Summary
Analytical ultracentrifugation (AUC) is a powerful technique for the characterization of hydrodynamic and thermodynamic properties. The intent of this article is to demonstrate the utility of sedimentation velocity (SV) studies to obtain hydrodynamic information for G-quadruplex systems and to provide insights into one part of this process, namely, data analysis of existing SV data. An array of data analysis software is available, mostly written and continually developed by established researchers in the AUC field, with particularly rapid advances in the analysis of SV data. Each program has its own learning curve and this article is intended as a resource in the data analysis process for beginning researchers in the field. We discuss the application of three of the most commonly used data analysis programs, DCDT+, Sedfit and SedAnal, to the interpretation of SV data obtained in our laboratory on two G-quadruplex systems.
Keywords: analytical ultracentrifugation, sedimentation velocity, sedimentation coefficient, frictional ratio, hydrodynamic, solution conformation, G-quadruplex DNA, data analysis, DCDT+, Sedfit, SedAnal
1. Introduction
The technique of analytical ultracentrifugation (AUC) was developed by Svedberg in 1923 when he equipped a rapidly rotating centrifuge with an optical detection system to observe sedimenting particles and applied his method to determine particle size distributions of gold sols. Use of the technique for the characterization of biochemical systems, especially proteins and their complexes, became widespread from 1950 to 1980 with the ability to determine sedimentation coefficients, molar masses and shape information (1). However, its use declined substantially in the following decade because of a lack of new instrumentation for digital data acquisition and the adoption of alternative, less demanding, techniques including gel electrophoresis and size exclusion chromatography. The reemergence of modern AUC instrumentation in the early 1990s occurred concomitantly with significant advancements in data analysis methods making possible a comprehensive characterization of hydrodynamic and thermodynamic properties using AUC studies (1–3).
Sedimentation velocity, SV, is one of two major AUC approaches, the other being sedimentation equilibrium, SE (1–8). SV experiments are performed at high rotor speeds, up to 60,000 rpm, resulting in the establishment of a sample concentration boundary that moves towards the bottom of the centrifuge cell over time. The rate of movement of the boundary is observed through the acquisition of optical scans (absorbance, interference or fluorescence) as a function of radial position every few minutes over the course of several hours. The rate of movement of the concentration boundary as a function of time yields the sedimentation coefficient, s. The rate of spreading of the concentration boundary allows an estimation of the translational diffusion coefficient, D, which opposes sedimentation. Molecular weight, M, can be estimated from the ratio of s and D. SE experiments are performed at much lower rotor speeds and run for much longer time periods in order to establish an equilibrium between sedimentation and diffusion. The exponential sample concentration distribution that results can be fit by non-linear least squares methods to thermodynamic equations to yield molecular weights, association constants and stoichiometries of systems. It is essential to perform SV experiments before equilibrium analysis in order to assess sample heterogeneity and polydispersity which could complicate analysis of SE data. With recent advances in the analysis of SV data these experiments alone are often informative enough to provide the information being sought.
This article focuses on the application of selected data analysis methods to the hydrodynamic characterization of G-quadruplex structures using sedimentation velocity ultracentrifugation. Significant literature exists concerning the appropriate design of AUC experiments, protocols for performing the experiments and methods of data analysis (1–29). A number of comprehensive reviews exist (1–8, 30). This article will not attempt to reproduce information that exists more expansively elsewhere and the reader is referred to these original papers. The intent is two-fold - to illustrate the utility of SV and to provide helpful insights into one small part of this process, namely, data analysis of existing SV data of G-quadruplex systems. To this end we will present SV data obtained in our laboratory on two G-quadruplex systems and provide commentary on our path through the data analysis process. A host of data analysis software is currently available for the analysis of sedimentation data, mostly written and continually developed by established researchers in the AUC field, with particularly rapid advances in the analysis of SV data. Examples of available software include Sedfit, Sedphat, SedAnal, DCDT+, Heteroanalysis, Svedberg and UltraScan (31–37). This article is focused solely on the analysis of SV data with a discussion of analysis using three of these programs, DCDT+, Sedfit and SedAnal.
2. Materials
The following programs are needed:
DCDT+ (http://www.jphilo.mailway.com/dcdt+.htm) (34)
Sedfit (http://www.analyticalultracentrifugation.com/download.htm) (31)
SedAnal (http://sedanal.bbri.org/) (33)
XLGraph (http://www.jphilo.mailway.com/download.htm) (38)
WinMatch (http://www.biotech.uconn.edu/auf/?i=aufftp) (39)
Sednterp (http://www.jphilo.mailway.com/download.htm) (40)
3. Methods
3.1. Introduction to the Analysis Approaches
Important differences exist between the analysis methods implemented in these programs and it is informative to include a brief discussion of the different approaches employed in the analysis of SV data. Early estimation of s-values was simply based on the measurement of the movement of the boundary midpoint over time (2). However, the boundary shape is subject to increasing diffusional broadening during the sedimentation experiment and the midpoint would only be free of diffusion and yield an accurate s-value for a single component sample (1). For multicomponent samples the midpoint yields only an average s-value which is insensitive to multiple solution species. The van Holde-Weischet method addressed this by dividing the sedimenting boundary into equal fractions, calculating an apparent s-value for each boundary fraction and extrapolating the apparent s-value at the same boundary fraction of each scan to infinite time to remove the effects of diffusion and yield the apparent s-value of that boundary region (1, 3, 4, 14–16). A plot of boundary fraction against extrapolated s-values yields an integral sedimentation coefficient distribution G(s) for the sample. For a homogeneous sample the G(s) distribution will be vertical since the extrapolated s-values will the same at all points in the boundary. For heterogeneous samples the G(s) distribution will have a positive slope with the fastest sedimenting species being present at the top of the boundary corresponding to the later boundary fractions and hence appearing at the top of the positive slope of the G(s) plot. The van Holde-Weischet method increases the resolution of components with similar s-values which is particularly helpful for complex mixtures giving a rapid assessment of sample heterogeneity and any interaction behavior. However, the removal of diffusional spreading from the sedimentation profiles means that the method cannot be used to determine diffusion coefficients or molecular weights. The method is available in Sedfit and UltraScan but does not form the basis of the analysis method in these programs.
The differential time-derivative dc/dt method, like the van-Holde Weischet method, provides a model-independent assessment of sedimentation behavior. However, the basis of the time-derivative method is very different (4, 17, 18). Closely spaced boundary scans are subtracted in pairs to yield a set of time-derivative dc/dt against radius profiles. The time-derivative dc/dt is then transformed into dc/ds, a differential apparent sedimentation coefficient distribution designated g(s*), and the radial position is transformed into an apparent sedimentation coefficient (s*). Subtraction of scans in the time-derivative dc/dt method results in a significant decrease in time-invariant noise. The g(s*) versus s* profile is similar in appearance to a chromatogram although the apparent s-value is far more informative than a retention time. Visually the g(s*) profile is more intuitive than the G(s) profile. A single component sample will yield a Gaussian-shaped curve with a sedimentation coefficient corresponding to the peak position and the diffusion coefficient corresponding to the peak width. Determination of both an s-value and D allows calculation of M (see Note 1). Inclusion of the effect of diffusion reduces the resolution compared to the G(s) method; however, some might argue that this situation is the reality and should be included. The model-independent nature of the method is a significant advantage of the approach and means that it is possible to obtain information from the sedimentation profiles of very complicated mixtures of species which would otherwise be extremely challenging to analyze if it was necessary to determine a suitable model before analysis (4). The time-derivative dc/dt method is the basis of the programs DCDT+ and implemented in SedAnal.
Around the time of the conception of the time-derivative dc/dt method computer programs were developed to fit SV data to approximate solutions of the Lamm equation, the underlying transport equation describing the SV process (4, 19, 20, 29). This approach is the basis of fitting of g(s*) data in the DCDT+ and Svedberg analysis programs. An alternative approach of using finite element solutions to the Lamm equation is used in the programs Sedfit, UltraScan and SedAnal (2, 4, 9, 21–26, 28) (UltraScan was written for Unix systems whereas all the other programs operate on the Windows platform). Both approaches allow the estimation of s-values, D, M, association constants and stoichiometries but require a model to be specified that best fits the sedimentation behavior of the sample under analysis, the selection of which can be difficult. DCDT+ allows the possibility of generating a model-independent g(s*) distribution prior to model-dependent fitting for accurate values of the aforementioned parameters whereas both Sedfit and SedAnal require the input of a model at the outset of analysis.
The finite element modeling method used in Sedfit and UltraScan uses a distribution of Lamm equation solutions to directly model the sedimentation boundary and yields a differential sedimentation coefficient distribution c(s) with the deconvolution of diffusion effects (21–26, 28). As previously mentioned the removal of diffusion information enhances the resolution and quantization of mixtures of solution components; however, the peak widths in a c(s) distribution are a function of the signal-to-noise ratio and the parameters employed by the maximum entropy regularization processes employed in Sedfit and have no physical meaning unlike the g(s*) distribution (4, 41). Also, it is important to be aware of the possibility of the generation of false peaks in the c(s) distribution where a good fit of the raw data cannot be obtained or inappropriate parameters are entered. Sedfit can model a wide range of sedimentation processes including associating systems, non-ideal sedimentation, the redistribution of salts, both very low mass (e.g. peptides) and high mass species (e.g. viruses), flotation as well as sedimentation, and can use data near the base of the cell where solutes accumulate (2, 4, 41). SedAnal contains a basic set of models that can be supplemented by an unlimited numbers of user-defined models incorporated using a separate ModelEditor program, whereas DCDT+ can fit g(s*) distributions for up to 5 discrete non-interacting species. Sedfit estimates a weight-average shape factor f/fo from the experimental data which forms the basis of the relationship between s- and D-values. The c(s) distribution in Sedfit can be converted to a c(M) distribution with the caveat of the assumption of f/fo which may lead to incorrect M-values for some species (2). Whenever a distribution of species is present the meaning of f/fo is complicated.
Having introduced the background to the various approaches used in the analysis of SV data we will now discuss the application of three data analysis programs, DCDT+, Sedfit and SedAnal, to the interpretation of SV data obtained in our laboratory for G-quadruplexes. The user is referred to the program manuals, references, help files, tutorials and courses applicable to these programs. These provide comprehensive instruction as to the operation of each analysis program. This information would be too expansive to reiterate here; instead we will attempt to provide useful comments at various stages of the data analysis process to expand upon the information provided by the program authors and assist researchers in the application of these programs to the analysis of their own experimental data. Data presented are absorbance scans obtained on the Beckman XL-A instrument, small differences in the analysis setup are necessary for interference scans but the same overall analysis strategies would apply.
3.2. DCDT+
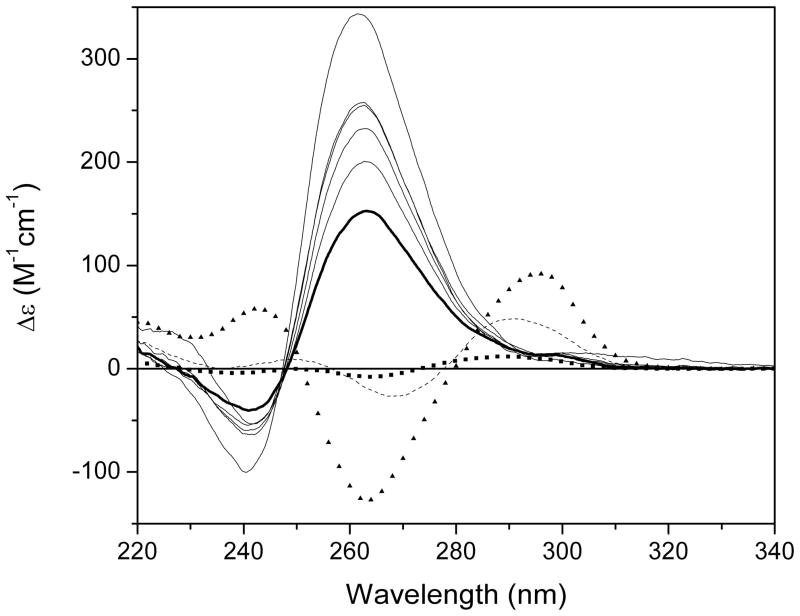
SV data was obtained for the G-quadruplex series GnT4Gn where n = 2 – 10. It has been reported that G4T4G4 forms a hairpin dimer structure in sodium solution (42, 43) and we hypothesized that with higher n increased numbers of G-quadruplex stacks would form within a dimeric quadruplex structure. To investigate this we performed circular dichroism, CD, and AUC studies. CD studies indicated that both “parallel” and “anti-parallel” G-quadruplex forms were apparent for n = 2 – 4 with only “parallel” forms present for n = 5 – 10; there was also an increase in ellipticity with increase in G-tract length (44) (Figure 1). While CD studies provided useful conformational information they can not provide an assessment of the molecularity or hydrodynamic properties of the samples and, to this end, AUC studies were undertaken. We will highlight here only some of this work in order to provide a relevant system for the demonstration of the utility of AUC studies for the characterization of G-quadruplexes. SV studies were performed using a Beckman Coulter XL-A instrument at 60,000 rpm (the current limit of the instrument) because of the expected low molecular weights of the samples (4.9 kDa for n = 2 to 15.5 kDa for n = 10) (see Note 2). Because of the large number of samples involved in the study, data were initially collected for a single concentration (A260 = 0.8) of each sample (3 samples per rotor = 3 SV runs). Scans were recorded with no time interval between scans with a run typically started towards the end of the day and run overnight. This resulted in more scans than were necessary (with a number of scans collected after the sample had pelleted) but ensured complete sedimentation of all solution contents and allowed the selection of appropriate scans to be made during the data analysis process.
Figure 1.
CD spectra of GnT4Gn (n = 2 – 10) in BPES buffer (6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM NaCl, pH 7.0). Both “parallel” and “anti-parallel” G-quadruplex forms were apparent for n = 2 – 4 (G2T4G2, squares; G3T4G3, dashed black line; G4T4G4, triangles). A switch to only “parallel” forms occurs at n = 5 with an increase in ellipticity with increase in G-tract length (G5T4G5, thick black line; G6T4G6 – G10T4G10 black lines).
-
1
Load SV data into DCDT+ according to the software directions. Inspect raw data for initial observations/impressions about the nature of the sample. Look for the rate of depletion of signal at the meniscus, the rate of movement of the boundary, the rate of depletion of the plateau region and the number of steps or inflection points evident in the boundary (this becomes more difficult to see over time as the effects of diffusion become more apparent).
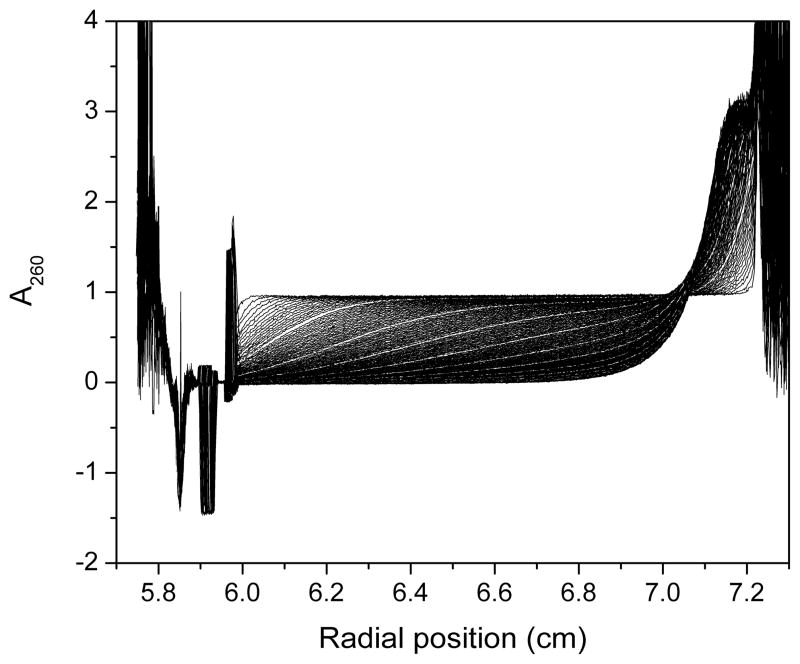
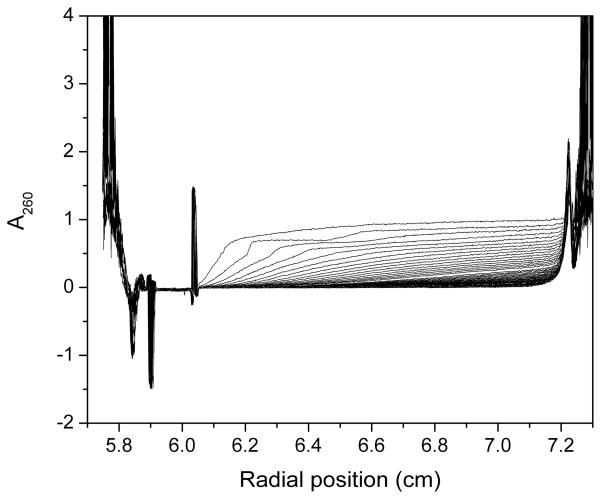
These general observations provide information about sample heterogeneity, the number of sedimenting species and their relative mass, density and shape. Inspection of the entire raw data set for G2T4G2 revealed that at 60,000 rpm the sample had cleared the meniscus region but had not sedimented to the bottom of the cell, even after overnight data collection (Figure 2). This indicated that the sample either had a low molecular weight, low density and/or a less compact shape. By G4T4G4 the sample sedimented to the bottom of the centrifuge cell indicating that the sample had a higher molecular weight, higher density and/or a more compact shape. Raw data for G5T4G5 through G10T4G10 exhibited larger spacing between scans with obvious “steps” in the earlier scans (Figure 3). This indicated the presence of multiple faster sedimenting solution components.
Figure 2.
Sedimentation velocity scans for G2T4G2 collected on the Beckman XL-A at 60,000 rpm at A260 ~ 0.8 in a 1.2 cm cell. The sample does not sediment to the bottom of the cell even after overnight data collection indicating the presence of small sedimenting species.
Figure 3.
Sedimentation velocity scans for G7T4G7 collected on the Beckman XL-A at 60,000 rpm at A260 ~ 0.8 in a 1.2 cm cell. Large spacing between scans and obvious “steps” are evident indicating the presence of multiple faster sedimenting solution components.
-
2
Set sample meniscus position indicator and left and right fitting limits.
When prompted we accept the default of setting the meniscus to the maximum of the meniscus peak. The left fitting limit is set to a position just clear of the meniscus. The right fitting limit is set to a position which will be at the end of the plateau region when scans from the middle of the run are selected. Fine adjust of all fitting limits is performed in step 3.
-
3
Use the auto adjust button to provide an initial selection of scans for transformation into the g(s*) distribution. Adjust the number and position of scans in the run as appropriate. Check that the scan selection is appropriate for the peak broadening limit. Make final adjustments.
This selection is usually quite good but it is important to use the slider controls to investigate other regions of the data. Move the run position slider to investigate earlier and later scans in the run to look for other sedimenting species (see Note 3). Once an initial range of scans has been selected encompassing the solution species of interest, visually inspect both the dc/dt distribution and the raw data scans then adjust the scan number slider and the run position slider in turn to optimize the selection of scans. The ultimate goal is to select raw scans that are in the middle of the run with a flat, zero signal after the meniscus peak and a flat plateau region after the sedimentation boundary. Check that the left and right fitting limits are in the flat meniscus and plateau regions and that the meniscus position indicator is still at the maximum of the meniscus peak. The dc/dt distribution should be centered in the window with a fairly symmetrical Gaussian-like shape tailing off to flat, zero regions on either side of the peak. (A non-zero region on the right-hand side of the peak might indicate the presence of aggregates in the sample).
By moving the run position and scan number sliders the peak broadening limit value changes. This is a numerical assessment of whether too many scans are being averaged to generate the dc/dt distribution. Including scans covering too much boundary movement causes the Δc/Δt values that are calculated to be a poor approximation of the true derivative dc/dt. The result is excessive peak broadening of the distribution and loss of resolution between species. The trade off is that decreasing the number of scans to prevent a peak from being broadened too much and ensuring good resolution results in a decrease in the signal-to-noise (The help section of the DCDT+ program contains useful information on the peak broadening limit). Having adjusted the scan number slider check the position of the scans within the run in both the dc/dt and raw data windows. Make final adjustments checking the meniscus indicator and the left and right fitting limit indicators (see Note 4).
-
4
Set the range of s-values for transformation to the g(s*) distribution.
Trim the data points at the extreme edges of the distribution in order to have flat, zero baseline in these regions. Depending on the nature of the sample this may not always be possible (e.g. non-zero baseline at higher s-values could indicate aggregation) but bad data should certainly be set outside the range for subsequent g(s*) transformation.
-
5
Transform the data into a g(s*) distribution. Normalize. Convert to s*20,w.
Normalization sets the area under the g(s*) distribution to a value of 1. It is important to run different concentrations of a sample and compare the normalized g(s*) distributions to provide information about any concentration-dependent properties (e.g. association) of the sample. DCDT+ has a convenient option to prepare a g(s*) overlay graph for this purpose. Conversion of a solution dependent s-value to that observed in water at 20 °C (s*20,w) accounts for solvent contributions to the sedimentation coefficient (see Note 5). This allows the comparison of data obtained under different experimental conditions. For a concentration series an extrapolation of s*20,w values can be made to zero concentration to yield an s-value which is solely a property of the macromolecule (4). The “Show integrals over distribution” button is a useful option to display number, weight, z- and z+1-average s values at both the time of analysis and extrapolated to t = 0.
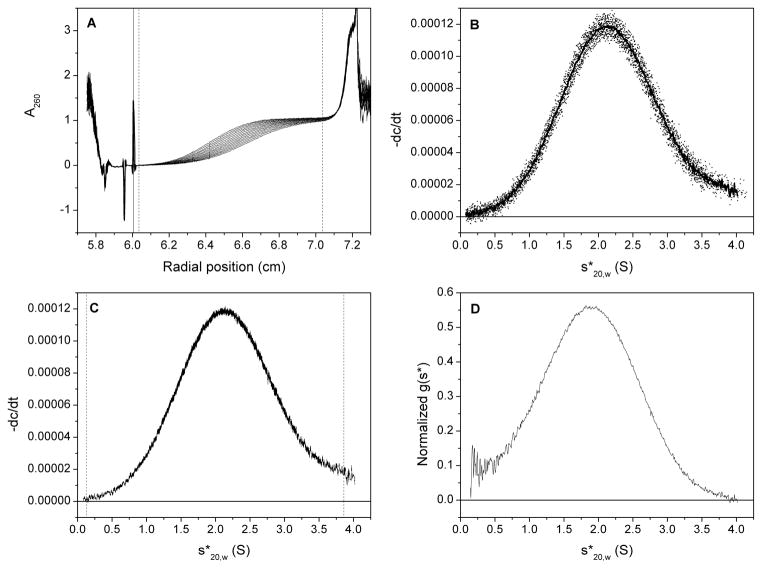
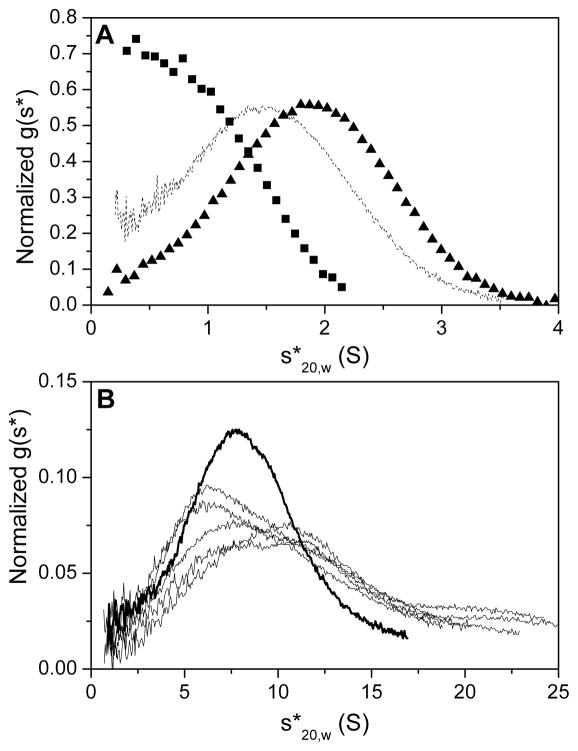
Figure 4 depicts steps 2 – 5 for the G-quadruplex sample G4T4G4. The resulting g(s*) distribution for all nine G-quadruplex samples is shown in Figure 5. It is apparent that there are two distinct groups of samples: discrete g(s*) distributions centered around low s-values are observed for G2T4G2 through G4T4G4, with a noticeably smaller s-value for G2T4G2; extremely broad distributions of significantly higher s-values are apparent for G5T4G5 through G10T4G10. These observations are in accordance with those made from an examination of the raw data scans; initial examination of the raw data is useful to provide important clues as to the nature of the sample for subsequent steps in the analysis process. The nature of the g(s*) distribution for G5T4G5 and higher indicates the presence of multiple higher order species and is a significant break in sample behavior from the discrete distributions obtained for G4T4G4 and lower.
Figure 4.
Analysis steps using the DCDT+ software. (A) Scans selected for calculation of g(s*) distribution (the meniscus position is depicted by the black vertical line, and the left and right fitting limits by the dashed vertical lines); (B) dc/dt curves from pair-wise subtraction of selected scans (the average curve is shown by the thick black line); (C) selection of s-range for g(s*) transformation (left and right limits are shown by the vertical dashed lines); (D) normalized and corrected g(s*) distribution.
Figure 5.
Normalized g(s*) distribution curves for G2T4G2 – G10T4G10. (A) Discrete s-distributions are revealed for G2T4G2 through G4T4G4 with the distribution centered ~ 1 S for G2T4G2 (squares) and ~ 2 S for G3T4G3 and G4T4G4 (dashed black line and triangles, respectively). (B) A switch in sedimentation behavior is observed for G5T4G5 and higher (G5T4G5, thick black line; G6T4G6 – G10T4G10 black lines) with a broad distribution of multiple faster sedimenting species.
We typically pause analysis using DCDT+ having generated a model-independent g(s*) distribution and some initial conclusions about the nature of a sample. We then proceed with a c(s) analysis using Sedfit before entering the final stages of the analysis. It is possible to export from DCDT+ at the g(s*) stage (or earlier) by right-clicking in the appropriate graph window and selecting either a picture file or a data/text file and appropriate data subsets.
3.3. Sedfit
-
1
Load data into Sedfit according to the software directions.
Although Sedfit analyzes scans from the entire SV run it is simply not appropriate to import every single scan collected. Current versions of Sedfit have a color-coded gradation from black to red to indicate that a good range of scans has been imported. It is recommended to import scans such that an even color gradation is obtained with 1/3rd of the scans black/blue, 1/3rd green/yellow and 1/3rd orange/red. For example, if a lot of scans are imported after the sample has completely sedimented these scans will all overlay at the end of the run and there will be very little orange/red coloration. In this situation it would be necessary to determine when the sample has completely sedimented and exclude later scans from the analysis. For a slower sedimenting species it will be possible to increase the interval between imported scans as there will be smaller differences between subsequent scans than there would be for a faster sedimenting sample. We typically use on the order of 100 scans for analysis which might mean, for example, importing every third scan from a 300 scan dataset. However, this is not a hard-and-fast rule and it is important to examine different combinations of scans to ensure that appropriate scans are selected that represent the observed sedimentation behavior. To aid in the process of scan selection we employ XLGraph and WinMatch (38, 39). XLGraph is used to examine all scans and identify bad scans that will not be included in the analysis (this is also useful for DCDT+ but less of an issue given the smaller number of scans selected for analysis). WinMatch is used to assess when the sample has completely sedimented. A group of scans are loaded and a plot made of the differences between each scan and the last scan of the group, in this way the scan number can be identified where there are no further changes in sedimentation.
-
2
Similarly to DCDT+, positions are marked for the meniscus and the left and right fitting limits. Sedfit also has a cell bottom position indicator and limits for both the meniscus and cell bottom that need to be adjusted (see Note 5).
The cell bottom position is typically marked close to a radial position of 7.2 cm and is visualized as a distinct break from a high absorbance signal corresponding to material piling up at the bottom of the cell to a lower absorbance noisy region corresponding to the material of the centerpiece. The meniscus and cell bottom limits are the upper and lower limits within which the meniscus and bottom can move when these positions are allowed to float during fitting. It is possible to accept default values for these limits if there is uncertainty about their placement or use the same values from DCDT+.
-
3
Subtract all systematic noise.
-
4
Select continuous c(s) distribution.
For a first pass analysis we select the continuous c(s) distribution. This model is strictly only correct for mixtures of non-interacting, ideally sedimenting species and other more suitable models can be selected in the later stages of fitting if appropriate.
-
5
Input appropriate parameter values.
Values for the meniscus and bottom are taken from the positions selected in step 2 and should not be changed. A typical range of s-values is from 0.5 to 10 S. We typically select a resolution to give a spacing of 0.1 S, so for a 0.5 to 10 S range the resolution would be 96. For absorbance data, radial independent and time independent noise are typically not fitted (unchecked). The baseline is typically floated (checked). The typical value of 1.2 for the frictional ratio is fixed (unchecked). The confidence level is set to 0.68 (1 standard deviation).
-
6
Execute the Run command.
The run command optimizes the linear parameters (baseline, RI noise, TI noise, loading concentrations and size distributions) to provide better starting guesses for subsequent fitting (Fit command – step 7). A simulation of the sedimentation process is made with the entered parameters and compared to the experimental data. At this stage the residuals and rmsd will appear indicating the similarity between the simulated and experimental data and an initial c(s) distribution will be displayed. Adjustment of the parameters or fitting limits can be made. For example, large residuals at the base of the cell would indicate that the right fitting limit should be adjusted to account for possible back diffusion extending further back into the cell than the position of the right fitting limit. Also, partial peaks may appear at the extreme ends of the c(s) distribution. This could indicate that there might be a sedimenting species outside of the range of s-values selected for analysis. If this is the case the s-range should be extended. If the rmsd is unchanged and the partial peak height increases this indicates that the partial peak is an artifact and does not correspond to a smaller sedimenting species; the s-range can be reset to the original range. Alternatively, if the rmsd is reduced and the partial peak height decreases then there is a smaller sedimenting species and the extended range should be used. If any adjustments are made after the initial Run command, the Run command should be executed again. It should be noted that the appearance of this initial c(s) distribution will not necessarily be close to the final c(s) distribution obtained after the Fit function.
-
7
Execute the fit command.
Having optimized the linear parameters, the meniscus, bottom and frictional ratio parameters are checked to be fitted and the fit command performed to optimize the nonlinear parameters that have been marked to be fitted. An initial round of fitting is performed with the simplex algorithm before switching to the Marquardt-Levenberg algorithm and then a final round of the simplex algorithm. This is continued until there is no further decrease in rmsd.
-
8
Integrate the final c(s) distribution.
Integration reveals important information about the peaks in a c(s) distribution. Unlike DCDT+, peaks in the c(s) distribution are not adjusted to s20,w; however, this information is calculated for each peak using the integration function and can be accessed by clicking on each of the molecular weight boxes that appear on each peak. The molecular weights are calculated using a weight-average best-fit frictional ratio and as such they may not be accurate for all solution species (see Note 7). These molecular weights can be graphically represented by transformation into a c(M) distribution. Similarly to DCDT+, the c(s) distribution should be normalized for a comparison of a concentration series. In this version of Sedfit there is no function to do this and so we export the data to a convenient graphing program for normalization.
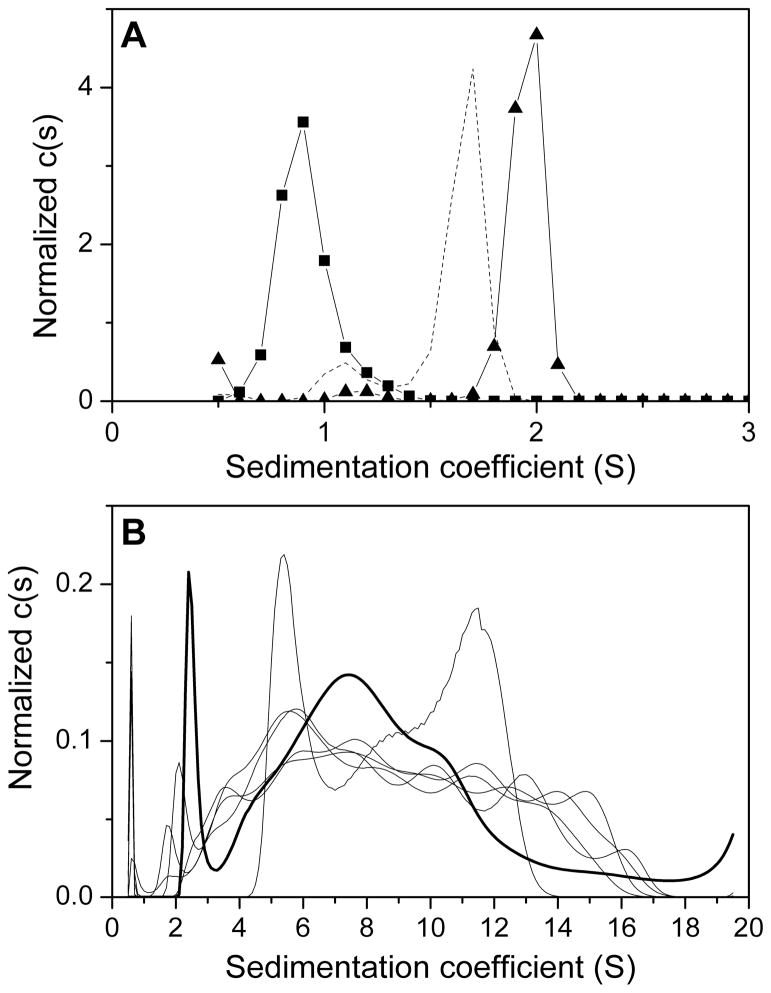
At this stage a comparison of the g(s*) and c(s) distributions for all G-quadruplexes can be made (Figures 5 and 6). The g(s*) and c(s) distributions show similar trends. Broad distributions are apparent for G5T4G5 through G10T4G10 suggesting multiple higher order sedimenting species. Discrete s-distributions are revealed for G2T4G2 through G4T4G4 with Sedfit revealing a second minor peak for G3T4G3 and G4T4G4. Estimation of M-values using Sedfit suggested the minor peak to correspond to monomer strand molecular weight and the major peak to dimer molecular weight. Peaks with relatively close s-values are not resolved in DCDT+ because of the inclusion of diffusion, but could be revealed by fitting to a two species model (see section 3.4).
Figure 6.
Normalized c(s) distribution curves for G2T4G2 – G10T4G10. (A) A single peak ~ 1 S is apparent for G2T4G2 (squares). For G3T4G3 and G4T4G4 (dashed black line and triangles, respectively) a major peak ~ 1.5 – 2 S and a minor peak ~ 1 S are observed indicating the presence of two sedimenting species. (B) Broad distributions of multiple faster sedimenting species (similar to the g(s*) distribution) are observed for G5T4G5 – G10T4G10 (G5T4G5, thick black line; G6T4G6 – G10T4G10 black lines).
It is informative to calculate a theoretical s-value for a sphere using the Svedberg equation and compare this to the experimental observed values. The s-value for a sphere will represent the maximum s-value for a sample of a given molecular weight because of the fact that a sphere has a minimum frictional coefficient and thus represents the shape of the fastest possible sedimentating species for that molecular weight. The comparison of a calculated s-value for a sphere and the s-value observed experimental will provide information about the molecularity of a given sample. This process has been outlined by Lebowitz et al. (2). As an example, G4T4G4 in Figure 6 shows a major peak corresponding to s20,w = 2.02 S and M = 7.9 kDa (close to the expected dimer M of 7.6 kDa) and a minor peak corresponding to s20,w = 1.23 S and M = 3.8 kDa (monomer strand M = 3.8 kDa). For G4T4G4, assuming v̄ = 0.55 mL/g; ρ = 1.00712 g/mL; and strand M = 3.8 kDa, would yield s-value (sphere) = 1.59 S. This compares to experimental s-values from Sedfit of 1.23 S and 2.02 S indicating that the 2.02 S peak cannot correspond to a monomer as it exceeds the maximum s-value for a monomer molecular weight. Calculation of the s-value (sphere) for a dimer gives 2.52 S and it is likely that the 2.02 S peak corresponds to a dimer. The ratio of ssphere/s20,w gives the weight-average shape factor f/fo which supports the assumption of a dimer with a moderately extended shape (f/fo = 1.25). This indicates that the two peaks in the c(s) distribution correspond to a monomer ~ 1.23 S and a dimer ~ 2.02 S with f/fo ~ 1.3 (similar to the minimized frictional ratio from Sedfit). Examination of the g(s*) and c(s) distributions in this way provides a useful starting point for model-dependent analysis.
3.4. SedAnal
-
1
Load data into SedAnal according to the software directions.
Having performed detailed analysis using DCDT+ and Sedfit, the same meniscus, fitting limits and scans from DCDT+ and the same cell bottom from Sedfit are used.
-
2
Select the appropriate model.
The number of points between the meniscus and base is set to 800 for the highest accuracy (for a single concentration of an uncomplicated system this is an appropriate value; this can be set to a lower value of 200 to speed up initial rounds of analysis for more complicated systems). The maximum iterations are set to 2000 since, typically, the fit has minimized before reaching the maximum (this can be increased to the limit of 9999 when fitting requires greater numbers of iterations). Starting values for molecular weight and sedimentation coefficient are taken from the previous DCDT+ and Sedfit analyses. Density and partial specific volume are required (see Note 5) to calculate the density increment, 1 −v̄ ρ. Mass extinction coefficient is the extinction coefficient per gram of sample. For nucleic acids, a molar extinction coefficient is more typical and can be converted to a mass extinction coefficient by dividing by molecular weight. A number of researchers use nearest neighbor extinction coefficients (routinely supplied by oligonucleotide manufacturers) but we prefer to measure these experimentally. The mass extinction coefficient must be multiplied by 1.2 to account for the 1.2 cm pathlength of the instrument. The loading concentration is calculated from the sample absorbance using a known extinction coefficient before loading.
-
3
Select the simplex fitting option for initial rounds of analysis and then switch to the Levenberg-Marquardt algorithm.
-
4
For initial fitting, fix density increment and mass extinction and allow other parameters to float. Assess the minimized values and standard deviation.
A good model should have a low standard deviation (typically values in the range of 0.003 can be obtained with good quality optics) and rational minimized parameters. The model can be tested further by fixing and floating other parameters. If the model is correct, similar parameter values should be obtained.
Two examples will be presented for G2T4G2 and G4T4G4 to illustrate different fitting strategies.
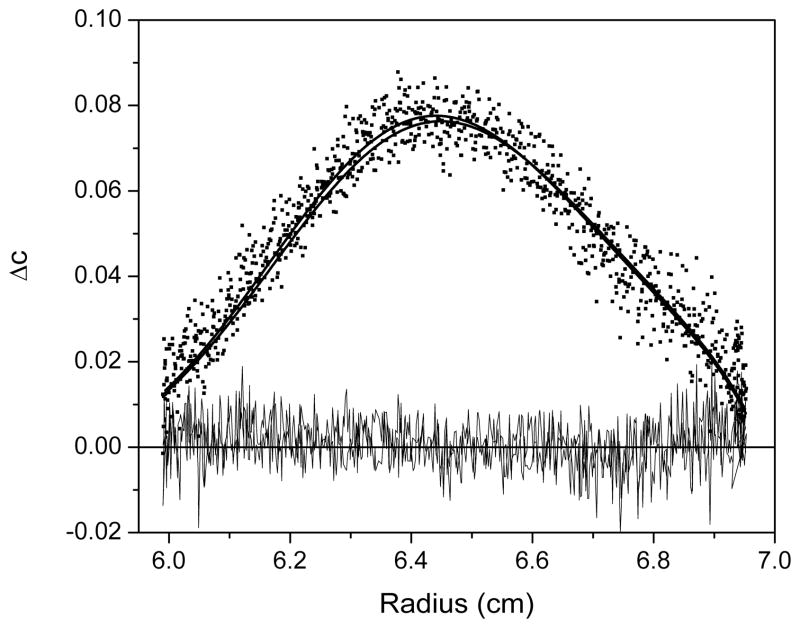
For G2T4G2, a single species model is selected and the results of the fitting are shown in Figure 7. The model describes the data well with the data points evenly spaced about the fitted lines and the residuals are randomly dispersed about zero; the minimized values for M, s and loading concentration are consistent with g(s*) and c(s) analyses and close to expected values for the monomer and the standard deviation is low (0.00393).
Figure 7.
A plot of the minimized fit to a single species model using SedAnal for G2T4G2. Data are plotted as ΔC versus radius where ΔC is the difference between pairs of closely spaced boundary scans. The data are shown as black squares and the best fit as thick black lines with the residuals (thin black lines) randomly distributed about zero.
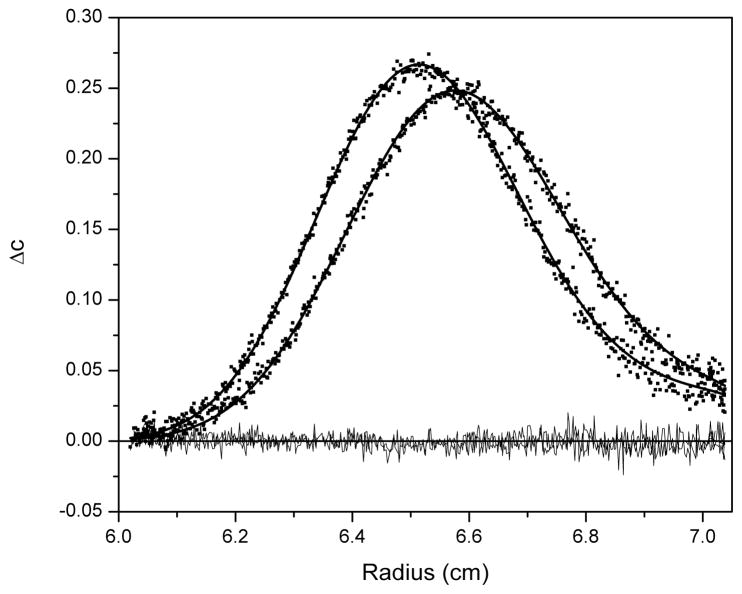
For G4T4G4, Sedfit indicated the presence of the two sedimenting species with a major peak ~ 2.02 S suggesting a dimeric species and a minor peak ~ 1.23 S of monomeric species; however, the minor peak was estimated to be on the order of only 5 % of the total solution content. Fitting to three different models was attempted with SedAnal: (1) monomer-dimer equilibrium model; (2) non-interacting two species model; and (3) single species model. An example of the fitting is shown in Figure 8 for the non-interacting two species model. Fitting to a monomer-dimer equilibrium model (setting K to a starting value of 1 x 105 M−1) returned an s-value ~ 2 S and an M-value close to dimer for one species and an s-value ~ 4 S and M-value close to tetramer for the second species with a good rmsd (0.00387). Analysis with a non-interacting two species model (setting their ratio at a starting value of 5 %) returned s-values close to the experimental c(s) values and M-values close to monomer and dimer with approximately 5 % of monomer and a standard deviation of 0.00385 for this limited dataset (Figure 8). It is impossible to assess the integrity of the two models by fitting to a single dataset for the sample. The monomer-dimer equilibrium model returns a second species ~ 4 S which is obviously not apparent in either the g(s*) or c(s) distributions but would not be expected to be apparent for a kinetically mediated equilibrium. A comprehensive concentration series is necessary to reveal the behavior of the system. Plots of the normalized g(s*) and c(s) distributions should superimpose for a non-interacting system but should shift to larger species with increasing loading concentration for self-associating systems. For self-associating systems it is informative to investigate the kinetics of association through fitting of koff; more information can be found in a recent article by Correia and Stafford (45). A thorough analysis of the solution behavior of this sample is beyond the scope of this introductory tutorial to sedimentation velocity and the reader is referred to the appropriate literature to attempt these studies (11, 45–54). The final model fitting attempted was to a single species model which might be appropriate given the low percentage of monomer estimated by c(s) analysis. The final fit from this model was also good returning an s-value ~ 2 S and an M-value close to dimer. The standard deviation was close to that for the other two models (0.00391). For this sample there is little to choose between the three models and a more comprehensive analysis is required as outlined above.
Figure 8.
A plot of the minimized fit to a non-interacting two species model using SedAnal for G4T4G4. Data are plotted as ΔC versus radius where ΔC is the difference between pairs of closely spaced boundary scans. The data are shown as black squares and the best fit as thick black lines with the residuals (thin black lines) randomly distributed about zero.
Model fitting can also be performed using DCDT+ and Sedfit. DCDT+ does not offer the range of models incorporated into Sedfit and SedAnal but can be used to fit up to 5 discrete, non-interacting species; for the three examples highlighted above DCDT+ would have utility. DCDT+ also has the option of fitting for either molecular weight or diffusion. A discrete species Lamm equation model would be a useful starting point using Sedfit. Of course, the example data highlighted in this article only represents the starting point for any thorough analysis of sedimentation procedure. It is useful to run the same sample at different concentrations and rotor speeds to reveal the presence of, for example, an interacting system, conformational changes or hydrodynamic nonideality. In the absence of macromolecular association a decrease in sedimentation coefficient with concentration can provide information about the asymmetry of a molecule (4). However, an increase in s-value with concentration is indicative of self-association; if association is present, stability of the complex can be determined by the effect of dilution on s-value. A recent tutorial article by Correia and Stafford illustrates the rigor that is needed to correctly characterize SV data for a relatively simple equilibrium monomer-dimer system (45).
While sedimentation coefficients, frictional ratios, diffusion coefficients and sample molecularity can provide useful information about the structural nature of a sample, determination of these properties is a long way from providing the absolute structure of a molecule. One final process that can be of utility in the characterization of the hydrodynamic properties of a sample is to compare experimentally determined sedimentation coefficients with those calculated from published structures and determine if the structures are consistent with the solution conformation of a molecule (4). This approach has been used successfully in our laboratory to assess the solution conformation of the 22nt human telomere sequence in both sodium and potassium solutions (55). Hydrodynamic properties were calculated using “bead models” from the published atomic-level structures for the crystallographic potassium propeller structure and the NMR determined sodium basket structure (56–59). Comparison of distributions of calculated sedimentation coefficients with experimentally determined c(s) distributions revealed striking differences between calculated and experimental s-values for the potassium form but not for the sodium form and suggested a hydrodynamically more compact conformation in potassium solution than that observed in the crystal form (Figure 9). Calculated diffusion coefficients showed the same trend with close similarity to the experimentally determined value for the sodium form and an experimentally determined diffusion coefficient representing a more hydrodynamically compact structure compared to that calculated from the crystal structure. The use of hydrodynamic parameters calculated from atomic structures has therefore proved to be of great utility in discerning the solution structure of G-quadruplexes.
Figure 9.
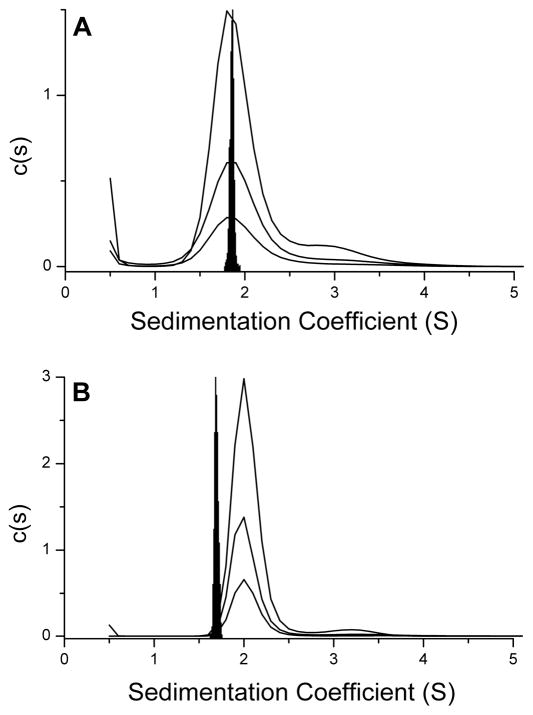
Comparison of distributions of calculated sedimentation coefficients with experimentally determined c(s) distributions. Data for three different loading concentrations (5.1, 2.55, 1.275 μM strand) are shown by the black lines. The black distributions denote calculated sedimentation coefficients using “bead models” from the published atomic-level structure for the 22nt human telomere in sodium solution. Significant differences are apparent between calculated and experimental s-values for the potassium form (panel B) but not for the sodium form (panel A) and suggest a hydrodynamically more compact conformation in potassium solution than that observed in the crystal form. Reproduced with permission from Li, J., Correia, J. J., Wang, L., Trent, J. O., and Chaires, J. B. (2005) Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 33, 4649–59.
Acknowledgments
We thank Jack Correia for countless helpful discussions. This work was supported by National Cancer Institute grant CA35635 to J.B.C.
Abbreviations
- AUC
analytical ultracentrifugation
- SV
sedimentation velocity
- SE
sedimentation equilibrium
- s
sedimentation coefficient
- s20
w, sedimentation coefficient corrected to a temperature of 20 °C and to the density and viscosity of pure water
- S
Svedberg unit equal to 10−13 sec
- D
translational diffusion coefficient
- M
molecular weight
- v̄
partial specific volume
- ρ
density
- CD
circular dichroism
Footnotes
Determination of molecular weight is typically more accurate using SE; therefore, in our laboratory we determine s-values using SV and M-values from SE experiments and then calculate diffusion coefficients using the Svedberg equation.
Runs at 60,000 rpm were performed with graphite-filled epoxy centerpieces from Spin Analytical, Inc. (SedVel60K). Beckman Coulter, the manufacturer of the modern AUC instruments XL-A or XL-I, supply Epon charcoal-filled centerpieces rated to 42,000 rpm. We routinely run these at 45,000 or 50,000 rpm without problems.
It might not be possible to capture multiple species with the selection of one set of data scans. In these cases it would be necessary to analyze these separately with the selection of difference ranges of scans. Judicial scan selection might also be a useful strategy to remove a complicating species (e.g. an aggregate) from the analysis.
When running a concentration series for a particular sample use the same scan selection for all concentrations.
Conversion to s*20,w requires input of solution density, viscosity and the partial specific volume of the sample. Ideally these values should be determined experimentally (density and viscosity can vary even between batches of the same buffer) using a density meter and a kinematic viscometer (2); however, in the absence of access to a suitable experimental setup they can be calculated using the program Sednterp (40). For proteins, the partial specific volume can be calculated with good accuracy from its amino acid composition which can easily be imported as one or three letter codes in a text file or from a databank. For DNA, a value of 0.55 mL/g is typically assumed (60). A recent article by Hellman et al. addressed the issue of estimation of partial specific volume for G-quadruplex DNA (61).
Depending on the nature of the sample, position of the right fitting limit might be a little different between DCDT+ and Sedfit. For Sedfit where scans are used for the entire run the limit might be set a little lower to exclude the upward curvature towards the base of the cell. This curvature would extend further into the cell for a sample with a small s-value that might not completely sediment to the bottom of the cell (similar to that seen for G2T4G2). However, for DCDT+ the limit is set to correspond to the end of the plateau region for scans selected in the middle of the run and this might be at a higher radial position than the situation for Sedfit. These differences are typically very small for samples that sediment completely to the bottom of the cell.
The frictional coefficient, f, provides a measure of the shape of a molecule (62). For a molecule with the same molecular weight, a compact structure will have a smaller f-value than a more elongated structure. For a given molecule, reference is made to a smooth, compact sphere which has the minimum surface area in contact with solvent and hence the minimal frictional coefficient, designated fo. fo is calculated from the Stokes equation with the particle radius defined as that of a sphere. The frictional ratio, f/fo, is a shape factor providing a measure of the maximal shape asymmetry from that of a sphere. The frictional ratio can be determined by the ratio of the experimental s-value to the maximal s-value calculated for a sphere of the same molecular weight under the same solution conditions (see section 3.3). The frictional ratio has a theoretical minimum value of 1 for a perfectly spherical molecule, a value of 1.2 is commonly accepted as a reasonable starting value during analysis for moderate molecular asymmetry. It is important to evaluate minimized values of frictional ratios to assess if they reasonably represent the expected solution structure. Unfortunately, little is known about frictional ratios for DNA; it is possible to generate some information about a molecule’s shape from published atomic structures or through molecular modeling; Sednterp and Sedfit also offer convenient shape calculators.
References
- 1.Hansen JC, Kreider JI, Demeler B, Fletcher TM. Analytical Ultracentrifugation and Agarose Gel Electrophoresis as Tools for Studying Chromatin Folding in Solution. Methods. 1997;12:62–72. doi: 10.1006/meth.1997.0448. [DOI] [PubMed] [Google Scholar]
- 2.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Sci. 2002;11:2067–79. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen JC, Lebowitz J, Demeler B. Analytical Ultracentrifugation of Complex Macromolecular Systems. Biochemistry. 1994;33:13155–63. doi: 10.1021/bi00249a001. [DOI] [PubMed] [Google Scholar]
- 4.Laue T. Biophysical studies by ultracentrifugation. Curr Opin Struc Biol. 2001;11:579–83. doi: 10.1016/s0959-440x(00)00250-5. [DOI] [PubMed] [Google Scholar]
- 5.Howlett GJ, Minton AP, Rivas G. Analytical ultracentrifugation for the study of protein association and assembly. Curr Opin Chem Biol. 2006;10:430–36. doi: 10.1016/j.cbpa.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Cole JL, Hansen JC. Analytical Ultracentrifugation as a Contemporary Biomolecular Research Tool. J Biomol Tech. 1999;10:163–76. [PMC free article] [PubMed] [Google Scholar]
- 7.Ralston G. Introduction to Analytical Ultracentrifugation. Beckman Instruments, Inc; Fullerton, CA: 1993. [Google Scholar]
- 8.Laue TM, Stafford WF., III Modern Applications of Analytical Ultracentrifugation. Annu Rev Biophys Biomol Struct. 1999;28:75–100. doi: 10.1146/annurev.biophys.28.1.75. [DOI] [PubMed] [Google Scholar]
- 9.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–43. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-Distribution Analysis of Proteins by Analytical Ultracentrifugation: Strategies and Application to Model Systems. Biophys J. 2002;82:1096–111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown PH, Balbo A, Schuck P. Characterizing Protein-Protein Interactions by Sedimentation Velocity Analytical Ultracentifrugation. Curr Protoc Immunol. 2008;81:18.15.1–18.15.39. doi: 10.1002/0471142735.im1815s81. [DOI] [PubMed] [Google Scholar]
- 12.Balbo A, Schuck P. Analytical Ultracentrifugation in the Study of Protein Self-association and Heterogeneous Protein-Protein Interactions. In: Golemis EA, Adams PD, editors. Protein-Protein Interactions: A Molecular Cloning Manual. Cold Spring Harbor Laboratory Press; Woodbury, NY: 2005. pp. 253–77. [Google Scholar]
- 13.Balbo A, Brown PH, Schuck P. Experimental Protocol for Sedimentation Velocity Analytical Ultracentrifugation. 2008 doi: 10.1002/0471142735.im1815s81. Available from http://www.analyticalultracentrifugation.com/svprotocols.htm. [DOI] [PubMed]
- 14.Demeler B, Saber H, Hansen JC. Identification and Interpretation of Complexity in Sedimentation Velocity Boundaries. Biophys J. 1997;72:397–407. doi: 10.1016/S0006-3495(97)78680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Holde KE, Weischet WO. Boundary analysis of sedimentation-velocity experiments with monodisperse and paucidisperse solutes. Biopolymers. 1978;17:1387–403. [Google Scholar]
- 16.Carruthers LM, Schirf VR, Demeler B, Hansen JC. Sedimentation Velocity Analysis of Macromolecular Assemblies. Methods Enzymol. 2000;321:66–80. doi: 10.1016/s0076-6879(00)21187-7. [DOI] [PubMed] [Google Scholar]
- 17.Stafford WF. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 18.Stafford WF. Analysis of Reversibly Interacting Macromolecular Systems by Time Derivative Sedimentation Velocity. Methods Enzymol. 2000;323:302–25. doi: 10.1016/s0076-6879(00)23371-5. [DOI] [PubMed] [Google Scholar]
- 19.Philo JS. A Method for Directly Fitting the Time Derivative of Sedimentation Velocity Data and an Alternative Algorithm for Calculating Sedimentation Coefficient Distribution Functions. Anal Biochem. 2000;279:151–63. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 20.Philo JS. An Improved Function for Fitting Sedimentation Velocity Data for Low-Molecular-Weight Solutes. Biophys J. 1997;72:435–44. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuck P, MacPhee CE, Howlett GJ. Determination of Sedimentation Coefficients for Small Peptides. Biophys J. 1998;74:466–74. doi: 10.1016/S0006-3495(98)77804-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuck P, Millar DB. Rapid Determination of Molar Mass in Modified Archibald Experiments Using Direct Fitting of the Lamm Equation. Anal Biochem. 1998;259:48–53. doi: 10.1006/abio.1998.2638. [DOI] [PubMed] [Google Scholar]
- 23.Schuck P. Sedimentation Analysis of Noninteracting and Self-Associating Solutes Using Numerical Solutions to the Lamm Equation. Biophys J. 1998;75:1503–12. doi: 10.1016/S0006-3495(98)74069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuck P. Size-Distribution Analysis of Macromolecules by Sedimentation Velocity Ultracentrifugation and Lamm Equation Modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuck P, Rossmanith P. Determination of the Sedimentation Coefficient Distribution by Least-Squares Boundary Modeling. Biopolymers. 2000;54:328–41. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 26.Schuck P. Sedimentation Equilibrium Analysis of Interference Optical Data by Systematic Noise Decomposition. Anal Biochem. 1999;272:199–208. doi: 10.1006/abio.1999.4172. [DOI] [PubMed] [Google Scholar]
- 27.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-Aided Interpretation of Analytical Sedimentation Data For Proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, U.K: 1992. pp. 90–125. [Google Scholar]
- 28.Schuck P, Demeler B. Direct Sedimentation Analysis of Interference Optical Data in Analytical Ultracentrifugation. Biophys J. 1999;76:2288–96. doi: 10.1016/S0006-3495(99)77384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philo J. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–46. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 30.Cole JL, Lary JW, Moody TP, Laue TM. Analytical Ultracentrifugation: Sedimentation Velocity and Sedimentation Equilibrium. In: Correia JJ, Detrich HW, editors. Methods in Cell Biology, Vol. 84, Biophysical Tools for Biologists: Volume 1, In Vitro Techniques. Academic Press; San Diego, CA: 2008. pp. 143–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck P. SEDFIT (version 11.3b) National Institutes of Health; Bethesda, MD: 2008. Available from http://www.analyticalultracentrifugation.com/download.htm. [Google Scholar]
- 32.Schuck P. SEDPHAT (version 5.14) National Institutes of Health; Bethesda, MD: 2008. Available from http://www.analyticalultracentrifugation.com/sedphat/download.htm. [Google Scholar]
- 33.Sherwood P, Stafford W. SedAnal, sedimentation analysis software (version 5.03) Boston Biomedical Research Institute; Watertown, MA: 2008. Available from http://sedanal.bbri.org/ [Google Scholar]
- 34.Philo JS. DCDT+ (version 2.1.0.28333) John S. Philo; Thousand Oaks, CA: 2007. Available from http://www.jphilo.mailway.com/dcdt+.htm. [Google Scholar]
- 35.Cole JL, Lary JW. Analytical Ultracentrifugation Facility. Biotechnology Bioservices Center, University of Connecticut; Storrs, CT: 2006. HeteroAnalysis (version 1.1.33) Available from http://www.biotech.uconn.edu/auf/?i=aufftp. [Google Scholar]
- 36.Philo JS. SVEDBERG (version 6.39) John S. Philo; Thousand Oaks, CA: 2001. Available from http://www.jphilo.mailway.com/svedberg.htm. [Google Scholar]
- 37.Demeler B. UltraScan (version 9.9 (Revision 714)) Borries Demeler and The University of Texas Health Science Center; San Antonio, San Antonio, TX: 2008. Available from http://www.ultrascan.uthscsa.edu/ [Google Scholar]
- 38.Philo JS. XLGraph (version 3.21) John S. Philo; Thousand Oaks, CA: 2003. Available from http://www.jphilo.mailway.com/download.htm. [Google Scholar]
- 39.Yphantis D, Lary JW. Program For Intercomparison of Ultracentrifuge Data Files (version 0.99.0039) David A. Yphantis and Jeffrey W. Lary; Mansfield Center, CT: 1999. WinMatch V0.99 for Win95 + WinNT. Available from http://www.biotech.uconn.edu/auf/?i=aufftp. [Google Scholar]
- 40.Hayes DB, Laue T, Philo J. Sedimentation Interpretation Program (version 1.09) University of New Hampshire; Durham, NH: 2006. Available from http://www.jphilo.mailway.com/download.htm. [Google Scholar]
- 41.Philo JS. DCDT+ Home Page. John S. Philo; Thousand Oaks, CA: Available from http://www.jphilo.mailway.com/dcdt+.htm. [Google Scholar]
- 42.Shafer RH, Smirnov I. Biological Aspects of DNA/RNA Quadruplexes. Biopolymers. 2001;56:209–27. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 43.Scaria PV, Shire SJ, Shafer RH. Quadruplex structure of d(G3T4G3) stabilized by K+ or Na+ is an asymmetric hairpin dimer. Proc Natl Acad Sci USA. 1992;89:10336–40. doi: 10.1073/pnas.89.21.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mekmaysy C. Masters Thesis. University of Louisville; Louisville, KY: 2007. The Structure and Stability of G-quadruplexes; p. 109. [Google Scholar]
- 45.Correia JJ, Stafford WF. Extracting equilibrium constants from kinetically limited reacting systems. Methods Enzymol. 2009:455. doi: 10.1016/S0076-6879(08)04215-8. in press. [DOI] [PubMed] [Google Scholar]
- 46.Correia JJ. Analysis of Weight Average Sedimentation Velocity Data. Methods Enzymol. 2000;321:81–100. doi: 10.1016/s0076-6879(00)21188-9. [DOI] [PubMed] [Google Scholar]
- 47.Kegeles G, Cann J. Kinetically controlled mass transport of associating-dissociating macromolecules. Methods Enzymol. 1978;48:248–70. doi: 10.1016/s0076-6879(78)48014-0. [DOI] [PubMed] [Google Scholar]
- 48.Gelinas AD, Toth J, Bethoney KA, Stafford WF, Harrison CJ. Mutational Analysis of the Energetics of the GrpE.DnaK Binding Interface: Equilibrium Association Constants by Sedimentation Velocity Analytical Ultracentrifugation. J Mol Biol. 2004;339:447–58. doi: 10.1016/j.jmb.2004.03.074. [DOI] [PubMed] [Google Scholar]
- 49.Stafford WF. Protein-Protein and Ligand-Protein Interactions Studied by Analytical Ultracentrifugation. In: Schriver JW, editor. Methods in Molecular Biology, Vol 490, Protein Structure, Stability, and Interactions. Humana Press; Totowa, NJ: 2009. pp. 83–113. [DOI] [PubMed] [Google Scholar]
- 50.Kegeles G. Pressure-jump light-scattering observations of macromolecular interaction kinetics. Methods Enzymol. 1978;48:308–20. doi: 10.1016/s0076-6879(78)48017-6. [DOI] [PubMed] [Google Scholar]
- 51.Cox DJ. Calculation of simulated sedimentation velocity profiles for self-associating solutes. Methods Enzymol. 1978;48:212–42. [Google Scholar]
- 52.Cann JR. Measurement of protein interactions mediated by small molecules using sedimentation velocity. Methods Enzymol. 1978;48:242–48. doi: 10.1016/s0076-6879(78)48013-9. [DOI] [PubMed] [Google Scholar]
- 53.Cann JR. Ligand binding by associating systems. Methods Enzymol. 1978;48:299–307. doi: 10.1016/s0076-6879(78)48016-4. [DOI] [PubMed] [Google Scholar]
- 54.Cann JR, Kegeles G. Theory of Sedimentation for kinetically controlled dimerization reactions. Biochemistry. 1974;13:1868–74. doi: 10.1021/bi00706a015. [DOI] [PubMed] [Google Scholar]
- 55.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–59. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García de la Torre J. Hydration from hydrodynamics. General considerations and applications of bead modelling to globular proteins. Biophys Chem. 2001;93:159–70. doi: 10.1016/s0301-4622(01)00218-6. [DOI] [PubMed] [Google Scholar]
- 57.García de la Torre J, Huertas ML, Carrasco B. Calculation of Hydrodynamic Properties of Globular Proteins from Their Atomic-Level Structure. Biophys J. 2000;78:719–30. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandes MX, Ortega A, López Martínez MC, García de la Torre J. Calculation of hydrodynamic properties of small nucleic acids from their atomic structure. Nucleic Acids Res. 2002;30:1782–88. doi: 10.1093/nar/30.8.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Byron O. Hydrodynamic bead modeling of biological macromolecules. Methods Enzymol. 2000;321:278–304. doi: 10.1016/s0076-6879(00)21199-3. [DOI] [PubMed] [Google Scholar]
- 60.Bloomfield VA, Crothers DM, Tinoco I., Jr . Nucleic Acids: Structures, Properties and Functions. University Science Books; Sausalito, CA: 2000. p. 359. [Google Scholar]
- 61.Hellman LM, Rodgers DW, Fried MG. Phenomenological Partial Specific Volumes for G-Quadruplex DNAs. Eur Biophys J. 2009 doi: 10.1007/s00249-009-0411-7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stafford WF, Schuster TM. Hydrodynamic Methods. In: Glasel JA, Deutscher MP, editors. Introduction to Biophysical Methods for Protein and Nucleic Acid Research. Academic Press, Inc; San Diego, CA: 1995. pp. 111–45. [Google Scholar]