Abstract
Motivation: The development of the omics technologies such as transcriptomics, proteomics and metabolomics has made possible the realization of systems biology studies where biological systems are interrogated at different levels of biochemical activity (gene expression, protein activity and/or metabolite concentration). An effective approach to the analysis of these complex datasets is the joined visualization of the disparate biomolecular data on the framework of known biological pathways.
Results: We have developed the Paintomics web server as an easy-to-use bioinformatics resource that facilitates the integrated visual analysis of experiments where transcriptomics and metabolomics data have been measured on different conditions for the same samples. Basically, Paintomics takes complete transcriptomics and metabolomics datasets, together with lists of significant gene or metabolite changes, and paints this information on KEGG pathway maps.
Availability: Paintomics is freely available at http://www.paintomics.org.
Contact: aconesa@cipf.es
1 INTRODUCTION
Biological research in the post-genomics era has been characterized by the extensive use of omics technologies. The general availability of transcriptomics, proteomics and metabolomics platforms, together with the development of user-friendly data analysis solutions (Da Wei Huang and Lempicki, 2008; Medina et al., 2010) has boosted the adoption of high-throughput approaches towards the understanding of the relationships between the genome and the phenotype. Integrated approaches that combine transcriptome, proteome and metabolome profiling have gained popularity and have proven to provide novel insights in the understanding of the biological systems (Cho et al., 2008; Heijne et al., 2005; Kolbe et al., 2006). A first approach to the interpretation of complex omics experiments is the joined visualization of the data on templates that collect previous knowledge. Graphical display is an effective tool to assist human reasoning and when different layers of biomolecular activity are presented in the context of the biological pathways they operate, much can be gained at the interpret ability of these large datasets.
To date, the number of bioinformatics tools that offer integrated visualization of omics datasets is limited. KaPPa-View (Tokimatsu et al., 2005) and MapMan (Thimm et al., 2004) are plant-specific tools that display metabolite and transcript levels on predefined pathway blocks. The MassTRIX software (Suhre and Schmitt-Kopplin, 2008) translates NMR spectra into metabolic compounds and maps them into KEGG pathways together with genome information. The tool is specially suited for exploring the metabolic repertoire of sequenced genomes but does not incorporates gene expression measurements. A recent development is ProMeTra (Neuweger et al., 2009) that accepts pre-computed and custom-made pathway maps in scalable vector graphics (SVG) format and is able to display dynamics data. The application is restricted microbial genomes and offers direct access to different omics experimental databases. Although these tools make an interesting use of visualization strategies, we found that available resources are either restricted to specific biological domains and/or have limitations for representing omics measurements.
We have developed Paintomics to provide a simple but effective resource for integrated visualization in genomics studies where transcriptomics and metabolomics data are generated on the same set of samples. Basically, the application accepts gene expression and metabolite quantifications and displays data on KEGG maps. The main distinctive features of Paintomics are: painted KEGG maps supported for a large range of organisms; joint visualization of different types of omics data, displaying both significant and non-significant changes; computation of pathway enrichment based on both transcriptomics and metabolomics data; interactive images with link-outs to KEGG info and experimental values; and easy to download mapped data for further analysis. Paintomics is available at http://www.paintomics.org.
2 THE PAINTOMICS APPLICATION
Paintomics is a platform-independent web application built on Perl and Python scripts running on an Apache web server. A simple web-form requires users to upload gene expression and metabolite concentration files, optionally provide lists of significant features, and indicate the organism under study. Paintomics directly supports over 100 top species of different biological kingdoms and offers user the possibility to request any other organism present in the KEGG database. Once data are submitted, Paintomics parses input files to match gene identifiers and metabolite names to the KEGG database. Generally, Paintomics will accept EntrezGene ID, although for a number of species different identifiers are supported. Regarding metabolites, ambiguity frequently exists for the assignment of supplied compound data and KEGG metabolite names. In this case, the user is prompted to manually assign compounds to KEGG descriptions or let the application choose the closest name(s). In the next step, the application will show matching results for the submitted data. A general summary is presented with the number of genes and metabolites of the selected species present in the current KEGG database version, the number of features matched by the input files and how many of those were labeled as significant. This summary gives users a broad feeling of the coverage achieved by the submitted data. Additionally, a per-pathway table is presented with matched and significant figures for both genes and metabolites together with the P-value of pathway enrichment based on the Fisher's exact test. This table is sortable by any of its columns to facilitate browsing of the pathways according to the user main interests. The user can then select which specific pathways to paint.
Pathways are painted locally at the Paintomics server overlaying the users data on the KEGG image templates using SVG technology. At each matched feature, a box is painted with as many sections as columns present in the input files, each section colored according to its correponding expression or concentration value. This aids in the visualization of different samples (conditions or a time course) on the same image. Significant features are highlighted by a black box and gene names are shown for all proteins present in the indicated organism. Images retain KEGG link-outs to gene and compound records to fully benefit from up-to-date pathway information. For browsers supporting SVG technology, additional interactivity is available such as box enlargement and display of actual numerical values on mouse pass-over. Images can be downloaded in different formats with a simple click, as well as all matching information used to paint each specific pathway. In order to offer reliable information, Paintomics database is automatically updated on a monthly basis.
3 USE CASE
To illustrate the usage and knowledge discover facilitated by Paintomics, we used the tool to analyse a recent study of Arabidopsis thaliana that surveyed metabolomic and transcriptomcis changes on Arabidopsis leaves in response to manipulation of the thiol-disulfide status (Kolbe et al., 2006). This study aimed at the understanding of the role of redox signals in the regulation of metabolic processes. Gene expression and metabolic data were downloaded from the publication site. The dataset contained 6390 named genes and 90 metabolites comprising sugars, amino acids and organic acids. Significant genes were selected as those with at least a 2-fold expression change, while the list significant metabolites was directly obtained from the author's analysis.
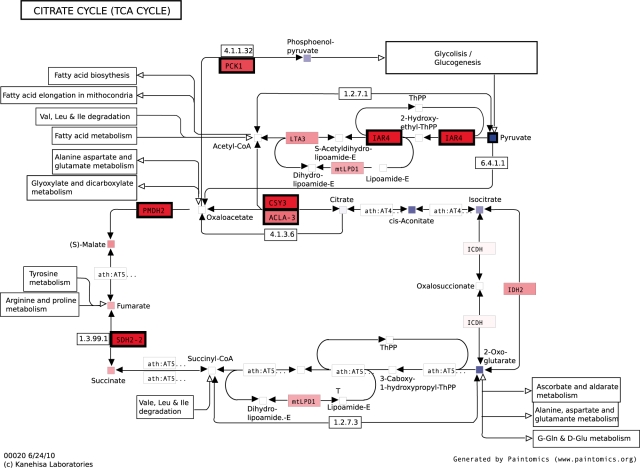
Paintomics was fed with data files and run with default parameters, selecting A.thaliana at the species check-box. A total of 60 pathways where obtained with at least one matching entry. From the visual analysis of these maps, conclusions on the co-regulation of transcripts and metabolites can be drawn. For example, citric acid map shows the down-regulation of the first part of the pathway (citrate, isocitrate, cis-asconitate and 2-oxoglutarate), while metabolites on the second part (malate, fumarate and succinate) had increased levels (Fig. 1). These changes were accompanied by the significant upregulation of genes on this second half of the cycle such as citrate synthase (CSY3), malate dehydrogenase (PMDH2) and succinate dehydrogenase (SDH2-2), pointing to a coordinated activity of genes and metabolites. Connections between the tricarboxylic acid cycle (TCA) and the pyruvate metabolism additionally reveal a decrease in pyruvate and phosphoenol pyruvate levels upon DTT treatment, which was accompanied by a significant upregulation of the pyruvate dehydrogenase (IAR4), malate dehydrogenase (IDH2) and phosphoenolpyruvate carboxilase (PCK1) enzymes that catalyze the conversion of these compounds toward acetyl-CoA, oxolacetate and finally malate. Interestingly, this pattern of metabolite balance at the TCA cycle was also observed by the authors although their transcriptomics analysis did not reveal any significant changes of genes in these pathways. These results led authors to postulate that changes in fluxes and metabolite concentrations in these pathways were most likely due of post-translational mechanisms. The integrated visualization offered by Paintomics did reveal the coordinated state of transcript and metabolite levels.
Fig. 1.
Painted citrate cycle map for the Arabidopsis DTT treatment example. Reduced levels of metabolites are found at the first part of the cycle (blue-colored metabolites), while increased concentrations are found on the second part of the pathway (red-colored metabolites). Black entry boxes represent significant regulation.
ACKNOWLEDGEMENTS
We thank the IT team of the Bioinformatics and Genomics Department of the Centro de Investigaciones Príncipe Felipe for their help. We also thank the support of the National Institute of Bioinformatics (www.inab.org) and the CIBER de Enfermedades Raras, both initiatives of the ISCIII, MICINN.
Funding: Spanish Ministry of Science and Innovation (MICINN) (grants BIO2008-05266-E BIO2008-04212 and CEN-20081002); GVA-FEDER (PROMETEO/2010/001); Red Tematica de Investigacion Cooperativa en Cancer (RTICC), ISCIII, MICINN (grant RD06/0020/1019, in part).
Conflict of Interest: none declared.
REFERENCES
- Cho K, et al. Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. J. Proteome Res. 2008;7:2980–2998. doi: 10.1021/pr800128q. [DOI] [PubMed] [Google Scholar]
- Da Wei Huang B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Heijne W, et al. Systems toxicology: applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Exp. Rev. Proteomics. 2005;2:767–780. doi: 10.1586/14789450.2.5.767. [DOI] [PubMed] [Google Scholar]
- Kolbe A, et al. Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol. 2006;141:412. doi: 10.1104/pp.106.081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina I, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;1:4. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuweger H, et al. Visualizing post genomics data-sets on customized pathway maps by ProMeTra – aeration-dependent gene expression and metabolism of Corynebacterium glutamicum as an example. BMC Syst. Biol. 2009;3:82. doi: 10.1186/1752-0509-3-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Schmitt-Kopplin P. MassTRIX: mass translator into pathways. Nucleic Acids Res. 2008;36:W481. doi: 10.1093/nar/gkn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Tokimatsu T, et al. KaPPA-View. A web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138:1289. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]