Abstract
Background:
MicroRNAs (miRs) have an important role in lung carcinogenesis and progression. Single-nucleotide polymorphisms (SNPs) in genes involved in miR biogenesis may affect miR expression in lung tissue and be associated with lung carcinogenesis and progression.
Methods:
We analysed 12 SNPs in POLR2A, RNASEN and DICER1 genes in 1984 cases and 2073 controls from the Environment And Genetics in Lung cancer Etiology (EAGLE) study. We investigated miR expression profiles in 165 lung adenocarcinoma (AD) and 125 squamous cell carcinoma tissue samples from the same population. We used logistic and Cox regression models to examine the association of individual genotypes and haplotypes with lung cancer risk and with lung cancer-specific survival, respectively. SNPs-miR expression associations in cases were assessed using two-sample t-tests and global permutation tests.
Results:
A haplotype in RNASEN (Drosha) was significantly associated with shorter lung cancer survival (hazard ratio=1.86, 95% CI=1.19–2.92, P=0.007). In AD cases, a SNP within the same haplotype was associated with reduced RNASEN mRNA expression (P=0.013) and with miR expression changes (global P=0.007) of miRs known to be associated with cancer (e.g., let-7 family, miR-21, miR-25, miR-126 and miR15a).
Conclusion:
Inherited variation in the miR-processing machinery can affect miR expression levels and lung cancer-specific survival.
Keywords: microRNA biogenesis, polymorphism, lung cancer, survival
MicroRNAs (miRs) are small non-coding RNAs that bind to the target transcript in the 3′-UTR and can inhibit the translation of proteins and destabilise their target mRNA (Baek et al, 2008). miRs are predicted to regulate ∼30% of the human genome (Lewis et al, 2005) including genes in stress resistance, fat metabolism, cell proliferation and apoptosis pathways (Ambros, 2003). Polymorphisms in miR genes or in genes involved in miR biogenesis may affect miR-mediated cell regulation (Mishra and Bertino, 2009; Clague et al, 2010). miR biogenesis includes generation of a primary transcript (pri-miR) under RNA polymerase II (PolR2A); excision of a stem-loop structure by the nuclear RNaseIII enzyme (Drosha) to generate the pre-miR; transportation of the pre-miR to the cytoplasm and processing by another RNaseIII enzyme (Dicer) into a ∼22-base mature duplex RNA (Bartel, 2004). An alteration in any step during the maturation process could affect miR production. Impaired miR processing and maturation has been shown to enhance cellular transformation and tumourigenesis (Kumar et al, 2008). Given the mounting evidence implicating miRs in lung cancer development and progression (Yanaihara et al, 2006; Kumar et al, 2008; Landi et al, 2010), we investigated the role of single-nucleotide polymorphisms (SNPs) in the RNA-mediated interference machinery involved in miR maturation in lung cancer.
Materials and methods
We performed SNP genotyping and miR expression profiling using blood and tumour tissue samples from the Environment And Genetics in Lung cancer Etiology (EAGLE) study (Landi et al, 2008), including 2100 primary lung cancer cases and 2120 population controls, frequency matched on age, sex and residence, all Caucasians, enrolled in the Lombardy region of Italy in 2002–2005. Institutional review boards of the enrolling hospitals and National Cancer Institute approved the study and participating subjects signed an informed consent.
Genomic DNA was isolated from blood samples from 1984 cases and 2073 controls and used to genotype 12 SNPs (Table 1) covering different haplotype blocks in POLR2A, RNASEN (Drosha) and DICER1 (Figure 1). Genotyping was performed at the Core Genotyping Facility, NCI, using TaqMan assays (http://snp500cancer.nci.nih.gov). Duplicate quality-control samples (2%) showed 100% agreement in all assays. Subjects with at least a 90% genotype call-rate (1946 cases, 1982 controls) were included in the final analyses. All SNPs passed the Hardy–Weinberg equilibrium test among controls (P⩾0.26).
Table 1. SNPs in the RNA-mediated interference machinery and correlation with miR expression.
|
All samples
|
AD samples
|
SQ samples
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Position | Alleles | MAF | Ref. | MA | miR | P-value | Ref. | MA | miR | P-value | Ref. | MA | miR | P-value |
| RNASEN | rs642321 | Chr5:31436760 | G:A | 0.21 | 168 | 108 | 20 | 0.125 | 101 | 59 | 15 | 0.205 | 67 | 49 | 10 | 0.365 |
| RNASEN | rs3805516 | Chr5:31456427 | T:C | 0.22 | 163 | 111 | 25 | 0.082 | 97 | 62 | 11 | 0.295 | 66 | 49 | 17 | 0.155 |
| RNASEN | rs493760 | Chr5:31472797 | A:G | 0.29 | 144 | 132 | 3 | 0.851 | 77 | 82 | 3 | 0.841 | 67 | 50 | 4 | 0.795 |
| RNASEN | rs640831 | Chr5:31495192 | C:A | 0.34 | 123 | 152 | 30 | 0.056 | 68 | 92 | 56 | 0.007 | 55 | 60 | 1 | 0.985 |
| RNASEN | rs7735863 | Chr5:31522297 | C:T | 0.10 | 223 | 49 | 9 | 0.393 | 130 | 26 | 5 | 0.656 | 93 | 23 | 9 | 0.429 |
| RNASEN | rs10520985 | Chr5:31560322 | C:T | 0.46 | 78 | 193 | 10 | 0.351 | 47 | 109 | 10 | 0.335 | 31 | 84 | 2 | 0.946 |
| DICER1 | rs1209904 | Chr14:94633465 | G:A | 0.26 | 139 | 137 | 4 | 0.757 | 80 | 79 | 20 | 0.123 | 59 | 58 | 2 | 0.944 |
| DICER1 | rs2297730 | Chr14:94648428 | A:G | 0.08 | 230 | 45 | 12 | 0.276 | 139 | 19 | 9 | 0.393 | 91 | 26 | 6 | 0.626 |
| POLR2A | rs8065577 | Chr17:7325898 | G:C | 0.24 | 165 | 112 | 2 | 0.929 | 100 | 60 | 3 | 0.840 | 65 | 52 | 3 | 0.877 |
| POLR2A | rs7217258 | Chr17:7336273 | A:G | 0.28 | 145 | 132 | 5 | 0.658 | 87 | 73 | 4 | 0.746 | 58 | 59 | 2 | 0.944 |
| POLR2A | rs2071504 | Chr17:7346661 | C:T | 0.10 | 215 | 58 | 3 | 0.852 | 123 | 34 | 2 | 0.928 | 92 | 24 | 9 | 0.420 |
| POLR2A | rs6761 | Chr17:7358387 | T:C | 0.33 | 123 | 151 | 1 | 0.982 | 75 | 83 | 2 | 0.927 | 48 | 68 | 2 | 0.945 |
Abbreviations: miR=microRNAs; SNP=single-nucleotide polymorphism.
Gene name, rsID, chromosomal location, major : minor alleles and minor allele frequency (MAF) of the studied SNPs are reported in the first five columns. The remaining columns show the results for the analysis of correlation between each SNP and the expression of 199 human miRs using all samples and restricted to adenocarcinoma (AD) and squamous cell carcinoma (SQ) lung cancer tissue samples. For each SNP and analysis type, we reported the number of subjects homozygous for the most common allele (used as reference and indicated with ‘Ref.’), the number carrying one or two minor variant alleles (indicated with ‘MA’), the number of miRs individually correlated with the SNP at a significant level 0.05 (indicated with ‘miR’), and the global P-value based on 10 000 permutations for the association of the SNP with the miR profile. Bold indicates a significant correlation (global P-value <0.01).
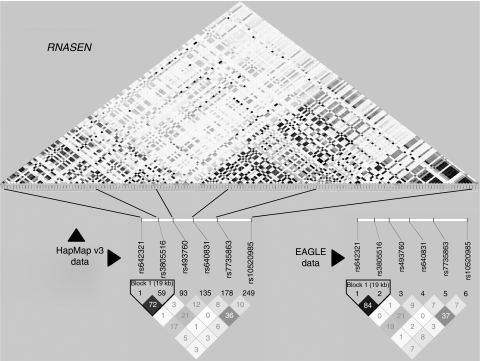
Figure 1.
SNPs coverage for the Drosha gene. SNPs data available from the HapMap v3 database for the chromosomal region corresponding to the Drosha gene. The six SNPs studied in this report are shown in the insets, and linkage disequilibrium (r2) data from HapMap are compared with data in controls from the EAGLE population showing very similar patterns between the two datasets.
The miR expression data were derived from formalin-fixed paraffin-embedded (FFPE) tissue samples in 165 lung adenocarcinoma (AD) and 125 squamous cell carcinoma (SQ) cases from EAGLE, who had not undergone chemotherapy or radiation therapy before tissue collection. Lung cancer histology and the presence of malignant cells in the FFPE tissue blocks were ascertained by the EAGLE local pathologists and were reviewed by a pathologist from the NCI. We excluded tissue blocks with mixed histologies or low frequency of malignant cells. miR expression profiles were obtained using a custom-made two-channel oligo array. The miRs represented on the array, data pre-processing, quality control procedures and selection of the analysed 199 human miRs were described previously (Landi et al, 2010). Array results were also confirmed by qRT–PCR using Taqman miRNA assays (Applied Biosystems, Foster City, CA, USA) in 49 samples from EAGLE that had sufficient tumour miR remaining after the array analysis (Landi et al, 2010). In addition, we analysed the association between RNASEN/rs640831 and RNASEN gene expression in non-involved lung tissue from 45 AD patients from EAGLE using data from an Affymetrix Chip HG U133A (Affymetrix Inc., Santa Clara, CA, USA).
We tested single SNP and haplotype associations with lung cancer risk in all 3928 subjects and with survival in the 1946 lung cancer cases. In single SNP analyses, homozygosity for the more frequent allele among controls was defined as the reference group and both additive and dominant models were considered. In the haplotype analyses, the most common haplotype was defined as the reference group. The associations between variant genotypes and risk of lung cancer were estimated by odds ratios and their 95% confidence intervals using unconditional logistic regression, adjusted for categories of age, sex, residence, cumulative smoking dose (pack-years), smoking intensity (cigarettes per day), and years-since-quitting smoking. We also performed subgroup analyses by smoking status (never/ever) and major histology types and analyses restricted to patients with resectable tumours (stage I, II and IIIA). Lung cancer-specific survival was defined as time from diagnosis of lung cancer to time of death due to lung cancer. Patients who were still alive at the time of last follow-up (n=439) or died for causes unrelated to lung cancer (n=98) were censored in the analyses. The association of SNPs and haplotypes with survival-time adjusted for age, sex, stage, and smoking status was estimated by fitting Cox proportional hazards model (Cox, 1972) in patients overall and separately for AD and SQ.
We evaluated the association between miR expression and SNPs within a dominant model in 290 cases using the t-test statistic for each miR-SNP combination. For each SNP we counted the number of significant (P⩽0.05) miR-SNP associations (ns) and then computed a global permutation P-value to evaluate the significance of the association between the SNP and the global miR expression profile. The permutation test for each SNPi (i=1, 2, …, 12) was performed by repeating 9999 times the 199 miRj−SNPip (j=1, 2, …, 199) association t-tests, where each time SNPip was a random permutation across subjects of the original SNPi (P=1, 2, …, 9999) data and the number of significant miRj−SNPip associations (nsp) was recorded for each permutation. The global P-value was then defined as one plus the number of times in which nsp was at least as large as ns (numerator) divided by the total number of permutations plus one (i.e., denominator=10 000). Finally, we evaluated the association between RNASEN mRNA expression and the rs640831 SNP with a dominant model in 45 AD cases using the t-test statistic.
To account for the fact that tests were conducted for 12 different SNPs and 3 haplotypes, we considered 0.01 as P-value threshold for statistical significance. All analyses were implemented and performed using the R-project (v2.10) statistical package (http://www.r-project.org/index.html) with the exception of the haplotype analysis, conducted using the THESIAS program (Tregouet and Garelle, 2007).
Results
None of the investigated polymorphisms in POLR2A, RNASEN and DICER1 showed significant association with lung cancer risk or lung cancer survival either overall or by subgroups of histology or smoking status. Analyses based on additive and dominant models gave similar results (Supplementary Materials 1–8). However, we found that a RNASEN haplotype, GTAATC (frequency=2%), was significantly associated with lung cancer-specific reduced survival compared with the most common haplotype GTACCT (frequency=30%) among all cases with hazard ratio (HR)=1.86, 95% CI=1.19–2.92 and P=0.007. Similar results were obtained in AD and SQ histological subtypes (HRs=2.33, 3.27; 95% CIs=1.04–5.25, 1.44–7.43; and P=0.041, 0.005, respectively) and among ever smoker patients (HR=1.81; 95% CI=1.16–2.82; P=0.009). This haplotype-survival association was not observed among the 133 never smoker patients (HR=0.82; 95% CI=0.33–2.06; P=0.679). When we repeated this analysis among the 821 patients with early stage resectable tumours (stage=I, II and IIIA), the association between RNASEN haplotype GTAATC and reduced lung cancer-specific survival was even stronger (HR=2.36, 95% CI=1.36–4.09 and P=0.002). Results were also significant in patients with resectable AD and resectable SQ separately (HRs=1.95, 3.99; 95% CIs=1.00–3.81, 1.75–9.11; and P=0.05, 0.001, respectively).
We further studied whether the 12 SNPs were associated with expression of mature miRs in lung cancer tissue for all samples and for AD and SQ separately (Table 1). In AD patients, RNASEN/rs640831, included in the GTACCT haplotype, was associated with the expression of 56 miRs (global P=0.007, Table 2). On average, for subjects who inherited this SNP, 37 miRs were upregulated and 19 miRs were downregulated in comparison to subjects with the consensus genotype. miRs with tumour suppressor potential (e.g., let-7 family) and miRs with oncogenic or metastatic potential (e.g., miR-21 (Zhu et al, 2008), miR-126 (Crawford et al, 2008) and miR-15a (Cimmino et al, 2005)), were among those with altered expression in the carriers.
Table 2. miRs significantly correlated with RNASEN SNP rs640831 in AD patients.
| MicroRNAs | Unique ID | P-value | Fold change | Mean in Ref. (log2) | Mean in MA (log2) |
|---|---|---|---|---|---|
| Upregulated | |||||
| has-miR-30b | MIMAT0000420 | 0.0003 | 1.72 | −2.60 | −1.82 |
| hsa-miR-25 | MIMAT0000081 | 0.0011 | 1.57 | −2.07 | −1.41 |
| hsa-miR-92 | MIMAT0000092 | 0.0013 | 1.51 | −4.07 | −3.48 |
| hsa-let-7g | MIMAT0000414 | 0.0016 | 1.68 | −2.81 | −2.06 |
| hsa-miR-21 | MIMAT0000076 | 0.0016 | 1.89 | −3.39 | −2.47 |
| hsa-miR-200c | MIMAT0000617 | 0.0020 | 1.47 | 0.90 | 1.46 |
| hsa-miR-106a | MIMAT0000103 | 0.0020 | 1.45 | −4.02 | −3.48 |
| hsa-miR-30c | MIMAT0000244 | 0.0022 | 1.68 | −2.65 | −1.90 |
| hsa-miR-30a-5p | MIMAT0000087 | 0.0025 | 1.48 | −1.18 | −0.62 |
| hsa-let-7b | MIMAT0000063 | 0.0025 | 1.50 | −1.22 | −0.64 |
| hsa-let-7f | MIMAT0000067 | 0.0031 | 1.65 | −2.74 | −2.02 |
| hsa-miR-181a | MIMAT0000256 | 0.0031 | 1.38 | −0.90 | −0.43 |
| hsa-miR-20b | MIMAT0001413 | 0.0036 | 1.38 | −3.52 | −3.06 |
| hsa-miR-103 | MIMAT0000101 | 0.0036 | 1.37 | −1.60 | −1.14 |
| hsa-miR-98 | MIMAT0000096 | 0.0038 | 1.62 | −2.33 | −1.63 |
| hsa-let-7c | MIMAT0000064 | 0.0040 | 1.53 | −1.76 | −1.15 |
| hsa-miR-20a | MIMAT0000075 | 0.0045 | 1.45 | −4.64 | −4.11 |
| hsa-miR-26a | MIMAT0000082 | 0.0070 | 1.65 | −1.01 | −0.29 |
| hsa-miR-29a | MIMAT0000086 | 0.0071 | 1.52 | −1.82 | −1.22 |
| hsa-miR-126 | MIMAT0000445 | 0.0074 | 1.55 | 2.23 | 2.86 |
| hsa-miR-17-5p | MIMAT0000070 | 0.0096 | 1.33 | −3.71 | −3.30 |
| hsa-miR-107 | MIMAT0000104 | 0.0104 | 1.30 | −1.54 | −1.16 |
| hsa-miR-19b | MIMAT0000074 | 0.0127 | 1.41 | −4.20 | −3.70 |
| hsa-let-7a | MIMAT0000062 | 0.0182 | 1.40 | −2.05 | −1.57 |
| hsa-miR-106b | MIMAT0000680 | 0.0198 | 1.27 | −3.32 | −2.98 |
| hsa-let-7i | MIMAT0000415 | 0.0199 | 1.39 | −2.15 | −1.68 |
| hsa-miR-200b | MIMAT0000318 | 0.0205 | 1.46 | 1.67 | 2.21 |
| hsa-miR-143 | MIMAT0000435 | 0.0205 | 1.18 | 0.88 | 1.12 |
| hsa-miR-26b | MIMAT0000083 | 0.0210 | 1.39 | −1.46 | −0.98 |
| hsa-miR-15a | MIMAT0000068 | 0.0244 | 1.33 | −2.04 | −1.62 |
| hsa-miR-30d | MIMAT0000245 | 0.0281 | 1.28 | −0.79 | −0.44 |
| hsa-miR-93 | MIMAT0000093 | 0.0285 | 1.30 | −3.36 | −2.98 |
| hsa-miR-23a | MIMAT0000078 | 0.0348 | 1.26 | 0.36 | 0.69 |
| hsa-miR-125a | MIMAT0000443 | 0.0377 | 1.21 | 0.68 | 0.96 |
| hsa-miR-22 | MIMAT0000077 | 0.0415 | 1.30 | 0.55 | 0.93 |
| hsa-miR-146b | MIMAT0002809 | 0.0422 | 1.36 | −4.46 | −4.02 |
| hsa-miR-429 | MIMAT0001536 | 0.0432 | 1.36 | −0.43 | 0.02 |
| Downregulated | |||||
| hsa-miR-452 | MIMAT0001635 | 0.0003 | 0.88 | 0.18 | 0.00 |
| hsa-miR-370 | MIMAT0000722 | 0.0024 | 0.90 | 0.41 | 0.26 |
| hsa-miR-122a | MIMAT0000421 | 0.0037 | 0.89 | 0.14 | −0.03 |
| hsa-miR-130b | MIMAT0000691 | 0.0044 | 0.92 | −0.11 | −0.23 |
| hsa-miR-510 | MIMAT0002882 | 0.0047 | 0.91 | 0.22 | 0.08 |
| hsa-miR-188 | MIMAT0000457 | 0.0065 | 0.91 | 0.18 | 0.04 |
| hsa-miR-509 | MIMAT0002881 | 0.0117 | 0.86 | −0.40 | −0.62 |
| hsa-miR-198 | MIMAT0000228 | 0.0134 | 0.90 | 0.43 | 0.28 |
| hsa-miR-485-5p | MIMAT0002175 | 0.0134 | 0.90 | 0.81 | 0.65 |
| hsa-miR-518c* | MIMAT0002847 | 0.0145 | 0.89 | −0.01 | −0.17 |
| hsa-mir-610 | MIMAT0003278 | 0.0162 | 0.83 | 1.01 | 0.73 |
| hsa-miR-488 | MIMAT0002804 | 0.0166 | 0.91 | −0.76 | −0.90 |
| hsa-miR-453 | MIMAT0001630 | 0.0353 | 0.93 | 0.38 | 0.28 |
| hsa-mir-628 | MIMAT0003297 | 0.0405 | 0.89 | −0.14 | −0.30 |
| hsa-miR-432 | MIMAT0002814 | 0.0420 | 0.89 | −0.02 | -0.20 |
| hsa-mir-623 | MIMAT0003292 | 0.0429 | 0.88 | −0.05 | −0.24 |
| hsa-miR-299-3p | MIMAT0000687 | 0.0473 | 0.92 | 0.05 | −0.07 |
| hsa-miR-524* | MIMAT0002849 | 0.0473 | 0.93 | 0.07 | −0.03 |
| hsa-miR-383 | MIMAT0000738 | 0.0498 | 0.91 | 0.83 | 0.70 |
Abbreviations: AD=adenocarcinoma; miR=microRNA; SNP=single-nucleotide polymorphism.
The 56 miRs significantly correlated with RNASEN/rs640831 in AD patients are listed ranking by P-value of each SNP-miR correlation. The analysed miR data is a miR expression intensity ratio between the examined miR and the reference EBV cell line, followed by median normalisation and log2 base transformation (i.e., a negative value indicates a ratio between 0 and 1). For each miR we also reported the fold change of the expression ratio for minor allele carriers (indicated with ‘MA’) compared with major allele homozygotes (indicated with ‘Ref.’), and the expression ratio means in the two compared groups. miRs whose expression has been associated with lung cancer in previous studies are shown in bold. The asterisk (*) symbol after a miR label designates a complementary miR.
We validated the microarray results by qRT–PCR for 4 of the 56 miRs significantly associated with RNASEN/rs640831 in 49 EAGLE lung tumour samples. As shown in Supplementary Figure 1, the correlation was highly significant (P=0.001, <0.0001, <0.0001, and 0.002 for let-7g, let-7f, miR-26a and miR-107, respectively). As expected, the correlation between the microarray and qRT–PCR data was inverse as qRT–PCR values are measured in terms of number of measurement cycles needed to reach a certain expression level: the lower the number of cycles the higher the detected expression level. In addition, the association between the expressions as measured by qRT–PCR and RNASEN/rs640831 was qualitatively concordant with the microarray-based results (inverse association in the 24 AD but not in the 23 SQ cases).
Finally, to further elucidate our finding of a correlation between the RNASEN/rs640831 and the miR expression profile among AD cases, we tested the association between RNASEN gene expression and the rs640831 polymorphism in non-involved lung tissue of 45 AD patients from EAGLE. The 25 AD patients carrying one or two rs640831 minor variants showed a significantly lower mRNA expression than the 20 AD patients homozygous with the more frequent allele (fold change=0.87, P=0.013).
Discussion
We have observed (i) an association between lung cancer survival and a haplotype in RNASEN, particularly, among early stage patients and (ii) a differentially expressed miR profile and RNASEN gene expression by RNASEN/rs640831 status in lung tissue. Carrying the minor variant A vs the common variant C in RNASEN/rs640831 contributed to the survival association for the RNASEN haplotype GTAATC compared with the haplotype GTACCT. Our results are consistent with the combined effect of multiple genetic markers within a haplotype as better representing the impact of the genetic locus on disease progression than individual markers (Johnson et al, 2001; Crawford and Nickerson, 2005). This is the first evidence that inherited variation in the miR-processing machinery, more specifically in RNASEN, might affect survival from lung cancer. Previous studies have shown that low RNASEN gene expression was associated with survival in oesophageal cancer patients (Sugito et al, 2006) and, suggestively, with reduced survival in non-small-cell lung carcinoma patients (Karube et al, 2005). Our findings provide a possible genetic basis for the previous reports. The most frequent variant in RNASEN was associated with miR expression changes and with lower RNASEN mRNA expression in AD. Several of these miRs have been previously reported to be associated with lung cancer survival in the EAGLE study (Landi et al, 2010) and other lung cancer studies (Markou et al, 2008; Yu et al, 2008; Raponi et al, 2009). An analogous global modification of miR profile due to changes in Drosha transcript expression level has been observed in cervical cancer (Muralidhar et al, 2007). A similar finding was not observed in our analysis restricted to cases with SQ histology, suggesting that the mechanism by which RNASEN/rs640831 affects miR expression is specific to AD lung tissues. Such observation is consistent with previous studies showing that protein levels of genes involved in the endogenous miR machinery differ between lung AD and SQ histology subtypes (Chiosea et al, 2007). It is also possible that the relatively small sample size of the SQ affected the results.
In contrast to the survival analysis that was based on a large sample (n∼2000), allowing for exploration of several models, including haplotypes, the miR expression analysis was based on a smaller sample size (n∼300, of which 125 were SQ). Consequently, only the most informative SNP (MAF=34%) could be adequately tested with the miR profile. In contrast, we were not able to detect possible effects of the full haplotype in association with the miR profile, because this test would have required hundreds of samples with miR expression data. Further functional studies and/or larger miR profiling studies are necessary to confirm our findings.
In conclusion, in the largest study of SNPs in genes involved in miR biogenesis and lung cancer to date, an inherited variant in the RNASEN gene, coding for the Drosha enzyme, was associated with reduced expression of the Drosha gene and changes in the expression of miRs involved in many cancer-related processes. A haplotype in Drosha that included the inherited RNASEN variant and five additional variants was associated with poor lung cancer survival, particularly, among early stage patients. Drosha gene variants may affect miR-processing machinery including miR maturation and expression level, which may consequently affect the function of miRs in transcript and protein translation regulation. These variants in Drosha may have potential prognostic implications in lung cancer.
Acknowledgments
This research was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, DHHS, Bethesda, MD and by a 2006 NCI Director's Innovation Award to MTL. We thank the EAGLE participants and study collaborators listed on the EAGLE website (http://eagle.cancer.gov/).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113: 673–676 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S (2007) Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res 67: 2345–2350 [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102: 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague J, Lippman SM, Yang H, Hildebrandt MA, Ye Y, Lee JJ, Wu X (2010) Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol Carcinog 49: 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D (1972) Regression models and life tables (with discussion). J Roy Stat Soc B 4: 187–220 [Google Scholar]
- Crawford DC, Nickerson DA (2005) Definition and clinical importance of haplotypes. Annu Rev Med 56: 303–320 [DOI] [PubMed] [Google Scholar]
- Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP (2008) MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun 373: 607–612 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di GG, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29: 233–237 [DOI] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T (2005) Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 96: 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 105: 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, Lubin JH, Goldin L, Alavanja M, Morgan G, Subar AF, Linnoila I, Previdi F, Corno M, Rubagotti M, Marinelli B, Albetti B, Colombi A, Tucker M, Wacholder S, Pesatori AC, Caporaso NE, Bertazzi PA (2008) Environment and genetics in lung cancer etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health 8: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, Tucker MA, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM, Wang E (2010) MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 16: 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES (2008) Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem 54: 1696–1704 [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Bertino JR (2009) MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 10: 399–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N (2007) Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol 212: 368–377 [DOI] [PubMed] [Google Scholar]
- Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG (2009) MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res 69: 5776–5783 [DOI] [PubMed] [Google Scholar]
- Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Ando T, Mori R, Takashima N, Ogawa R, Fujii Y (2006) RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res 12: 7322–7328 [DOI] [PubMed] [Google Scholar]
- Tregouet DA, Garelle V (2007) A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics 23: 1038–1039 [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198 [DOI] [PubMed] [Google Scholar]
- Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13: 48–57 [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18: 350–359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.