Abstract
Recent studies have demonstrated that miR-205 has a role in both normal development and cancer, however conflicting reports on its function illustrate the complexity of its regulation and targets. Additionally, miR-205 was found to be highly expressed in stem cell-enriched populations from the mouse mammary gland, and thus may play a function in normal mammary stem cell maintenance. The role that miR-205 plays in tumor formation and metastasis is likely context-dependent as reports have indicated that it may function as either a tumor suppressor or an oncogene. The role that miR-205 plays in directing stem cell fate is still unknown.
Keywords: miR-205, breast cancer, mammary gland development, stem cells
miR-205 in Normal Mammary Gland Development
The mammary gland originates as an invaginated structure from the embryonic epidermis formed during fetal development. At birth the mammary gland consists of a rudimentary ductal structure, and only upon the stimulation by estrogen, progesterone and growth hormones during puberty does ductal branching and differentiation occur.1 The epithelium and the stroma make up the two tissue compartments of the mammary gland. The stroma is the connective tissue layer, composed of mostly adipocytes in addition to fibroblasts, blood vessels, neurons and hematopoietic cells. The mammary gland epithelium is organized into two layers comprised of inner secretory luminal cells, which form ductal or alveolar cells, and outer basal myoepithelial cells, which are the contractile cells responsible for forcing milk produced by the alveoli through to ducts. Normal mammary gland stem cells are thought to reside within the basal cell compartment.2 Throughout the reproductive cycle, the adult mammary gland undergoes significant morphological changes comprised of lobuloalveolar growth (pregnancy), differentiation/secretion (lactation) and apoptosis (involution). The capacity of the mammary gland to regenerate throughout successive cycles of lactation and involution is due to this small population of mammary gland stem cells.3
Analysis of mouse mammary glands from various stages of pregnancy indicated that miR-205 may be developmentally regulated.4 These studies showed that in the adult virgin miR-205 expression was restricted to the myoepithelial cell layer, which contains both myoepithelial cells and basal stem cells, and expression in both luminal and basal epithelium increased during pregnancy and lactation, with another increase in expression during late involution. Likewise, normal human mammary gland samples showed miR-205 expression in ductal and lobular myoepithelial/basal cell compartments.5 Cell culture models of normal mammary epithelial cells also indicated that miR-205 was highly expressed in the “progenitor-like” subpopulation based on FACS purification of AldefluorhiSca-1hi or Sca-1+ cells.6,7 Several groups identified normal mouse mammary stem cell-enriched populations that are capable of repopulating the mammary gland based on CD24 (heat stable antigen), Sca-1 (Stem cell antigen-1; encoded by Ly6a), CD29 (β1-integrin) and CD49f (α6-integrin) cell surface marker expression. These three populations contain a mix of myoepithelial and basal cells, with an estimated 1 in 2000 enrichment for mammary gland stem cells. These stem cell-enriched populations were isolated by the profiles of Lineage(Lin)−CD24+/loSca-1−,2 Lin−CD29hiCD24+,8 or Lin−CD49fhiCD24med (also referred to as the mammary repopulating unit, or MRU).9 Analysis of miR-205 in the normal adult mammary gland revealed high expression of miR-205 in these three stem cell-enriched populations, in addition to high expression in the myoepithelial cell population (CD49floCD24lo)6 (Fig. 1). These data suggest a role for miR-205 in mouse mammary gland stem cells. While miRNAs important in directing the fate of somatic stem cells of many tissues have been identified,10–13 the role that miRNAs play in normal mammary gland stem cells is only just starting to be elucidated. Additionally, other studies have found miR-205 expression to be localized specifically to epithelial cells of the developing eye,14 suggesting an epithelial-specific role for miR-205. Based on miR-205 expression data in mammary stem cell populations6 and its regulation during pregnancy and involution,4 this miRNA may play a role in mammary epithelial stem cell maintenance and differentiation.
Figure 1.
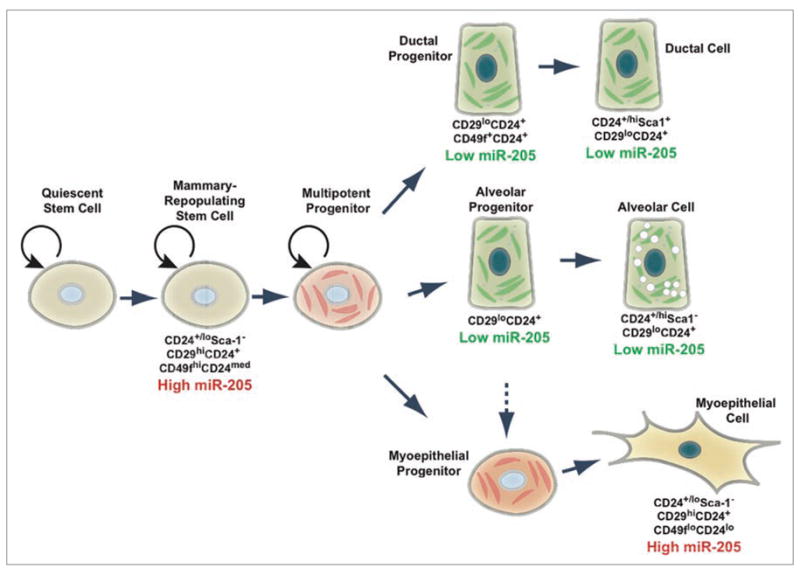
miR-205 expression in mammary gland cells. Cells comprising the mammary gland can by identified by their cell surface marker profiles.3 High miR-205 expression is found in the stem cell-enriched populations: CD24+/loSca-1−, CD29hiCD24+ and CD49fhiCD24med (also referred to as the MRU population) and the myoepithelial cell populations: CD24+/loSca-1−, CD29hiCD24+ and CD49floCD24lo. Low miR-205 expression is in the ductal (CD24+/hiSca1+, CD29loCD24+ and CD49f+CD24+) and alveolar (CD24+/hiSca1− and CD29loCD24+) populations.
miR-205 in Cancer
Because miRNAs are important for directing stem cell fate, altered miRNA expression may induce the transformation of a stem or progenitor cell to a cancer stem cell. Cancers of different origins have unique miRNA signatures. These tumors can not only be distinguished by microarray profiling, but their unique miRNA profiles can be used to identify their respective tissue of origin,15,16 implying that there are not only tissue-specific miRNA profiles, but these profiles are inherited by developing tumors. These tumor-specific differences may prove to be useful prognostic and predictive factors.
Altered miR-205 expression has been found in many clinical samples of solid tumors compared to normal tissue, as well as many cancer cell lines. High expression of miR-205 was found in head and neck cancer cell lines17 and squamous cell carcinoma cell lines,18 as well as clinical samples of metastatic head and neck squamous cell carcinomas.19 However, other studies demonstrated clinical samples of head and neck squamous cell carcinomas expressing low levels of miR-205 is associated with increased recurrence and poor prognosis.20 In clinical bladder samples, miR-205 was also found to be significantly upregulated compared to normal tissue,21 while low expression of miR-205 and members of the miR-200 family was associated with invasive bladder cancers compared to non-invasive bladder cancers.22 Clinical prostate cancer samples also showed down-regulation of miR-205 relative to normal adjacent tissue, and loss of miR-205 was associated with prostate cancer progression.23 Additionally, miR-205 was highly expressed in cervical cancer cell lines as well as clinical cervical cancer samples compared to normal cervical tissue.24 High expression of miR-205 was found in nearly 65% of the patients studied with non small cell lung cancers.25 Further classification of non small cell lung cancers into squamous cell carcinomas and adenocarcinomas could be achieved based on miR-205 expression, where miR-205 is highly expressed in squamous cell carcinomas but not in adenocarcinomas.26,27 From these expression studies, it could be hypothesized that miR-205 is expressed at higher levels in cancer subtypes with more epithelial characteristics and a better prognosis and is lower in those that are more invasive with a poorer prognosis.
miR-205 in Breast Cancer
Breast cancers also display this heterogeneity of miR-205 expression. Based on microarray expression experiments, miR-205 was found to be high in ER+PR+Her2+ breast cancer relative to other subtypes.28 Comparative genomic hybridization analysis found a genomically amplified region on human chromosome 1 that contains miR-205 is in some human breast cancers.29 However, other studies observed high miR-205 expression in ER−PR−Her2− tumors, but low expression in other tumor types, and low miR-205 expression in metastatic breast cancer cell lines.5 Likewise, other studies found low miR-205 expression in clinical samples of metastatic breast compared to non-meta-static cancers.30
This last finding is consistent with recent studies indicating that miR-205 is a negative regulator of the epithelial-to-mesenchymal transition (EMT), an early process in metastasis, and expression of miR-205 is lost in mesenchymal breast cancer cell lines.31 These studies indicate a negative feedback loop that regulates EMT where miR-205, in addition to members of the miR-200 family, silence the EMT-inducing transcriptional repressors Zeb1 and Zeb2, while induction of EMT via TGFβ results in loss of miR-205 and the miR-200 family members.31 Furthermore, a metastatic miRNA expression signature in human breast cancers included decreased expression of miR-205, in addition to mis-expression of miR-200 family members, as well as several other miRNAs, associated with metastasis.32 While miR-205 and the miR-200 family have overlapping functions in the regulation of EMT via Zeb1 and Zeb2, they are not functionally redundant. In both normal mammary gland stem cells (MRUs) and breast cancer stem cells, members of the miR-200 family were significantly downregulated relative to other cell populations,33 while miR-205 expression was elevated in normal MRUs.6 This suggests a balance between the expression miR-200 family members and miR-205 and implies that there must be unique roles for these miRNAs. Interestingly, in normal development miR-205 is observed in the endoderm and ectoderm, but not the mesoderm, during gastrulation of chick embryo, a process that is regulated by EMT.34
miR-205: Oncogene or Tumor Suppressor?
The role that miR-205 plays in breast cancer is likely specific to the tumor subtype as well as the cell of origin and the stage of tumor progression. While expression of miR-205 seems to be epithelial-specific, its expression is enriched in mammary epithelial stem cell populations.6 Thus, miR-205 may not only be involved in normal cell function (proliferation, differentiation and growth), its mis-regulation may contribute to cancer initiation, progression and metastasis. Depending on the study, miR-205 was found to be either up or downregulated in breast cancers. Overexpression of miR-205 in prostate cancer cell lines and breast cancer cell lines lead to inhibition of growth,35,36 while overexpression in normal mammary epithelial cells lead to increased growth and colony forming ability, in addition to expansion of the progenitor cell population defined by Sca-1.6 Since miR-205 overexpression affects progenitor cell expansion, it may be important in the etiology of cancer. To date, targets of miR-205 include the tumor suppressors PTEN6 and SHIP2,18 the oncogenes HER3,36 E2F1, E2F5 and PKCε,35 the pro-metastatic genes Zeb1 and Zeb2,31 and the angiogenic factor VEGFA37 (Fig. 2). Other miRNAs have been shown to function as either tumor suppressors or oncogenes depending on the cell type in which they are expressed. For example, miR-155 may function as a tumor suppressor in pancreatic cancers,38 while in B-cell lymphomas it has oncogenic activity,39 so this situation is not unique to miR-205. While it is unclear whether mis-regulation of miR-205 is an initiating or merely a modulating event in conjunction with oncogene activation or loss of tumor suppressors in cancer, loss of miR-205 expression in breast cancer may be an event leading to metastasis, and the result of miR-205 mis-expression may be cell type-specific. Most likely, miR-205 directs cell fate decisions and may not be acting as an oncogene or a tumor suppressor.
Figure 2.
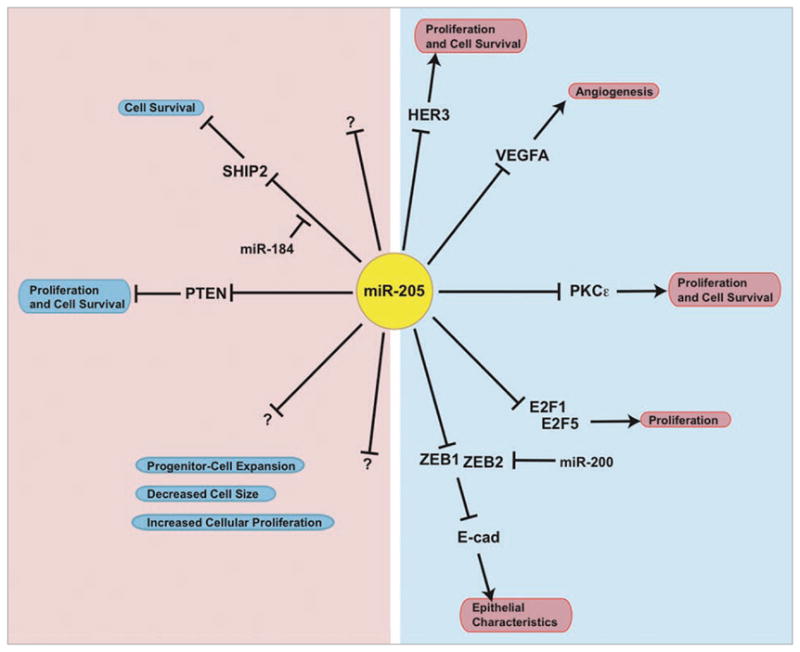
miR-205 in epithelial cells. Model of context-specific effects of miR-205 on cellular phenotype. On the left side, miR-205 can act as an oncogene and/or affect cell fate in mammary stem cells and keratinocytes by targeting tumor-suppressors like PTEN and SHIP2, respectively. In other cellular contexts, miR-205 can act as a tumor-suppressor and/or affect epithelial characteristics. Other targets of miR-205 include the oncogenes HER3, E2F1, E2F5 and PKCε, the pro-metastatic genes Zeb1 and Zeb2, and the angiogenic factor VEGFA (right side). There are also many unknown targets of miR-205 that may regulate proliferation, cell size and progenitor cell expansion.
Overall, there is likely a tightly controlled balance of miR-205 expression in epithelial cells. Normal mammary epithelial cells would maintain relatively high levels of miR-205, compared to non-epithelial cells. These levels of miR-205 are sufficient to decrease the transcription factors Zeb1 and Zeb2, which would subsequently result in the expression of E-cadherin and cell polarity factors, thus conferring epithelial characteristics.31 Regulation of PTEN expression by miR-205 allows for activation of the PI3K/Akt pathway, downstream targets of which include cell survival and proliferation factors.6 If miR-205 is lost, however, the Zeb1 and Zeb2 transcription factors are upregulated. The ZEB transcription factors are then able to bind to the E-box elements of the E-cadherin gene, as well as cell polarity genes, resulting in repression E-cadherin, loss of cell polarity, and loss of cell-cell junctions.40 These molecular changes can result in an EMT. In corneal epithelial cells, a balance between miR-205 and another miRNA has been shown in a unique mechanism, where miR-184 competes for the binding of miR-205 to the SHIP2 3′UTR to maintain SHIP2 levels required for epithelial characteristics specific to eye function.18
The Future for miR-205
These functional studies described rely mainly on ectopic overexpression of miR-205 in cell culture models. Generation of loss of function mouse models for miR-205 by a genetic technique will be required to establish the role of miR-205 in normal development and cancer. It has also been shown that miR-205 may confer tumor suppressor activity in tumors and cancer cell lines.35,36 Hence, generating miR-205 overexpression and loss of function mouse breast cancer models may demonstrate the mechanisms by which miR-205 mis-regulation contributes to cancer initiation and progression. In these studies it will be important to investigate the role of miR-205 mis-regulation in different mammary epithelial populations because its effects may vary depending on the cellular context.
Acknowledgments
S.B.G. was supported by a Department of Defense Breast Cancer Program Predoctoral Fellowship (DAMD W81XWH-06-1-0716), J.I.H. was supported by a Komen Post-doctoral fellowship PDF0707744, and J.M.R. was supported by grants from the NIH (CA-16303).
Abbreviations
- FACS
fluorescent activated cell sorting
- Sca-1
stem cell antigen-1
- lin
lineage
- MRU
mammary repopulating unit
- ER
estrogen receptor
- PR
progesterone receptor
- EMT
epithelial-to-mesenchymal transition
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/11837
References
- 1.Wagner KU, Smith GH. Pregnancy and stem cell behavior. J Mammary Gland Biol Neoplasia. 2005;10:25–36. doi: 10.1007/s10911-005-2538-1. [DOI] [PubMed] [Google Scholar]
- 2.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaMarca HL, Rosen JM. Minireview: hormones and mammary cell fate—what will I become when I grow up? Endocrinology. 2008;149:4317–21. doi: 10.1210/en.2008-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avril-Sassen S, Goldstein LD, Stingl J, Blenkiron C, Le Quesne J, Spiteri I, et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics. 2009;10:548. doi: 10.1186/1471-2164-10-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell sub-populations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 6.Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010;123:606–18. doi: 10.1242/jcs.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 9.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 10.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 11.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–77. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–84. [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci USA. 2008;105:19300–5. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher AM, Heaford AC, Trask DK. Detection of metastatic head and neck squamous cell carcinoma using the relative expression of tissue-specific mir-205. Transl Oncol. 2008;1:202–8. doi: 10.1593/tlo.08163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–45. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Wszolek MF, Rieger-Christ KM, Kenney PA, Gould JJ, Silva Neto B, Lavoie AK, et al. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol Oncol. 2009 doi: 10.1016/j.urolonc.2009.08.024. In press. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–76. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 26.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–9. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 27.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–7. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 28.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel bio-marker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol. 2007;8:214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 31.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 32.Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 33.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–65. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 35.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, et al. miR-205 Exerts tumor-suppressive functions in human prostate through downregulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–95. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 36.Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–48. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 39.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Grunsven LA, Michiels C, Van de Putte T, Nelles L, Wuytens G, Verschueren K, et al. Interaction between Smad-interacting protein-1 and the corepressor C-terminal binding protein is dispensable for transcriptional repression of E-cadherin. J Biol Chem. 2003;278:26135–45. doi: 10.1074/jbc.M300597200. [DOI] [PubMed] [Google Scholar]


