the meta command of STATA and for mathematical reasons, cells with zero frequencies were assumed to be 0.5 (as defaulted by the meta command).
Analysis conducted only for the AA Y179C, that is, WW.
Analysis conducted only for the GG G396D, that is, WW.
Including subjects with data for both Y179C and G396D.
This category includes 29 G396D GA and Y179C AG cases; 5 cases with either G396D GA or Y179C AG and any other pathogenic MUTYH mutation were excluded.
Colorectal cancer risk associated with mono-allelic MUTYH mutations
The results of the combined analysis demonstrate that there are no significant mono-allelic effects for either G396D or for combined MUTYH variants (Table 2). However, the specific Y179C variant was shown to increase risk of disease in the heterozygous state (OR=1.34; (95% CI: 1.00–1.80)) in the whole sample set and also when stratified by sex, male sex demonstrated a mono-allelic effect (OR=1.70; (95% CI: 1.06–2.73)). However, after Bonferroni correction, these mono-allelic effects did not remain significant.
The role of study population and HRT in modulating CRC risk
We hypothesised that origin of the data, that is, study population might modify the association between the genotype and CRC risk. However, there was no evidence for an interaction between study code and MUTYH genotypes (Supplementary Table 5). Similarly, HRT intake, a known risk factor for CRC (Chan et al, 2006; Theodoratou et al, 2008), might be influenced by genotype and therefore modulate female risk. Both the Scottish and Canadian datasets had recorded data on HRT intake and these were used to test for an interaction between HRT and MUTYH genotype. Across both datasets there was no evidence of any interaction between HRT and MUTYH genotype (Supplementary Table 6).
Meta-analysis of published and unpublished datasets
The results of a meta-analysis of published and unpublished datasets submitted to us, estimating the effect of the MUTYH whole gene defects demonstrated a pooled fixed bi-allelic effect of 10.8 (95% CI: 5.02–23.2) for the MM and a pooled fixed mono-allelic effect of 1.16 (95% CI: 1.00–1.34) for WM genotype (Table 3; Figures 1 and 2). Analysis of the specific variants by pooled meta-analysis demonstrated bi-allelic effects for both G396D and Y179C (OR=6.47 (95% CI: 2.33–18.0) and OR=3.35 (95% CI: 1.14–9.89), respectively) and in agreement with the logistic regression analysis results, Y179C variant also demonstrated a very similar pooled fixed mono-allelic effect of 1.34 (95% CI: 1.01–1.77; Tables 4 and 5; Supplementary Figures 1–4).
Table 3. Meta-analysis of studiesa.
|
Genotype (cases)
|
Genotype (controls)
|
WM vs WW | MM vs WW | |||||
|---|---|---|---|---|---|---|---|---|
| Study | WW | WM | MM | WW | WM | MM | OR (95% CI) | OR (95% CI) |
| Enholm et al (2003) | 994 | 5 | 4 | 424 | 0 | 0 | 4.70 (0.26, 85.10) | 3.84 (0.21, 71.51) |
| Kambara et al (2004) | 90 | 2 | 0 | 52 | — | 0 | 1.16 (0.10, 13.06) | NA |
| Wang et al (2004) | 432 | 10 | 2 | 309 | 4 | 0 | 1.79 (0.56, 5.75) | 3.58 (0.17, 74.79) |
| Zhou et al (2005) | 432 | 6 | 0 | 466 | 3 | 0 | 2.16 (0.54, 8.68) | NA |
| Peterlongo et al (2005) | 549 | 4 | 2 | 911 | 7 | 0 | 0.95 (0.28, 3.25) | 8.29 (0.40, 173.08) |
| Tomlinson I (2006)b | 662 | 15 | 1 | 197 | 2 | 0 | 2.23 (0.51, 9.84) | 0.89 (0.04, 22.04) |
| Moreno et al (2006) | 323 | 9 | 0 | 278 | 6 | 0 | 1.29 (0.45, 3.67) | NA |
| Colebatch et al (2006) | 859 | 11 | 2 | 473 | 5 | 0 | 1.21 (0.42, 3.51) | 2.75 (0.13, 57.49) |
| Koessler T (2007)c | 2198 | 37 | 9 | 2204 | 42 | 0 | 0.88 (0.57, 1.38) | 19.05 (1.11, 327.53) |
| Küry et al (2007) | 999 | 24 | 1 | 1100 | 21 | 0 | 1.26 (0.70, 2.27) | 3.30 (0.13, 81.18) |
| Balaguer et al (2007) | 1089 | 19 | 8 | 912 | 22 | 0 | 0.72 (0.39, 1.34) | 14.24 (0.82, 247.02) |
| SOCCS (2008)d | 3429 | 77 | 16 | 2993 | 57 | 0 | 1.18 (0.83, 1.67) | 28.80 (1.73, 480.32) |
| Avezzù et al (2008) | 435 | 2 | 2 | 246 | 1 | 0 | 1.13 (0.10, 12.54) | 2.83 (0.14, 59.19) |
| Cleary et al (2009) e | 3697 | 87 | 27 | 2758 | 43 | 1 | 1.51 (1.04, 2.18) | 20.14 (2.74, 148.32) |
| Lubbe et al (2009) f | 9043 | 198 | 27 | 4962 | 101 | 1 | 1.08 (0.84, 1.37) | 14.82 (2.01, 109.06) |
| M–H pooled effect (fixed) | 25231 | 506 | 101 | 18285 | 315 | 2 | 1.16 (1.00, 1.34) | 10.80 (5.02, 23.21) |
| P-value | 0.05 | <0.0005 | ||||||
| Heterogeneity | ||||||||
| P-value | 0.82 | 0.84 | ||||||
| I2 (95% CI) | 0 (0, 54) | 0 (0, 60) | ||||||
Abbreviations: CI=confidence interval; MM=mutated/mutated; NA=not available; OR=odds ratio; WM=wild type/mutant; WW=wild type/wild type.
This table presents the data as they were published. Two unpublished studies included.
Unpublished data obtained in 2006.
Unpublished data obtained in 2007.
Includes data from Farrington et al (2005), Tenesa et al (2006) and unpublished data from the SOCCS study obtained in 2008.
Includes data from Croitoru et al (2004).
Includes data from Webb et al (2006) and Fleischmann et al (2004).
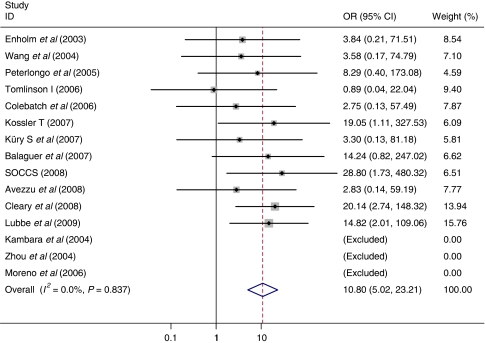
Figure 1.
Meta-analysis of studies comparing MUTYH MM vs WW. SOCCS data include Farrington et al (2005), Tenesa et al (2006) and unpublished data from the SOCCS study obtained in 2008; Cleary data include Croitoru et al (2004); Lubbe data include Webb et al (2006) and Fleischmann et al (2004). Unpublished studies included are Tomilson I (2006) and Koessler T (2007).
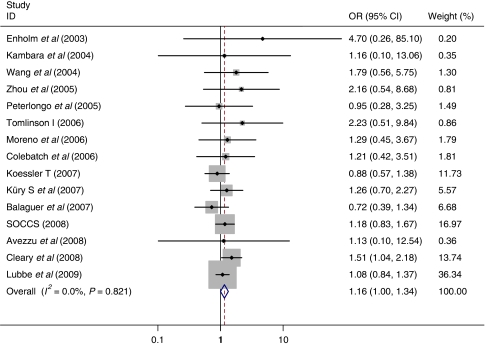
Figure 2.
Meta-analysis of studies comparing MUTYH WM vs WW. SOCCS data include Farrington et al (2005), Tenesa et al (2006) and unpublished data from the SOCCS study obtained in 2008; Cleary data include Croitoru et al (2004); Lubbe data include Webb et al (2006) and Fleischmann et al (2004). Unpublished studies included are Tomilson I (2006) and Koessler T (2007).
Table 4. Meta-analysis of studies for the G396D genotypesa.
|
G396D (cases)
|
G396D (controls)
|
GA vs GG | AA vs GG | |||||
|---|---|---|---|---|---|---|---|---|
| Study | GG | GA | AA | GG | GA | AA | OR (95% CI) | OR (95% CI) |
| Enholm et al (2003) | 994 | 4 | 1 | 424 | 0 | 0 | 3.84 (0.21, 71.51) | 1.28 (0.05, 31.50) |
| Kambara et al (2004) | 90 | 2 | 0 | 52 | — | 0 | Not enough data available | NA |
| Wang et al (2004) | 432 | 5 | 0 | 309 | 2 | 0 | 1.79 (0.35, 9.28) | NA |
| Zhou et al (2005) | 432 | 1 | 0 | 466 | 1 | 0 | 1.08 (0.07, 17.30) | NA |
| Peterlongo et al (2005) | 549 | 2 | 0 | 911 | 5 | 0 | 0.66 (0.13, 3.43) | NA |
| Tomlinson I (2006)b | 662 | 9 | 1 | 197 | 2 | 0 | 1.34 (0.29, 6.25) | 0.89 (0.04, 22.04) |
| Moreno et al (2006) | 323 | 9 | 0 | 278 | 6 | 0 | 1.29 (0.45, 3.67) | NA |
| Colebatch et al (2006) | 859 | 8 | 0 | 473 | 4 | 0 | 1.10 (0.33, 3.67) | NA |
| Koessler T (2007)c | 2198 | 25 | 2 | 2204 | 31 | 0 | 0.81 (0.48, 1.37) | 5.01 (0.24, 104.49) |
| Küry et al (2007) | 999 | 21 | 0 | 1100 | 16 | 0 | 1.45 (0.75, 2.79) | NA |
| Balaguer et al (2007) | 1089 | 15 | 1 | 912 | 20 | 0 | 0.63 (0.32, 1.23) | 2.51 (0.10, 61.75) |
| SOCCS (2008)d | 3429 | 56 | 8 | 2993 | 42 | 0 | 1.16 (0.78, 1.74) | 14.84 (0.86, 257.19) |
| Avezzù et al (2008) | 435 | 2 | 1 | 246 | 0 | 0 | 2.83 (0.14, 59.19) | 1.70 (0.07, 41.84) |
| Cleary et al (2009) e | 3697 | 63 | 11 | 2758 | 32 | 0 | 1.47 (0.96, 2.25) | 17.16 (1.01, 291.31) |
| Lubbe et al (2009) f | 9043 | 128 | 10 | 4962 | 75 | 0 | 0.94 (0.70, 1.25) | 11.52 (0.68, 196.69) |
| M–H pooled effect (fixed) | 25 231 | 350 | 35 | 18 285 | 236 | 0 | 1.07 (0.90, 1.26) | 6.47 (2.33, 17.97) |
| P-value | 0.44 | <0.0005 | ||||||
| Heterogeneity | ||||||||
| P-value | 0.74 | 0.73 | ||||||
| I2 (95% CI) | 0 (0, 55) | 0 (0, 68) | ||||||
Abbreviations: CI=confidence interval; MM=mutated/mutated; NA=not available; OR=odds ratio; WM=wild type/mutant; WW=wild type/wild type.
This table presents the data as they were published. Two unpublished studies included.
Unpublished data obtained in 2006.
Unpublished data obtained in 2007.
Includes data from Farrington et al (2005), Tenesa et al (2006) and unpublished data from the SOCCS study obtained in 2008.
Includes data from Croitoru et al (2004).
Includes data from Webb et al (2006) and Fleischmann et al (2004).
Table 5. Meta-analysis of studies for the Y179C genotypesa.
|
Y179C (cases)
|
Y179C (controls)
|
AG vs AA | GG vs AA | |||||
|---|---|---|---|---|---|---|---|---|
| Study | AA | AG | GG | AA | AG | GG | OR (95% CI) | OR (95% CI) |
| Enholm et al (2003) | 994 | 1 | 0 | 424 | 0 | 0 | 1.28 (0.05, 31.50) | NA |
| Kambara et al (2004) | 90 | 0 | 0 | 52 | — | 0 | Not enough data available | NA |
| Wang et al (2004) | 432 | 5 | 1 | 309 | 2 | 0 | 1.79 (0.35, 9.28) | 2.15 (0.09, 52.87) |
| Zhou et al (2005) | 432 | 3 | 0 | 466 | 2 | 0 | 1.62 (0.35, 9.28) | NA |
| Peterlongo et al (2005) | 549 | 2 | 0 | 911 | 2 | 0 | 1.66 (0.23, 11.81) | NA |
| Tomlinson I (2006)b | 662 | 6 | 0 | 197 | 0 | 0 | 3.88 (0.22, 69.10) | NA |
| Moreno et al (2006) | 323 | 0 | 0 | 278 | 0 | 0 | NA | NA |
| Colebatch et al (2006) | 859 | 3 | 0 | 473 | 1 | 0 | 1.65 (0.17, 15.93) | NA |
| Koessler T (2007)c | 2198 | 12 | 1 | 2204 | 11 | 0 | 1.09 (0.48, 2.48) | 3.01 (0.12, 73.88) |
| Küry et al (2007) | 999 | 3 | 0 | 1100 | 4 | 0 | 0.83 (0.18, 3.70) | NA |
| Balaguer et al (2007) | 1089 | 4 | 2 | 912 | 1 | 0 | 3.35 (0.37, 30.02) | 4.19 (0.20, 87.34) |
| SOCCS (2008)d | 3429 | 21 | 3 | 2993 | 15 | 0 | 1.22 (0.63, 2.38) | 6.11 (0.32, 118.34) |
| Avezzù et al (2008) | 435 | 0 | 0 | 246 | 1 | 0 | 0.19 (0.01, 4.65) | NA |
| Cleary et al (2009) e | 3697 | 15 | 5 | 2758 | 10 | 1 | 1.12 (0.50, 2.50) | 3.73 (0.44, 31.95) |
| Lubbe et al (2009) f | 9043 | 70 | 4 | 4962 | 26 | 1 | 1.48 (0.94, 2.32) | 2.20 (0.25, 19.64) |
| M-H pooled effect (fixed) | 25 231 | 145 | 16 | 18 285 | 75 | 2 | 1.34 (1.01, 1.77) | 3.35 (1.14, 9.89) |
| P-value | 0.04 | 0.03 | ||||||
| Heterogeneity | ||||||||
| P-value | 0.98 | 0.99 | ||||||
| I2 (95% CI) | 0 (0, 57) | 0 (0, 75) | ||||||
Abbreviations: CI=confidence interval; MM=mutated/mutated; NA=not available; OR=odds ratio; WM=wild type/mutant; WW=wild type/wild type.
This table presents the data as they were published. Two unpublished studies included.
Unpublished data obtained in 2006.
Unpublished data obtained in 2007.
Includes data from Farrington et al (2005), Tenesa et al (2006) and unpublished data from the SOCCS study obtained in 2008.
Includes data from Croitoru et al (2004).
Includes data from Webb et al (2006) and Fleischmann et al (2004).
Assessment of study publication bias
Funnel plots for both the mono- and bi-allelic effect were created to assess whether study size was significantly influencing the results. These plots appeared asymmetric, but the Harbord's test for small study effect demonstrated that this was not statistically significant (Supplementary Figures 5 and 6).
Discussion
This large meta-analysis study refines the estimates of CRC risk associated with mutations in the MUTYH gene to date. Bi-allelic carriers of the combined MUTYH mutations (MM) are associated with a 28-fold (95% CI: 6.95–115) increase in CRC risk from the logistic regression analysis. Bi-allelic carriers of the G396D variant and Y179C/G396D compound heterozygotes were also significantly associated with a similar increase in CRC risk (OR=23.1 (95% CI: 3.15–169) and 21.6 (95% CI: 2.94–159), respectively). Although the risk estimate was slightly lower from the overall larger pooled meta-analysis of published and unpublished datasets (OR=10.8 (95% CI: 5.02–23.2)), both G396D and Y179C variants demonstrated bi-allelic effects in this pooled analysis (OR=6.47 (95% CI: 2.33–18.0) and OR=3.35 (95% CI: 1.14–9.89), respectively). A marginal significant mono-allelic effect was demonstrated for the specific variant Y179C (OR=1.34 (95% CI: 1.00–1.80)) and indeed a marginally significant result was also observed in the pooled meta-analysis for MUTYH WM (OR=1.16 (95% CI: 1.00–1.34)) and Y179C variant alone, 1.34 (95% CI: 1.01–1.77). The increased bi-allelic risk of CRC varied when stratified for age and sex but none of the differences were significant, although when stratified by sex, males showed a marginal significant mono-allelic effect for Y179C (OR=1.70; 95% CI: 1.06–2.73). The results from this large dataset indicate that the two variants may be acting mechanistically differently; G396D appears to be a true example of recessive Mendelian disease, whereas the results for Y179C are more complex and there is therefore some argument against combining the two variants. However, the results from the Y179C/G396D compound heterozygotes analysis demonstrates an increase in risk similar to G396D bi-allelic carriers, suggesting that the two variants are complementary and analysis of combined MUTYH mutations as historically performed, appears appropriate to assess risk for the whole gene. The rarity of the Y179C allele has made it difficult to truly assess its effect on disease risk, however the large numbers analysed in this report have resulted in the demonstration that both bi-allelic and mono-allelic Y179C variants are associated with disease risk.
The study population did not appear to modulate disease risk and although the study replicated the reported decrease in disease risk in MUTYH wild-type females associated with HRT intake (Chan et al, 2006; Theodoratou et al, 2008), we found no interaction with the MUTYH gene and its variants. Therefore, it is unlikely that HRT intake is an explanation for any sex variation in risk and other genetic factors may be involved in modifying CRC risk.
Evidence of a mono-allelic MUTYH effect on CRC has been reported in several case–control studies (Croitoru et al, 2004; Wang et al, 2004; Farrington et al, 2005; Zhou et al, 2005; Tenesa et al, 2006; Cleary et al, 2009) and family-based studies (Jenkins et al, 2006; Jones et al, 2009), but not in other studies (Kambara et al, 2004; Webb et al, 2006; Balaguer et al, 2007; Lubbe et al, 2009). Our large meta-analysis has demonstrated a marginal significant association for the specific variant Y179C, highlighting the possible increased phenotypic severity of this allele. This is in agreement with other studies and biochemical and model organism studies, which indicate that this variant shows an increased detrimental effect on protein function (Al-Tassan et al, 2002; Parker et al, 2005; Lubbe et al, 2009; Nielsen et al, 2009; D’Agostino et al, 2010). The pooled analysis of published studies and unpublished datasets submitted to us also indicated a marginally significant mono-allelic MUTYH effect, as well as a mono-allelic Y179C effect.
However, there are a number of caveats that need to be considered; if any of the studied datasets contain cases recruited because of the familial clustering of disease, there may be ascertainment bias, artificially inflating the number of MUTYH WM variant allele carriers; secondly the screening of the MUTYH gene has predominantly been performed on the two most common pathogenic variants Y179C and G396D – in some studies, the rest of the gene may be explored in cases with a heterozygous allele for these variants but not usually in the controls, hence there is an overall screening bias and bi-allelic carriers may well have been missed in both cases and controls.
The demonstration of a mono-allelic effect specifically for Y179C should be considered with further caution, as analysis of the control datasets for the Y179C allele demonstrated that it was not in Hardy–Weinberg equilibrium. This may be because of several factors, the rarity of the allele and the fact that both female control subjects with bi-allelic mutations carry Y179C variants. One of these control subjects was shown to have polyps on colonoscopy (Cleary et al, 2009) and may therefore be considered a case. The other is relatively young, less than 60 years old (Lubbe et al, 2009), so potentially may develop cancer over the next few years. However, in this large dataset, we have also shown that bi-allelic carriers of Y179C predisposes to an earlier onset of disease than G396D, consistent with previous reports (Lubbe et al, 2009; Nielsen et al, 2009) and highlights a severer disease phenotype of this variant.
In conclusion, inactivation of the MUTYH gene is a recessive risk factor for CRC, with possible modifying effects indicated by increased risk in cases with early age of onset, although not significantly different in the current dataset. An increased risk associated with mono-allelic MUTYH mutation is indicated, albeit small and not currently clinically relevant, and likely specific for the variant Y179C. Despite the size of this study it has not been possible to definitively establish whether there are significant age and sex effects of increasing disease risk for G396D and Y179C carriers. The evidence presented raises the possibility of a mono-allelic effect for Y179C, but the effect is low (OR 1.34; 95% CI: 1.00–1.80) and is sensitive to variations in population allele frequency because of the rarity of the variant (allele frequency 0.002), as well as potential issues of subgroup analysis and multiple testing (indeed the overall significance is lost after Bonferoni correction). Nonetheless, it does appear that this study is the first to demonstrate that the Y179C variant does impart an increased risk of CRC.
Acknowledgments
We thank all the study participants and the many colleagues world wide who contributed to recruitment. We are particularly grateful to Ruth Wilson, Rosa Bisset, Nicola Cartwright and Gisela Johnstone, and all those who contributed to recruitment, data collection and data curation for the Scottish COGS and SOCCS studies. This work was supported by grants from Cancer Research UK (C348/A3758, C348/A8896), Scottish Government Chief Scientist Office (K/OPR/2/2/D333, CZB/4/94, CZB/4/449); Medical Research Council (G0000657-53203); Centre Grant from CORE as part of the Digestive Cancer Campaign (http://www.corecharity.org.uk). ET is funded by a Cancer Research UK Fellowship C31250/A10107. The work at the Institute of Cancer Research is supported grants from Cancer Research UK (supported by Bobby Moore), Steven Lubbe is supported by a Cancer Research UK PhD Studentship. The work in Canada was made possible through collaboration and cooperative agreements with the Colon Cancer Family Registry and PIs. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating institutions or investigators in the Colon CFR, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government or the Colon CFR. The work in Cambridge (SEARCH) is funded through a programme grant from Cancer Research UK, TK is funded by the Foundation Dr Henri Dubois-Ferriere Dinu Lipatti. The work in France was supported by a regional Hospital Clinical Research Program, the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la LUtte contre le Cancer, the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer. The work in Stockholm was supported by The Swedish Cancer Society, The Swedish Research Council and The Stockholm Cancer Foundation. The work in Spain was supported by the Ministerio de Educación y Ciencia (SAF09-7319), Spanish Networks RTICCC (RD06/0020/1050, 1051), Instituto de Salud Carlos III grants (FIS PI03/0114 and FIS 05/1006 and CIBERESP).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP (2002) Inherited variants of MYH associated with somatic G:C → T:A mutations in colorectal tumors. Nat Genet 30(2): 227–232 [DOI] [PubMed] [Google Scholar]
- Avezzù A, Agostini M, Pucciarelli S, Lise M, Urso ED, Mammi I, Maretto I, Enzo MV, Pastrello C, Lise M, Nitti D, Viel A (2008) The role of MYH gene in genetic predisposition to colorectal cancer: another piece of the puzzle. Cancer Lett 268(2): 308–313 [DOI] [PubMed] [Google Scholar]
- Balaguer F, Castellví-Bel S, Castells A, Andreu M, Muñoz J, Gisbert JP, Llor X, Jover R, de Cid R, Gonzalo V, Bessa X, Xicola RM, Pons E, Alenda C, Payá A, Piqué JM (2007) Identification of MYH mutation carriers in colorectal cancer: a multicenter, case-control, population-based study. Clin Gastroenterol Hepatol 5: 379–387 [DOI] [PubMed] [Google Scholar]
- Chan JA, Meyerhardt JA, Chan AT, Giovannucci EL, Colditz GA, Fuchs CS (2006) Hormone replacement therapy and survival after colorectal cancer diagnosis. J Clin Oncol 24: 5680–5686 [DOI] [PubMed] [Google Scholar]
- Cleary SP, Cotterchio M, Jenkins MA, Kim H, Bristow R, Green R, Haile R, Hopper JL, LeMarchand L, Lindor N, Parfrey P, Potter J, Younghusband B, Gallinger S (2009) Germline MutY human homologue mutations and colorectal cancer: a multi-site case-control study. Gastroenterology 136(4): 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch A, Hitchins M, Williams R, Meagher A, Hawkins NJ, Ward RL (2006) The role of MYH and microsatellite instability in the development of sporadic colorectal cancer. Br J Cancer 95(9): 1239–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croitoru ME, Cleary SP, Di Nicola N, Manno M, Selander T, Aronson M, Redston M, Cotterchio M, Knight J, Gryfe R, Gallinger S (2004) Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst 96(21): 1631–1634 [DOI] [PubMed] [Google Scholar]
- D’Agostino VG, Minoprio A, Torreri P, Marinoni I, Bossa C, Petrucci TC, Albertini AM, Ranzani GN, Bignami M, Mazzei F (2010) Functional analysis of MUTYH mutated proteins associated with familial adenomatous polyposis. DNA Repair 9: 700–707 [DOI] [PubMed] [Google Scholar]
- Enholm S, Hienonen T, Suomalainen A, Lipton L, Tomlinson I, Kärjä V, Eskelinen M, Mecklin JP, Karhu A, Järvinen HJ, Aaltonen LA (2003) Proportion and phenotype of MYH associated colorectal neoplasia in a population-based series of Finnish colorectal cancer patients. Am J Pathol 163(3): 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington SM, Tenesa A, Barnetson R, Wiltshire A, Prendergast J, Porteous M, Campbell H, Dunlop MG (2005) Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am J Hum Genet 77(1): 112–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington SM, Tenesa A, Barnetson R, Wiltshire A, Prendergast J, Porteous M, Campbell H, Dunlop MG (2006) Colorectal cancer risk in monoallelic carriers of MYH variants. Am J Hum Genet 79(4): 771–772 [Google Scholar]
- Fleischmann C, Peto J, Cheadle J, Shah B, Sampson J, Houlston RS (2004) Comprehensive analysis of the contribution of germline MYH variation to early-onset colorectal cancer. Int J Cancer 109(4): 554–558 [DOI] [PubMed] [Google Scholar]
- Gismondi V, Meta M, Bonelli L, Radice P, Sala P, Bertario L, Viel A, Fornasarig M, Arrigoni A, Gentile M, Ponz de Leon M, Anselmi L, Mareni C, Bruzzi P, Varesco L (2004) Prevalence of the Y165C, G382D and 1395delGGA germline mutations of the MYH gene in Italian patients with adenomatous polyposis coli and colorectal adenomas. Int J Cancer 109(5): 680–684 [DOI] [PubMed] [Google Scholar]
- Jenkins MA, Croitoru ME, Monga N, Cleary SP, Cotterchio M, Hopper JL, Gallinger S (2006) Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: a population-based case-family study. Cancer Epidemiol Biomarkers Prevention 15(2): 312–314 [DOI] [PubMed] [Google Scholar]
- Jones N, Vogt S, Nielsen M, Christian D, Wark PA, Eccles D, Edwards E, Evans DG, Maher ER, Vasen HF, Hes FJ, Aretz S, Sampson JR (2009) Increased colorectal cancer incidence in obligate carriers of heterozygous mutations in MUTYH. Gastroenterology 137(2): 489–494 [DOI] [PubMed] [Google Scholar]
- Kambara T, Whitehall VL, Spring KJ, Barker MA, Arnold S, Wynter CV, Matsubara N, Tanaka N, Young JP, Leggett BA, Jass JR (2004) Role of inherited defects of MYH in the development of sporadic colorectal cancer. Genes Chromosomes Cancer 40(1): 1–9 [DOI] [PubMed] [Google Scholar]
- Küry S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, Lelièvre B, Olschwang S, Le Houérou C, Le Neel T, Faroux R, Ollivry J, Lafraise B, Chupin LD, Bézieau S (2007) The thorough screening of the MUTYH gene in a large French cohort of sporadic colorectal cancers. Genet Test 11(4): 373–379 [DOI] [PubMed] [Google Scholar]
- Lubbe SJ, Di Bernardo MC, Chandler IP, Houlston RS (2009) Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Oncol 27: 3975–3980 [DOI] [PubMed] [Google Scholar]
- Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, González S, Guino E, Capellà G, Canzian F (2006) Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 12: 2101–2108 [DOI] [PubMed] [Google Scholar]
- Nielsen M, Joerink-van de Beld MC, Jones N, Vogt S, Tops CM, Vasen HF, Sampson JR, Aretz S, Hes FJ (2009) Analysis of MUTYH genotypes and colorectal phenotypes in patients with MUTYH-associated polyposis. Gastroenterology 136(2): 471–476 [DOI] [PubMed] [Google Scholar]
- Parker AR, Sieber OM, Shi C, Hua L, Takao M, Tomlinson IP, Eshleman JR (2005) Cells with pathogenic biallelic mutations in the human MUTYH gene are defective in DNA damage binding and repair. Carcinogenesis 26(11): 2010–2018 [DOI] [PubMed] [Google Scholar]
- Peterlongo P, Mitra N, Chuai S, Kirchhoff T, Palmer C, Huang H, Nafa K, Offit K, Ellis NA (2005) Colorectal cancer risk in individuals with biallelic or monoallelic mutations of MYH. Int J Cancer 114(3): 505–507 [DOI] [PubMed] [Google Scholar]
- Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, Maynard J, Pigatto F, Shaw J, Cheadle JP (2003) Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet 362: 39–41 [DOI] [PubMed] [Google Scholar]
- Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, Thomas HJ, Tomlinson IP (2003) Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med 348(9): 791–799 [DOI] [PubMed] [Google Scholar]
- Tenesa A, Campbell H, Barnetson R, Porteous M, Dunlop M, Farrington SM (2006) Association of MUTYH and colorectal cancer. Br J Cancer 95(2): 239–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoratou E, Campbell H, Tenesa A, McNeill G, Cetnarskyj R, Barnetson RA, Porteous ME, Dunlop MG, Farrington SM (2008) Modification of the associations between lifestyle, dietary factors and colorectal cancer risk by APC variants. Carcinogenesis 29(9): 1774–1780 [DOI] [PubMed] [Google Scholar]
- Venesio T, Molatore S, Cattaneo F, Arrigoni A, Risio M, Ranzani GN (2004) High frequency of MYH gene mutations in a subset of patients with familial adenomatous polyposis. Gastroenterology 126(7): 1681–1685 [DOI] [PubMed] [Google Scholar]
- Wang L, Baudhuin LM, Boardman LA, Steenblock KJ, Petersen GM, Halling KC, French AJ, Johnson RA, Burgart LJ, Rabe K, Lindor NM, Thibodeau SN (2004) MYH mutations in patients with attenuated and classic polyposis and with young-onset colorectal cancer without polyps. Gastroenterology 127(1): 9–16 [DOI] [PubMed] [Google Scholar]
- Webb EL, Rudd MF, Houlston RS (2006) Colorectal cancer risk in monoallelic carriers of MYH variants. Am J Hum Genet 79(4): 768–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-L, Djureinovic T, Werelius B, Lindmark G, Sun XF, Lindblom A (2005) Germline mutations in the MYH gene in Swedish familial and sporadic colorectal cancer. Genetic Test 9: 147–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.