Abstract
Conserved vertebrate transcription factors (TFs) direct gene expression by binding to DNA regulatory regions. To explore the evolution of gene regulation, we experimentally determined the genome-wide occupancy of two TFs, CEBPA and HNF4A, in livers of multiple vertebrates. Although each TF displays highly conserved DNA binding preferences, most binding is species-specific, and aligned binding events present in all five species are rare. Regions near genes with expression levels dependent on a TF are often bound by the TF in multiple species, yet show no enhanced DNA sequence constraint. Binding divergence between species can be largely explained by sequence changes to the the bound motifs. Among the binding events lost in one lineage, only half are recovered by another binding event within 10 kilobases. Our results reveal large interspecies differences in transcriptional regulation and provide insight into their evolution.
The relationship between genetic sequence and transcriptional regulation is central to understanding species-specific biology, disease, and evolution (1). Identifying the divergence and conservation among functional regulatory elements is an important goal of comparative genomic research, and this is often done via DNA sequence comparisons using distant (2) and closely related species (3). Although both approaches have successfully identified conserved regulatory regions, the majority of transcription factor (TF) binding events change rapidly between closely related species, making them difficult to detect using DNA sequence alone (4-7). For instance, the experimentally-determined binding events for homologous TFs found in mouse and human livers are unlikely to align with each other (7), despite conservation of their functional targets (8) and global liver transcription (9). The evolution of mammalian transcriptional regulation remains largely unexplored beyond limited mouse-human comparisons.
We therefore identified the genome-wide binding of two transcription factors: (i) CEBPA, in livers of species representing five vertebrate orders: human (primate), mouse (rodent), dog (carnivora), short-tailed opossum (didelphimorphia), and chicken (galliformes), and (ii) HNF4A, in livers from human, mouse, and dog. Chromatin immunoprecipitation experiments were combined with high-throughput sequencing (ChIP-seq) using healthy, nutritionally unstressed adult liver from the heterogametic sex as a functionally and transcriptionally conserved homologous tissue type (8, 10) (Figure 1, Figure S1).
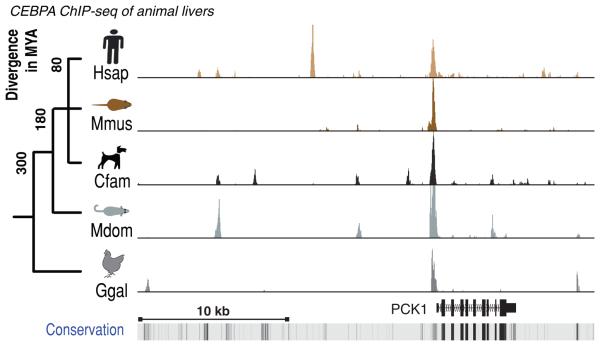
Figure 1.
CEBPA binding in vivo in livers isolated from five vertebrate species cross-mapped to the human PCK1 gene locus. A rare ultraconserved binding event is shown surrounded by species-specific and partially-shared binding events. On the left is the evolutionary tree of the five study species (Hsap=Homo sapiens; Mmus=Mus musculus; Cfam=Canus familiaris; Mdom=Monodelphis domesticus; Ggal=Gallus gallus), with their approximate evolutionary distance in millions of years (MY). The bottom track shows evolutionary conservation measured across 44 vertebrate species, and darker shading represents slower evolution.
CEBPA and HNF4A were selected as representative transcription factors within the liver-specific regulatory network because both are conserved and constitutively expressed with well-characterized target genes (10, 11). In addition, they represent distinct TF classes, and the DNA binding domains of each factor's orthologs are nearly identical among the study species (Figure S2).
The genomic TF occupancy data were reproducible between different individuals of the same species (Figure S3), and were validated using alternative antibodies (Figure S4). Using a mouse carrying a human chromosome we confirmed that genetic sequence, and not diet, lifestyle, or environment, is the primary determinant of liver-specific TF binding (Figure S5) (12). Given their greater evolutionary distance, contributions from non-genetic sources could be higher in opossum and chicken.
We identified TF-bound regions using a dynamic programming algorithm, and our results were robust to different peak-calling thresholds (Methods, Figure S6, Figure S7, Figure S8). To detect TF binding events shared among any combination of the five vertebrates, we used the Ensembl 12-way multi-species alignment (13), which incorporates approximately half of each species' genome into the global alignments. Our findings did not substantially change with an alternate methodology that used pairwise alignments performed using a separate algorithm (Methods, Figure S6, Figure S7, Figure S8).
Each transcription factor bound between 16,000 and 30,000 locations in each mammalian genome; CEBPA bound approximately half this number in the smaller chicken genome (Figure 2, Figure S6, Figure S7, Figure S9). For both factors, less than a quarter of bound regions were within three kilobases of known transcription start sites. Between 30% to 50% of the binding sites of the two transcription factors overlapped in the genome (Table S1). These overlapping sites did not exhibit substantially different characteristics in the conservation of underlying genetic sequence than the sites of CEBPA and HNF4A considered individually.
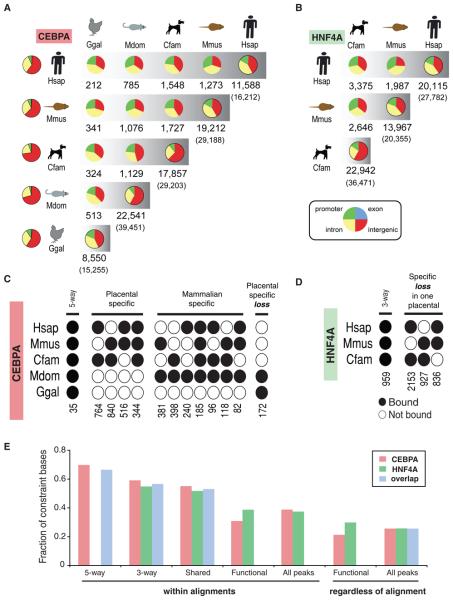
Figure 2.
Conservation and divergence of transcription factor binding. (A) For CEBPA and (B) HNF4A, the pair-wise distribution and numbers of binding events are shown as a pie chart distributed into: intergenic (red), intronic (yellow), exonic (blue), and promoter [TSS +/− 3kb] (green) regions. The left-most column contains the distributions of the bulk genomes. The right-most pie chart represents all binding events in each species with the total number of alignable peaks above the total peaks (in parentheses). (C,D) Multi-species CEBPA and HNF4A binding event analysis where black circles indicate binding in a given species. For instance, there are 764 regions bound by CEBPA only in dog and human (see also Figure S6, S7, S17, and Tables S2, S6). (E) The DNA sequence constraint beneath binding events was measured by average Genomic Evolutionary Rate Profiling (19) scores for peaks found: in all 5 species (5-way) among all the placental mammals (3-way), bound in any two species (Shared), within 10 kb of the TSS of functional targets (Functional), and all peaks.
For these two liver-specific transcription factors, binding events appear to be shared 10%-22% of the time between mammals from any two of the three placental lineages we profiled, separated by approximately 80 million years of evolution (Figure S6, Figure S7). This reveals a rapid rate of evolution in transcriptional regulation among closely related vertebrates. Nevertheless, the number of CEBPA and HNF4A transcription factor binding events shared between any two of our five study species is far greater than could have occurred by chance (Figure S10).
We used the genome-wide binding of CEBPA in opossum to test the hypothesis that regulatory regions have diverged substantially between eutherian and metatherian mammals (14). Opossum indeed showed dramatic changes in transcription factor binding, and only between 6-8 % of the genomic regions occupied by CEBPA in opossum liver align with CEBPA binding events also found in mouse, dog, and/or human liver. This divergence was even greater in chicken, which shared only 2% of CEBPA binding with human, demonstrating extensive and continuous rewiring of gene regulation during vertebrate evolution that corresponds to evolutionary distance.
Ultra-conserved noncoding regions are an intriguing discovery revealed by comparative genomic sequencing (15). We identified ultra-shared interactions between CEBPA and the vertebrate genome as binding events preserved over the 300 million years of evolution and thus found in aligned positions in all five species: human, mouse, dog, opossum, and chicken. Using our most stringent threshold, a set of 35 binding events were found to be shared by all five vertebrate species, and these binding events are almost invariably near genes central to liver-specific biology (Figure 2C, Table S2, Table S3, see also below). Although these ultra-shared binding events are close to important liver-specific genes, they make up less than 0.3% of the total CEBPA binding found in human.
About 250 direct functional HNF4A target genes have recently been identified using multiple independent methodologies in mouse and human, including perturbation analysis in both species (8). We experimentally identified a similar set of transcriptional target genes whose expression is dependent on CEBPA in adult mouse liver by using a conditional knock-out strategy (11). In mammals, the target genes for both transcription factors have a disproportionate fraction of binding events that are shared in at least two species (p-value > 1× 10−5) (Table S4). CEBPA binding near direct target genes did not overlap with the binding events shared by five species.
We further compared our results to a set of 53 regulatory sequences within known, authentic liver enhancers in human (Table S5) (16). Thirty-eight of these regulatory sequences were located within nine HNF4A-bound regions. CEBPA binding overlapped with five of these HNF4A-bound regions, and we also found five of the nine HNF4A binding events were bound by HNF4A in more than one species. Overall these findings suggest that functional targets are enriched for TF binding events found in multiple species.
Mammalian TF binding studies have suggested that functional enhancers show increased sequence constraint (17). As expected, the relatively few binding events shared among three or five species showed increased sequence constraint. The sequence constraint, evaluated using Genomic Evolutionary Rate Profiling (GERP) scores (19), in bound regions near functional targets was similar to that for all bound regions for both TFs and these results were robust to the method applied. Regions bound by both CEBPA and HNF4A have sequence constraint patterns similar to those found for each factor analyzed independently (Figure 2E, Figure S11). In sum, TF binding events near functional targets showed enhanced sharing between species, without a corresponding increase in sequence constraint.
DNA binding specificities of transcription factors show remarkable diversity and complexity (18), yet few studies have compared specificities of orthologous transcription factors among multiple species. The motifs we directly determined from experimental binding data showed that in vivo bound consensus sequences remain virtually unchanged during vertebrate evolution despite most binding events being species-specific (Figure 3A, Figure S12). Neither the quality of a bound motif, as determined by its similarity to the consensus, nor the regional ChIP enrichment, as measured by sequencing read depth, was correlated with the conservation of TF binding events (Figure S13).
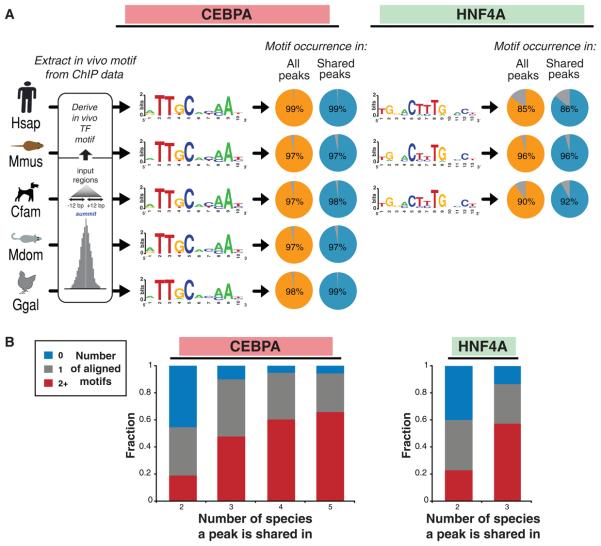
Figure 3.
DNA binding specificities of CEBPA and HNF4A are highly conserved during vertebrate evolution. (A) The known sequence motifs were identified de novo in each species interrogated (Methods), and found within almost all binding events (see Figure S12). (B) Multiple aligned motif occurrences are highly associated with binding events shared among three or more species. Peaks are categorized by the number of species they are shared in and the fraction of peaks with 0 (blue), 1 (grey), and 2 or more (red) aligned motifs are shown.
Searching for the sequence features that are associated with shared binding events, we discovered that binding events shared by more species contain more aligned motifs (Figure 4B). These shared regions represent examples of deeply conserved regulatory architecture featuring multiple motifs at specific sequence locations maintained through vertebrate evolution. The most conserved of these, the five-way ultra-shared sites, also exhibit the strongest sequence constraint (Figure 2E).
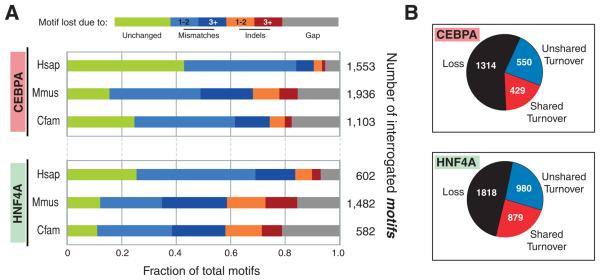
Figure 4.
Lineage-specific loss and turnover of transcription factor binding events. (A) The unbound regions in each placental mammal that align to regions showing TF binding in the other two placental mammals were collected, and the mechanisms by which the underlying motifs were disrupted were summarized. (B) Turnovers occurred near lineage-specific lost binding events approximately half the time; shared turnovers represent cases where a cluster of binding events likely occurred in a common ancestor (see text, Figure S16).
To explore the genetic mechanisms underlying the divergence of transcription factor binding, we identified potentially lost CEBPA and HNF4A binding events. A binding event was assumed to be lost if it was not present in one placental mammal, yet was experimentally found at aligned, orthologous regions in the other two placental mammals. Using parsimony, this situation is best explained by an ancestral TF binding event present before the mammalian radiation that was subsequently lost along one lineage.
The lost binding events were categorized by the sequence changes to the alignable binding motifs within the orthologus regions of the other species (Figure 4). Between 20 and 40% of the motifs associated with lineage-specific binding event losses were unchanged. These regions may represent cases of epigenetic redirection, yet-to-be characterized SNPs or indels, or loss of nearby genomic binding partners. A larger fraction of the absent binding events were associated with motifs whose disruption could be assigned to base pair substitutions, indels, and gaps in the alignment. Across all the vertebrate species, indels appear to be associated with loss of the underlying sequence motif a third as often as mismatches. A four-mammal analysis using opossum as an outgroup afforded similar results (Figure S14). Analogous mechanisms appear to explain species-specific gains of transcription factor binding events (Figure S15). Taken together, the steady accumulation of small changes in the genetic sequence appears to rapidly remodel thousands of transcription factor binding sites.
Approximately half of lineage-specific losses of TF binding showed evidence of nearby compensatory binding events (Figure 4B). A quarter of species-specific losses had a nearby (+/−10kb) gained binding event unique to the same lineage (unshared turnover), and an additional quarter of the losses had a nearby binding event that is shared in one or more other species (shared turnover) (Figure S16). The latter case suggests the existence of a cluster of binding events in the common ancestor. In both cases, the probability of finding a turnover decreased rapidly with distance from the loss (Figure S16), but a shared turnover was typically closer to the site of the loss than was an unshared turnover (p-value <1.0e-10 (CEBPA) and p-value <1e-15 (HNF4A)).
Understanding the evolutionary dynamics of transcription factor binding is essential to understanding the evolution of gene regulation. Many comparative genomics approaches assume that a multi-species alignment of a high quality motif is indicative of functionality (19-25). Our analysis of experimentally determined in vivo occupancy of two TFs in multiple vertebrates revealed apparent limitations to this model and a number of other insights about the complex relationship between genetic sequence, transcription factor binding, and genome regulation.
First, the vast majority of ChIP-identified transcription factor binding events are unique to each species; in mammals, the binding events that occur within species-specific, repetitive DNA are more common than conserved binding events. Second, ultrashared TF binding events, which are the functional counterpart of ultraconserved sequences, appear rarely in vivo among all five vertebrates. Third, only approximately half of binding events that are lost in one placental mammal yet present in at least two others are potentially recovered by nearby turnover events. Fourth, neither motif nor strength of TF binding correlate with conservation of a transcription factor's genomic occupancy. Alterations in the DNA binding specificity of CEBPA and HNF4A cannot account for rapid binding divergence, nor can species-specific environmental differences.
Nevertheless, comparing binding events within 10 kb of the transcription start site (TSS) of experimentally determined target genes of CEBPA and HNF4A has shown that binding events near these genes are more likely to be shared with other species, although this does not correspond to an increase in sequence constraint. In fact, the set of the ultra-shared, five-way binding events is entirely disjoint from the set of genes directly dependent on CEBPA in adult liver. For HNF4A, only 6% of binding events shared across three placental mammals (Figure 2D) are near the highest-quality functional target genes, namely, those genes that depend on HNF4A for proper expression in both mouse and human . Given that most TFs are active in multiple cell types (26), it is possible that the remaining shared sites are active in other tissues or other developmental stages. Indeed, the ultra-shared CEBPA binding events are uniformly found near liver-specific genes that would be expected to be upregulated upon liver organogenesis. Conversely, those binding events near functional targets in adult liver that are neither shared nor show signs of sequence constraint may represent lineage-specific regulatory interactions.
The preponderance of specific-specific binding and the rapid lineage-specific loss of binding events suggests that a sizeable majority of specific TF-DNA interactions could be evolving neutrally. Liver-specific TFs and subsequent gene expression are both highly conserved, the rapid gain and loss of binding events may be indicative of compensatory changes that maintain local concentrations of TF binding near functional targets (27). Indeed, a recent computational approach which uses a high concentration of TF binding motifs, regardless of their alignment, showed improved ability to predict regulatory interactions (28).
Despite the rapid gain and loss of TF binding events in mammals, tissue-specific gene regulation seems to be maintained by identifiable regulatory architectures that can be independent of sequence constraint.
Supplementary Material
Acknowledgments
We thank N. Matthews and J. Hadfield at the CRI Genomics Core, the CRI Bioinformatics Core, and W. Howat and the Histopathology Core, T. Davidge, S. Ballantyne, A. Enright, S. Wilder and J. Herrero (EBI). Work supported by the European Research Council Starting Grant, EMBO Young Investigator Award, Addenbrooke's Biomedical Research Centre, Hutchinson Whampoa (D.T.O), Swiss National Science Foundation (C.K.), University of Cambridge (D.S., M.D.W., D.T.O.), Cancer Research UK (D.S., M.D.W., G.B., C.K., D.T.O.), the Wellcome Trust [grant numbers WT062023 and WT079643] (B.B., P.F.) and EMBL (P.C.S., P.F.). ChIP-seq experiments were deposited into ArrayExpress under the accession number E-TABM-722. CEBPA KO gene expression experiments were deposited into ArrayExpress under the accession number E-MTAB-178.
Footnotes
AUTHOR CONTRIBUTIONS
D.S., M.D.W., D.T.O. designed experiments; D.S., M.D.W, C.K., S.W., C.P.M.-J. performed experiments; D.S., M.D.W., B.B., G.B., P.C.S., and P.F. analyzed the data; S.M., C.P.M.-J., I.T., A.M. provided tissues; D.S., M.D.W., B.B., I.T., P.C.S., P.F., and D.T.O. wrote the manuscript. P.F. and D.T.O. oversaw the work.
REFERENCES
- 1.Wray GA. Nat Rev Genet. 2007;8:206–16. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 2.Lenhard B, Sandelin A, Mendoza L, Engström P, et al. J Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, et al. Science. 2003;299:1391–4. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 4.Borneman AR, Gianoulis TA, Zhang ZD, Yu H, et al. Science. 2007;317:815–9. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 5.Dermitzakis ET, Clark AG. Mol Biol Evol. 2002;19:1114–21. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 6.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, et al. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odom DT, Dowell RD, Jacobsen ES, Gordon W, et al. Nat Genet. 2007;39:730–2. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boj SF, Servitja JM, Martin D, Rios M, et al. Diabetes. 2009;58:1245–53. doi: 10.2337/db08-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan ET, Quon GT, Chua G, Babak T, et al. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Jimenez CP, Kyrmizi I, Cardot P, Gonzalez FJ, Talianidis I. Mol Cell Biol. 2009 doi: 10.1128/MCB.00927-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatzis P, Kyrmizi I, Talianidis I. Mol Cell Biol. 2006;26:7017–29. doi: 10.1128/MCB.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, et al. Science. 2008;322:434–8. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard TJ, Aken BL, Ayling S, Ballester B, et al. Nucleic Acids Res. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, et al. Nature. 2007;447:167–77. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 15.Bejerano G, Pheasant M, Makunin I, Stephen S, et al. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 16.Portales-Casamar E, Kirov S, Lim J, Lithwick S, et al. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visel A, Blow MJ, Li Z, Zhang T, et al. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badis G, Berger MF, Philippakis AA, Talukder S, et al. Science. 2009;324:1720–3. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper GM, Stone EA, Asimenos G, NISC Comparative Sequencing Program et al. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margulies EH, Vinson JP, NISC Comparative Sequencing Program. Miller W, et al. Proc Natl Acad Sci U S A. 2005;102:4795–800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchette M, Bataille AR, Chen X, Poitras C, et al. Genome Res. 2006;16:656–68. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor J, Tyekucheva S, King DC, Hardison RC, et al. Genome Res. 2006;16:1596–604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, et al. Genome Res. 2007;17:760–74. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller W, Rosenbloom K, Hardison RC, Hou M, et al. Genome Res. 2007;17:1797–808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, et al. Proc Natl Acad Sci U S A. 2007;104:7145–50. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. Nat Rev Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 27.MacArthur S, Li XY, Li J, Brown JB, et al. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordân R, Narlikar L, Hartemink AJ. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkp1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.