Abstract
Covalent histone modifications play crucial roles in chromatin structure and genome stability. We previously reported biotinylation of lysine (K) residues in histones H2A, H3, and H4 by holocarboxylase synthetase, and demonstrated that K12-biotinylated histone H4 (H4K12bio) is enriched in repeat regions and participates in gene repression. The biological functions of biotinylation marks other than H4K12bio are poorly understood. Here, novel biotinylation site-specific antibodies against H3K9bio, H3K18bio, and H4K8bio were used in chromatin immunoprecipitation studies to obtain first insights into possible biological functions of these marks. Chromatin immunoprecipitation assays were conducted in human primary fibroblasts and Jurkat lymphoblastoma cells, and revealed that H3K9bio, H3K18bio, and H4K8bio are enriched in repeat regions such as pericentromeric alpha satellite repeats and long-terminal repeats while being depleted in transcriptionally active promoters in euchromatin. Transcriptional stimulation of the repressed interleukin-2 promoter triggered a rapid depletion of histone biotinylation marks at this locus in Jurkat cells, which was paralleled by an increase in interleukin-2 mRNA. Importantly, the enrichment of H3K9bio, H3K18bio, and H4K8bio at genomic loci depended on the concentration of biotin in culture media at nutritionally relevant levels, suggesting a novel mechanism of gene regulation by biotin.
Keywords: biotin, gene regulation, heterochromatin, histones, human
1. Introduction
Histones are DNA-binding proteins that mediate the folding of DNA into chromatin [1, 2]. In mammals, five major classes of histones have been identified: linker histone H1, and core histones H2A, H2B, H3, and H4. In chromatin, the core histones form octamers, each consisting of one H3-H3-H4-H4 tetramer and two H2A-H2B dimers. One hundred forty-six base pairs of DNA are wrapped around each octamer to form nucleosomal core particle. The DNA located between nucleosomal core particles is associated with histone H1 [1].
The N-termini of core histones protrude from the nucleosomal surface, and some regions in the C-termini and globular domains are also exposed at the nucleosomal surface [1, 2]. These regions are targets for covalent modifications of histones, e.g., acetylation and methylation [3]. Covalent modifications of histones play critical mechanistic roles in the epigenetic regulation of DNA repair [4], gene transcriptional activity [3], and mitotic and meiotic chromosome condensation [5]. For example, lysine (K)9-acetylated histone H3 (H3K9ac) is enriched in promoters in transcriptionally active genes[3], whereas K9 dimethylated histone H3 (H3K9me2) is associated with transcriptionally repressed chromatin such as satellite repeats in pericentromeric heterochromatin and retrotransposons [6, 7].
Previously, we reported the covalent binding of biotin to distinct lysine residues in core histones as a novel histone modification [8–12], mediated by holocarboxylase synthetase (HCS) [13, 14]. The following biotinylation sites were identified in vivo and in vitro by using mass spectrometry, immunoblot analysis, HCS knockdown models, radiotracer studies, and incubation of recombinant HCS with synthetic histone-based peptides [8–12]: K9 and K13, K125, K127 and K129 in histone H2A [11], K4, K9, K18, and perhaps K23 in histone H3 [10, 12], and K8 and K12 in histone H4 [9]. Preliminary evidence suggests that K5 and K16 in histone H4 also might be biotinylated [9, 15].
Multiple lines of evidence suggest that K12-biotinylated histone H4 (H4K12bio) is enriched in repeat regions such as pericentromeric alpha satellite repeats [16], telomeric repeats [17], and long terminal repeats (LTR) [18]. H4K12bio participates in the transcriptional repression of LTR [18], the transcriptionally competent interleukin-2 (IL-2) promoter [16], and the euchromatic sodium dependent multivitamin transporter (SMVT) promoter 1 [19] in human lymphocytes and mammalian cell lines. Importantly, the enrichment of H4K12bio in chromatin and its effect in gene repression depend on biotin in studies in free-living human subjects and cell cultures; these studies were conducted using doses of biotin that are nutritionally relevant [18, 19].
The biological functions for histone biotinylation marks other than H4K12bio are largely unknown. Preliminary evidence suggests that K8-biotinylated histone H4 (H4K8bio) might also be enriched in repeat regions and participate in gene repression [16]. These previous studies of H4K8bio provided somewhat inconclusive results, due to the cross-reaction of the probe anti-H4K8bio with H4K12bio [9, 16]. In recent studies, we generated novel antibodies to H3K9bio, H3K18bio, and H4K8bio that are highly specific for those marks. Subsequently, we expanded our initial specificity tests to demonstrate that anti-H3K9bio, anti-H3K18bio are highly specific for biotinylation sites and do not cross-react with histone marks such as methylation and acetylation (T. Kuroishi et al., in preparation). In this study, we used the new antibodies (i) to quantify the relative enrichment of these marks in heterochromatin repeats, and gene promoters in euchromatin and repressed, yet transcriptionally competent, genes; (ii) to determine whether the abundance of these marks depends on the concentration of biotin in cell culture media at nutritionally relevant levels; and (iii) to determine whether depletion of these marks is associated with a transcriptional activation of genes.
2. Materials and methods
2.1. Cell culture
Human lymphoblastoma (Jurkat) cells (clone E6-1; ATCC, Manassas, VA) were cultured in humidified atmosphere (37°C, 5% CO2) as described [20]. Culture medium was replaced with fresh medium (RPMI-1640; Thermo Scientific, Waltham, MA) every 48 h. Commercial RPMI-1640 contains 0.82 μmol/l biotin, which is three orders of magnitude greater than physiological biotin concentrations. Where indicated, Jurkat cells were cultured in the following biotin-defined media for 12 d before sample collection: 0.025 nmol/l of biotin (denoted “deficient”), 0.25 nmol/l of biotin (“physiological”), and 10 nmol/l of biotin (“pharmacological”). These concentrations represent levels of biotin observed in plasma from biotin-deficient adults, biotin-normal adults, and biotin-supplemented adults. Biotin-defined media were prepared as described [20]; biotin concentrations in media were confirmed by avidin-binding assay [20], and efficacy of biotin treatment was monitored at bi-weekly intervals by quantifying the abundance of biotinylated (holo-)carboxylases in Jurkat cells [20]. Biotinylation of carboxylases is a well-established marker for biotin status [21]. Holocarboxylases were visualized by gel electrophoresis [20], using fluorophore-conjugated streptavidin (Licor, Lincoln, NE) and an Odyssey Infrared Imaging system (Licor).
Jurkat cells are an excellent model to link epigenetic changes at the IL-2 promoter locus to altered transcriptional activity. In unstimulated Jurkat cells, the IL-2 gene is repressed, yet transcriptionally competent [20, Mascher, 1999 #2130]. If Jurkat cells are stimulated with phorbol-12-myristate-13-acetate (PMA) and phytohemagglutinin (PHA), the enrichment of H4K12bio at the promoter locus decreases, while IL-2 mRNA abundance increases [16]. In this study, IL-2 expression was activated with PMA and PHA as described [16].
In some experiments, biotinylation of histones was abrogated by mutation of HCS. In these experiments, fibroblasts from a human HCS-deficient patient (clone WG2215 cells; Montreal Children’s Hospital Cell Repository, Montreal, Canada) were compared to HCS wild-type fibroblasts IMR-90 cells (ATCC). Fibroblasts were cultured as recommended by ATCC, using biotin-defined Dulbecco’s modified Eagle’s medium (10 nmol/l biotin). Differences in HCS activity in WG2215 and IMR-90 fibroblasts were confirmed by probing biotinylated carboxylases as described above.
2.2. Antibodies
In previous studies, we generated rabbit polyclonal antibodies to biotinylation sites in histones H3 (H3K4bio, H3K9bio, H3K18bio) and H4 (H4K8bio) [9, 10]. Specificity tests were conducted to demonstrate target specificity. Anti-H3K9bio and anti-H3K18bio passed these tests, while anti-H3K4 cross-reacted with H3K9bio and H3K18bio, and anti-H4K8bio cross-reacted with H4K12bio. Thus, new batches of anti-H3K4bio and anti-H4K8bio were raised in rabbits, and all four antibodies were re-tested using synthetic peptides, histone bulk extracts, biotin-depleted histones, and histone/peptide competition studies as described for H4K12bio [9]. A recent report suggested that some commercial antibodies to biotinylated histones cross-react with acetylated histones [22]. Although we could not reproduce the findings from that report, we expanded our battery of antibody tests by including histone-based acetylated peptide. Based on these tests, our antibodies to H3K9bio, H3K18bio, and H4K8bio are biotinylation site specific; bind only to the targeted class of histones; do not cross-react with biotin-free histones; and do not cross-react with acetylated or methylated histones (T. Kuroishi et al., in preparation). In contrast, multiple preparations of anti-H3K4bio cross-reacted with H3K9bio and H3K18bio and, therefore, the H3K4bio mark was not considered in this study. Affinity-purified antibodies against the C-terminus in histone H3 (ab1791) and H3K9ac (ab10812) were obtained from Abcam (Cambridge, MA). Polyclonal rabbit anti-human H3K9me2 and polyclonal rabbit anti-human H4K12bio were generated in a commercial facility (Cocalico Biologicals, Reamstown, PA, USA) as described [9, 10]. H3K9ac is a marker for euchromatin, whereas H3K9me2 and H4K12bio are markers for heterochromatin [16, 23].
2.3. Micro chromatin immunoprecipitation (μChIP) assay
Using the antibodies described above, μChIP assays were conducted as described [24] with the following minor modifications. First, 120 ng of commercial affinity-purified anti-H3 and anti-H3K9ac or 7 μl of antisera to H3K9bio, H3K18bio, H3K9me2, H4K8bio, and H4K12bio were used per 100,000 Jurkat cells. If μChIP assays were conducted in fibroblasts, 2.5×106 cells were used and the amount of antibodies was increased accordingly. Second, chromatin was precipitated overnight in a rotator at a setting of 20 rpm. If H3K9ac was investigated, cross-linking and lysis buffers were supplemented with 20 mmol/l sodium butyrate to inhibit histone deacetylase [24]; other marks were studied without sodium butyrate. Total DNA from Jurkat cells was used as input control (denoted “input DNA”). Precipitation of chromatin with non-specific rabbit IgG (Santa Cruz; Santa Cruz, CA) was used as negative control and produced negligible amounts of DNA; gene-specific sequences were at the detection limit of quantitative real-time PCR (qRT-PCR). Therefore, values were not corrected for values obtained with non-specific IgG. The antibody against H3 was used to normalize for nucleosomal occupancy at genomic loci of interest.
2.4. Quantitative real-time polymerase chain reaction
DNA from μChIP assays was purified as described [24] and quantified by qRT-PCR [16], using the cycle threshold values to calculate the abundance of DNA [25]. PCR primer sequences for LTR15, LTR22, alpha satellite repeats in pericentromeric heterochromatin in chromosome 4, SMVT promoter 1, aldehyde dehydrogenase 5 (ADH5) promoter, and IL-2 promoter were described previously [16, 18, 19]; the following primers were used to PCR amplify a 101-bp fragment in the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter: forward = 5′-ATGACAAGCATGAGGCAGAG -3′ and reverse = 5′-CAACCAGGACCGTTAACCCTTTCT-3′. Green FastMix ROX (VWR; West Chester, PA) was used for qRT-PCR reactions. The relative enrichments of gene sequences by μChIP are expressed in units of % of input DNA that was precipitated with a given antibody [26, 27]. All values were normalized for the level of nucleosomal occupancy at the gene locus by using an antibody to the C-terminus in histone H3 as a probe [18].
The abundance of mRNA coding for LTR, SMVT, and IL-2 was quantified by qRT-PCR as described [16, 18, 19]. ABsolute QPCR SYBR Green fluorescein mix (ABgene, Rochester, NY) was used for quantification of mRNA.
2.5 Statistical analysis
The homogeneity of variances among groups was tested with Bartlett’s test; if variances were heterogenous, data were log transformed before further statistical analysis [28]. Significance of differences among more than two groups was tested by one-way analysis of variance and Fisher’s protected least significant difference procedure for post hoc testing. Student’s paired t test was used for pairwise comparisons. μChIP data from fibroblasts were analyzed by using the unpaired Wilcoxon’s rank-sum test because of the comparably large data variation in these samples. StatView 5.0.1 (SAS Institute, Cary, NC) was used to perform all calculations. Differences were considered significant if P < 0.05. Data are expressed as mean ± S.D.
3. Results
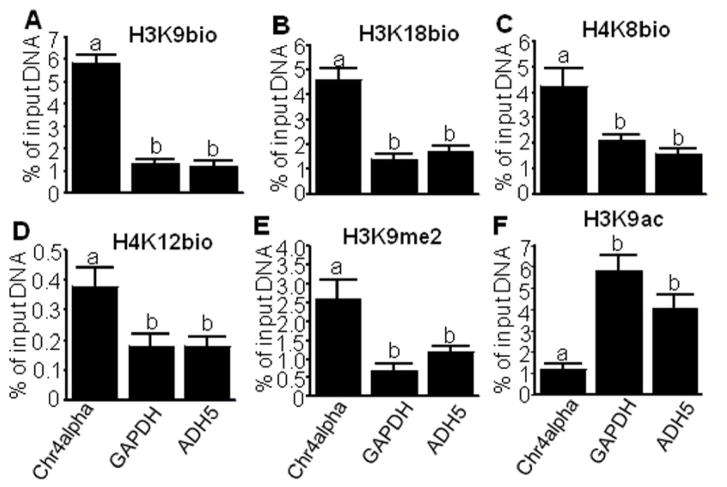
The enrichments of H3K9bio, H3K18bio, and H4K8bio in pericentromeric alpha satellite repeats (heterochromatin) greatly exceeded their enrichments in the promoters of the housekeeping genes GAPDH and ADH5 (euchromatin) in Jurkat cells (Fig. 1A–C). Previous studies revealed a similar pattern for the gene repression marker H4K12bio [16], and these patterns were confirmed here (Fig. 1D, positive control). Additional controls included the repression mark H3K9me2 and the activation mark H3K9ac. Both marks behaved as expected: the enrichment of H3K9me2 in satellite repeats exceeded those in housekeeping gene promoters (Fig. 1E), whereas the enrichment of H3K9ac in housekeeping genes exceeded that in satellite repeats (Fig. 1F). Similar observations were made for another repressed repeat element in human chromatin, i.e., LTRs (see below). These studies indicate that each of the histone biotinylation marks studies to data localize in repressed regions rather than transcriptionally activated regions.
Fig. 1.
Relative enrichment of histone biotinylation marks in heterochromatin and euchromatin in Jurkat cells. Chromatin was immunoprecipitated using antibodies against H3K9bio (panel A), H3K18bio (B), H4K8bio (C), H4K12bio (D), H3K9me2 (E), and H3K9ac (F). qRT-PCR was used to quantify sequences from alpha satellite repeats on chromosome 4 (Chr4alpha), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and aldehyde dehydrogenase 5 (ADH5) in the precipitated DNA. Bars denote the percent of input DNA from a given locus that was precipitated with antibodies (mean ± S.D., n = 4). a, bBars not sharing the same letter are significantly different (P < 0.05).
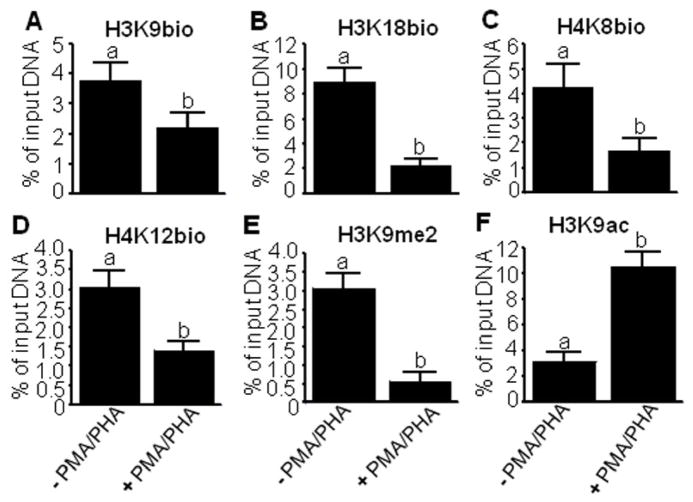
Consistent with a previous report [16], the new biotinylation marks were not only enriched in repeat elements in heterochromatin, but also in repressed, yet transcriptionally competent, gene promoters such as IL-2 in human lymphoblastoma Jurkat cells. As described above, the IL-2 promoter is repressed in unstimulated lymphoid cells, but is rapidly activated in response to stimulation with the mitogens PMA and PHA. Here, we compared the enrichment of histone marks before and after stimulation of Jurkat cells with mitogens. The abundance of IL-2 mRNA increased by 59 ± 1.2% in cells 2 h after addition of PMA and PHA compared to cells before stimulation (P < 0.01; n = 3). Based on this observation, we concluded that our experimental protocol to stimulate IL-2 expression successfully activated IL-2 expression. The same cells were used for subsequent μChIP experiments.
The enrichment of H3K9bio, H3K18bio, and H4K8bio at the IL-2 promoter was consistently greater in Jurkat cells before stimulation than after stimulation with PMA and PHA (Fig. 2A–C). The H4K12bio mark is known to exhibit a similar behavior [16] and was used as a positive control; the H4K12bio mark exhibited the expected decrease in enrichment in response to mitogenic stimulation (Fig. 2D). The repression mark H3K9me2 and the activation mark H3K9ac also showed the expected response in control experiments. The enrichment of H3K9me2 decreased at the IL-2 promoter in response to mitogenic stimulation (Fig. 2E), whereas the enrichment of H3K9ac increased (Fig. 2F). Taken together, these studies suggest that histone biotinylation in repressed regions are removed during gene activation. These studies also suggest that histone biotinylation marks are not only enriched in repeat regions, but also in transcriptionally competent gene promoters.
Fig. 2.
Relative enrichment of histone biotinylation marks in the transcriptionally inactive and active interleukin-2 promoter in Jurkat cells. Jurkat cells were collected before (“−PMA/PHA” = inactive IL-2 promoter) and after (“+PMA/PHA” = active promoter) stimulation with 50 μg/l PMA and 2 mg/l PHA for 2 h. Chromatin was immunoprecipitated using antibodies against H3K9bio (panel A), H3K18bio (B), H4K8bio (C), H4K12bio (D), H3K9me2 (E), and H3K9ac (F). qRT-PCR was used to quantify IL-2 promoter sequences in the precipitated DNA. Bars denote the percent of input DNA from the IL-2 promoter locus that was precipitated with antibodies (mean ± S.D., n = 4). a, bBars not sharing the same letter are significantly different (P < 0.05).
The enrichment of H3K9bio, H3K18bio, and H4K8bio at genomic target loci depends on biotin availability. Previous studies revealed three loci in which the enrichment of H4K12bio depends on the concentration of biotin in culture media and dietary biotin supply: LTR15, LTR22, and promoter 1 of the SMVT gene [18, 19]. If Jurkat cells were cultured in biotin-defined media, the enrichment of H3K9bio, H3K18bio, and H4K8bio at the LTR22 locus exhibited a dose-response relationship with regard to biotin availability (Fig. 3A–C). The enrichment pattern was similar for the repression mark H3K9me2 (Fig. 3D), presumably due to direct physical interactions between HCS and histone K9 methyltransferases [29]. Likewise, the positive control H4K12bio also showed the expected behavior [18], i.e., increased enrichment in response to biotin supplementation (data not shown). The enrichment of H3K9ac did not depend significantly on the concentration of biotin in culture media (data not shown). We repeated the experiments at two additional loci known to depend on biotin, LTR15 and SMVT promoter 1, and obtained the same patterns of enrichment as described for LTR22 (data not shown).
Fig. 3.
The enrichment of H3K9bio, H3K18bio, and H4K8bio at LTR22 depends on biotin availability in Jurkat cells. Cells were cultured in biotin-defined media for 12 d, and the relative enrichment of H3K9bio (panel A), H3K18bio (B), H4K8bio (C), and H3K9me2 (D) at the LTR22 locus was quantified by μChIP and qRT-PCR. Bars denote the percent of input DNA from the LTR22 locus that was precipitated with antibodies (mean ± S.D., n = 4). a, bBars not sharing the same letter are significantly different (P < 0.05).
In control experiments we demonstrated that biotin-defined culture conditions efficiently alter the cellular biotin status and that altered biotinylation of histones at LTR loci affect gene transcriptional activity. First, biotinylated carboxylases were probed in cell extracts by using streptavidin as a probe. The abundance of biotinylated pyruvate carboxylase, 3-methylcrotonyl-CoA carboxylase, and propionyl-CoA carboxylase depended on the biotin concentrations in culture media (Fig. 4A), consistent with previous studies in our laboratory [18, 20]. Second, the abundance of LTR transcripts was inversely linked to biotin in culture media (Fig. 4B). This is consistent with previous observations, suggesting that biotinylation of histones plays a role in the repression of retrotransposons [18].
Fig. 4.
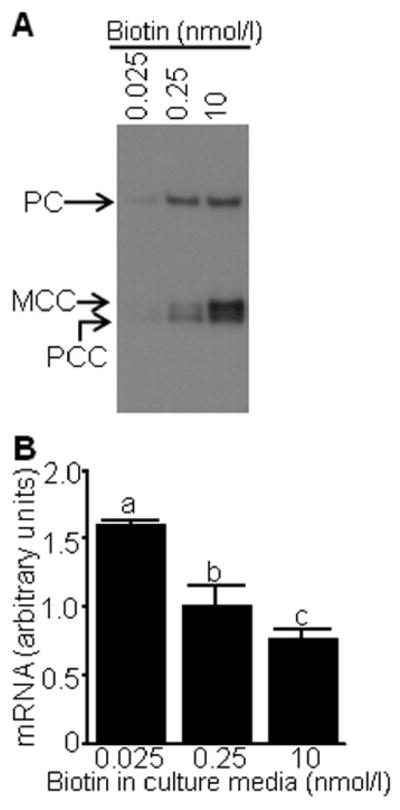
Abundance of biotinylated carboxylases and LTR transcripts in biotin-defined Jurkat cells. Cells were cultured in biotin-defined media for 12 d. Panel A: Streptavidin was used as a probe for biotinylated pyruvate carboxylase (PC), 3-methylcrotonyl-CoA carboxylase (MCC), and propionyl-CoA carboxylase (PC). Panel B: mRNA originating in the U5 region of LTRs was quantified by qRT-PCR. Values are means ± SD, n = 3. a, b, cBars not sharing the same letter are significantly different (P < 0.05).
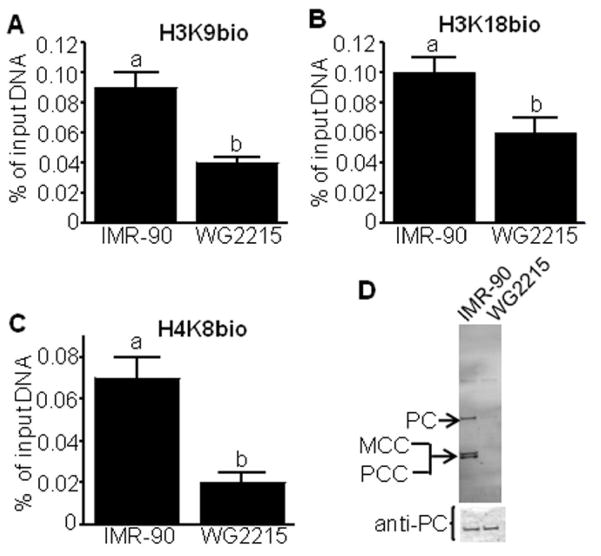
Mutation of the HCS gene abolished the enrichment of H3K9bio, H3K18bio, and H4K8bio at target loci. The enrichment of all three histone marks decreased by ~50% at the LTR22 locus in HCS mutant WG2215 fibroblasts compared with HCS wild-type IMR-90 fibroblasts (Fig. 5A–C); comparable results were obtained for LTR15 and SMVT promoter 1 (data not shown). The mutation of the HCS gene in WG2215 fibroblasts abolishes the biotinyl protein ligase activity of HCS compared with IMR-90 fibroblasts. If biotinylated carboxylases were probed with streptavidin, HCS wild-type fibroblasts produced strong signals for pyruvate carboxylase, 3-methylcrotonyl-CoA carboxylase, and propionyl-CoA carboxylase (Fig. 5D, upper panel); in contrast, no signal was detectable for HCS mutant WG2215 fibroblasts. The decreased abundance of biotinylated (holo-)carboxylases in WG2215 fibroblasts was not caused by altered apo-carboxylase expression, gel loading, or blotting efficiency, based on the observation that an antibody to PC produced a similar signal in extracts from IMR-90 and WG2215 fibroblasts (Fig. 5D, lower panel).
Fig. 5.
The enrichment of H3K9bio, H3K18bio, and H4K8bio at LTR22 depends on HCS activity in human fibroblasts. HCS mutant (“WG2215”) and wild-type (“IMR-90”) human fibroblasts were cultured in medium containing 10 nmol/l biotin. The relative enrichment of H3K9bio (panel A), H3K18bio (B), and H4K8bio (C) at the LTR22 locus was quantified by μChIP and qRT-PCR. Bars denote the percent of input DNA from the LTR22 locus that was precipitated with antibodies (mean ± S.D., n = 4). a, bBars not sharing the same letter are significantly different (P < 0.05). Panel D: Streptavidin was used as a probe for biotinylated pyruvate carboxylase (PC), 3-methylcrotonyl-CoA carboxylase (MCC), and propionyl-CoA carboxylase (PC).
4. Discussion
This study provides novel insights into possible biological functions of the new histone biotinylation sites H3K9bio, H3K18bio, and H4K8bio. We show for the first time that these three marks are enriched in pericentromeric heterochromatin and LTRs, i.e., transcriptionally repressed loci. The proposed role for histone biotinylation in gene repression is also consistent with the depletion of H3K9bio, H3K18bio, and H4K8bio at the IL-2 promoter coinciding with transcriptional activation of IL-2. Important mechanistic insights suggest that low activity of the histone biotinyl ligase HCS causes low levels of histone biotinylation at target loci in the human genome.
These observations are physiologically important because the abundance of H3K9bio, H3K18bio, and H4K8bio, and the corresponding repression of genes depend on the concentrations of biotin in cell culture media. The levels of biotin tested in this study span the range of concentrations observed in biotin-deficient, biotin-normal, and biotin-supplemented persons [30, 31], i.e., the observations reported here are nutritionally relevant. Importantly, phenotypes and increased risk for chromosomal abnormalities have been linked to low levels of histone biotinylation in previous studies. Findings from one such study suggest that low levels of histone biotinylation decreases life span and heat stress resistance in HCS knockdown Drosophila melanogaster [13]; findings in a subsequent study suggest that these phenotypes were not caused by decreased biotinylation of carboxylases but were genuine to decreased histone biotinylation [14]. Another study revealed that decreased enrichment of H4K12bio at LTRs in biotin-deficient or HCS-deficient cells de-represses LTRs, thereby enhancing frequency of retrotransposition events and chromosomal abnormalities [18]; similar findings were made in biotin supplementation studies in adults [18]. Based on the data reported here, we propose that H3K9bio, H3K18bio, and H4K8bio play similar roles in maintaining genome stability. We currently do not know if one of the novel biotinylation marks is more important for gene regulation than another, including the previously characterized H4K12bio [16]. Studies are currently underway in our laboratory in which we seek to map various histone biotinylation marks at high resolution by using ChIP-Seq protocols in human lymphoid cells. Previous studies suggest that biotinylation of histones depends on prior methylation of DNA, but not vice versa [18].
There are still many unknowns in the field of histone biotinylation. For example, while it appears that biotinylated histone H2A is less abundant than biotinylated histones H3 and H4 [8], the percentage of histones that are biotinylated remains to be determined. It has been proposed that one out of three molecules of histone H4 is biotinylated at K12 in human telomeres, based on ChIP assays and Southern blot analysis [17]. The abundance of histone biotinylation might be substantially lower for other genomic loci or biotinylation marks other than H4K12bio. A previous study suggests that H4K12bio is enriched in the transcriptionally repressed IL-2 promoter, but that no enrichment can be found in other regions of that gene [16]. Another report proposed that less than 0.1% of histones in bulk extracts are biotinylated [32], but that study relied on using [14C]biotin with low specific activity as a radiotracer. We are currently in the process of using customized [14]biotin and commercial [3H]biotin as a radiotracer for biotin; both compounds have a specific activity that is about 1000 times that of commercial [14C]biotin. We consistently detect biotinylated histones, albeit at low concentrations, in bulk histone extracts from human cells that are not due to contamination with other biotinylated proteins. We agree with Bailey et al. that the overall abundance of histone biotinylation marks is low [32], but we maintain our conclusion that biotinylated histones are comparably abundant in repressed regions. The relative abundance of epigenetic marks must not be confused with their biological importance. For example, only 2–3% of the cytosines in DNA are methylated and, yet, the importance of DNA methylation for gene silencing and gene imprinting is well recognized [33]. Likewise, phosphorylation of serine-14 in histone H2B becomes detectable only after programmed cell death is induced [34].
Ongoing projects in our laboratory address some of the uncertainties identified above. We are currently in the process of confirming biotinylation sites in histones isolated from human cells by using mass spectrometry; quantifying the relative abundance of biotinylated histones by using radiotracers of great specific radioactivity; and demonstrating the biological significance of histone biotinylation by linking biotinylation events to genome stability.
Abbreviations
- ADH5
aldehyde dehydrogenase 5
- GAPDH
glyceraldehyde3-phosphate dehydrogenase
- H3K4bio
K4-biotinylated histone H3
- H3K9bio
K9-biotinylated histone H3
- H3K18bio
K18-biotinylated histone H3
- H3K9me2
K9-dimethylated histone H3
- H3K9ac
K9-acetylated histone H3
- H4K8bio
K8-biotinylated histone H4
- H4K12bio
K12-biotinylated histone H4
- HCS
holocarboxylase synthetase
- K
lysine
- LTR
long terminal repeat
- μChIP
micro chromatin immunoprecipitation
- PHA
phytohemagglutinin
- PMA
phorbol-12-myristate-13-acetate
- qRT-PCR
quantitative real-time polymerase chain reaction
- SMVT
sodium-dependent multivitamin transporter
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945, DK077816, DK082476 and ES015206, USDA CSREES grant 2006-35200-17138, and by NSF grant EPS 0701892.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolffe A. Chromatin. 3. Academic Press; San Diego, CA: 1998. [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 5.Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. doi: 10.1016/s0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- 6.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 7.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–29. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 9.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–63. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 10.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, et al. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–59. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Reports. 2008;41:310–15. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J Nutr. 2006;136:2735–42. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camporeale G, Zempleni J, Eissenberg JC. Susceptibility to heat stress and aberrant gene expression patterns in holocarboxylase synthetase-deficient Drosophila melanogaster are caused by decreased biotinylation of histones, not of carboxylases. J Nutr. 2007;137:885–89. doi: 10.1093/jn/137.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew YC, Raza AS, Sarath G, Zempleni J. Biotinylation of K8 and K12 co-occurs with acetylation and mono-methylation in human histone H4. FASEB J. 2006;20:A610. [abstract] [Google Scholar]
- 16.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J Nutr Biochem. 2007;18:760–68. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wijeratne SS, Camporeale G, Zempleni J. K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts. J Nutr Biochem. 2010 doi: 10.1016/j.jnutbio.2009.01.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, et al. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J Nutr. 2008;138:2316–22. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J Nutr Biochem. 2008;19:400–08. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 21.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. Vol. 1. International Life Sciences Institute; Washington, D.C: 2006. pp. 314–26. [Google Scholar]
- 22.Healy S, Perez-Cadahia B, Jia D, McDonald MK, Davie JR, Gravel RA. Biotin is not a natural histone modification. Biochim Biophys Acta. 2009;1789:719–33. doi: 10.1016/j.bbagrm.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2007. pp. 191–209. [Google Scholar]
- 24.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature Protocols. 2008;3:1032–45. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Blanchard R, Gu X, Burnette A, Huang J, Zeng X. Genome-Wide, Promoter-Tilling, Real-Time PCR Assays for Analyzing Chromatin Immunoprecipitation Samples. [accessed 11/23/2009.];2007 www.sabiosciences.com/support_posters.php.
- 27.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SAS Institute. StatView Reference. 3. SAS Publishing; Cary, NC: 1999. [Google Scholar]
- 29.Hassan YI, Zempleni J. A novel, enigmatic histone modification: biotinylation of histones by holocarboxylase synthetase. Nutr Rev. 2008;66:721–25. doi: 10.1111/j.1753-4887.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 30.Mock DM, Lankford GL, Mock NI. Biotin accounts for only half of the total avidin-binding substances in human serum. J Nutr. 1995;125:941–46. doi: 10.1093/jn/125.4.941. [DOI] [PubMed] [Google Scholar]
- 31.Zempleni J, Helm RM, Mock DM. In vivo biotin supplementation at a pharmacologic dose decreases proliferation rates of human peripheral blood mononuclear cells and cytokine release. J Nutr. 2001;131:1479–84. doi: 10.1093/jn/131.5.1479. [DOI] [PubMed] [Google Scholar]
- 32.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal Biochem. 2008;373:71–77. doi: 10.1016/j.ab.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Li E, Bird A. DNA methylation in mammals. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 341–56. [Google Scholar]
- 34.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–17. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]